Abstract
Cone snail venoms provide an ideal resource for neuropharmacological tools and drug candidates discovery, which have become a research hotspot in neuroscience and new drug development. More than 1,000,000 natural peptides are produced by cone snails, but less than 0.1% of the estimated conotoxins has been characterized to date. Hence, the discovery of novel conotoxins from the huge conotoxin resources with high-throughput and sensitive methods becomes a crucial key for the conotoxin-based drug development. In this review, we introduce the discovery methodology of new conotoxins from various Conus species. It focuses on obtaining full N- to C-terminal sequences, regardless of disulfide bond connectivity through crude venom purification, conotoxin precusor gene cloning, venom duct transcriptomics, venom proteomics and multi-omic methods. The protocols, advantages, disadvantages, and developments of different approaches during the last decade are summarized and the promising prospects are discussed as well.
1. Introduction
Cone snails (Conus) are carnivorous mollusks from the Conidae family (Figure 1). They live in the tropical oceans around the world and hunt fish (piscivorous), worms (vermivorous), or molluscs (molluscivorous) for food, although they are slow-moving creatures [1]. Cone snails have evolved a full set of specialized envenomation apparatus to release bioactive venoms to compensate their slow movement for fast-moving prey, competitors, or/and predators [2,3]. Cone snail venom peptides are secreted by the epithelial secretory cells in the long and convoluted venom duct [2,4]. The venom is pushed by muscle peristalsis of venom bulb and loaded into the harpoon-like radula tooth for envenomation [5]. Due to the human casualties that are caused by cone snail stings in 1960s [6], these venoms first caught researcher’s interest in their toxicity and bioactivity.

Figure 1.
Representative Conus species native to Hainan China (shot by Cheng Li).
Early studies have confirmed that these bioactive venoms are a cocktail of neuroactive peptides, termed conopeptides or conotoxins, which can cause paralysis, shudder, and even death of the prey within seconds [1,5]. Subsequent research have revealed that conopeptides are able to selectively modulate voltage-gated ion channels [7] (Table 1), including sodium channels [8,9], potassium channels [10], and calcium channels [11,12], as well as ligand-gated ion channels (Table 1), such as nAChRs [13,14,15], serotonin receptor [16], NMDA receptor [17], GABA receptor [18], GPCRs [19] (α1-adrenoceptors [20,21], vasopressin receptor [22], neurotensin receptor [23]), and neurotransmitter transporters (noradrenaline transporter [21,24]), which are key targets for chronic diseases, like neuralgia [8,25,26], addiction [27], epilepsy [17,28], cancer [29], heart disease [30,31], and so on [32,33,34].

Table 1.
Target and clinical potential of representative conotoxins.
Additionally, the venom peptides show high selectivity and efficacy when interacting with the targets, resulting in minor side effects for disease treatment [35]. Hence, cone snail venoms provide an ideal resource for neuropharmacological tools and drug candidates screening, which have become a research hotspot in neuroscience and new drug development [36,37,38] (Table 1). For instance, an ω-conotoxin, named MVIIA (Ziconotide, Prialt) from Conus magus, which blocks voltage-gated calcium channels, has been approved by FDA for chronic pain treatment since 2004 [39,40]. At present, more than 10 conotoxins, including Xen2174 (MrIA) [41], CGX-1007 (Conantokin G) [17], CGX-1051 (κ-PVIIA) [42], ACV1 (Vc1.1) [43], and CGX-1160 (contulakin-G) [44] have marched into preclinical or clinical research stage, which present good prospects on conotoxin drug discovery.
There are more than 700 Conus species in the world [48] and each can secrete over 1000 conotoxins [49]. In particular, 3305 novel conopeptide precursors were discovered from one Conus episcopatus specimen [50]. Owning to small overlap of conopeptides between different Conus species [51], there are an estimated 1,000,000 natural peptides that are produced by cone snails. However, <0.1% of the estimated conopeptides has been characterized to date [36,52]. Therefore, high-throughput and sensitive methods are crucial for the discovery of novel conotoxins and the development of conotoxin-based drug screening from this enormous peptide reservoir.
In this review, the discovery methodology of novel conotoxins from mollusks Conus species has been summarized, which mainly focuses on obtaining full N- to C-terminal sequences, regardless of disulfide bond connectivity, through crude venom purification, conotoxin precusor gene cloning, venom duct transcriptomics, venom proteomics, and multi-omic method. The protocols, advantages, disadvantages, and developments of different approaches during the last ten years are overviewed and the promising prospects of these methods are discussed.
2. Diversity of Conotoxins
Conotoxins normally consist of 10 to 40 amino acid residues with 2 to 4 or more disulfide bonds. They are expressed as RNA-encoded precursor proteins, which are processed and transferred into mature peptides in the endoplasmic reticulum (ER) and Golgi apparatus. A typical conopeptide precursor is composed of a highly conserved ER signal region, a pro-region and a greatly variable mature peptide region [52]. Conotoxins can be classified into different gene superfamily categories, according to the similarities between the ER signal sequences [35].
Generally, conotoxin-encoding transcripts produce diverse precursors by hypermutation, fragment insertion/deletion, and mutation-induced premature termination [53]. One precursor can produce far more than one mature peptide because of various posttranslational modifications (PTMs) and variable peptide processing (VPP), which create the exponential diversity of conopetides [53,54]. For example, 20 different conopeptide variants on average for each precursor have been detected and characterized from venom duct transcriptomics of Conus marmoreus [49]. VPP refers to the C- and N-terminal truncations of the precursor by proteolytic cleavage at alternative sites [53,54]. These variations generated by interrupting, deleting, or elongating partial sequences and cysteine frameworks. It produced highly variable mature peptides or isoforms with multiple primary sequences [54].
PTMs are ubiquitous and play a key role in the structure and activity of conotoxins [55]. Many types of PTMs are found in the conotoxin maturation process, such as oxidative folding (disulfide bond formation, the most common PTMs), C-terminal amidation, hydroxylation of proline, valine, and lysine, carboxylation of glutamate, cyclization of N-terminal glutamine, glycosylation, sulfation, bromination, and residue epimerization, etc. [53,55]. These multi-diversification mechanisms, such as transcript variation, VPP and PTMs, explain how thousands of specific conopeptides are produced from such a limited gene precursors in a single Conus specie and reveal the reason for inter- and intra-specific variability [49,53,56].
3. Conotoxins Purified from Crude Venom
Conotoxins have been obtained by isolation from crude venom of cone snails since 1970s [57] (Figure 2). The envenomation apparatus of the snails was dissected first. Only about 10 to 50 µL crude venom could be squeezed from each snail specimen, or the dissected tissues were directly subjected to extraction. Sometimes, tens to hundreds of collected snails were dissected to obtain enough venom for conopeptide isolation. The sampling process is non-renewable. It is a waste of precious resource of cone snails, especially for the rare species.
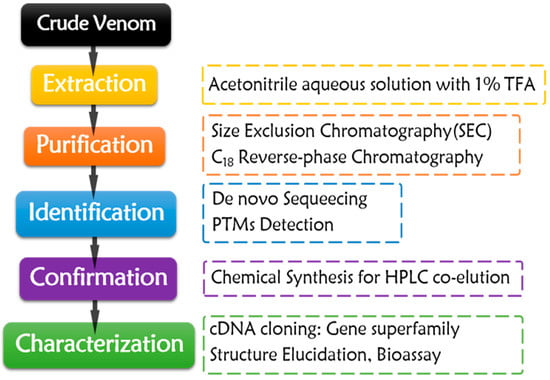
Figure 2.
Purification workflow of native conotoxins obtained from crude venom.
The purification process almost has remained constant for decades (Figure 2). Crude venom or dissected tissues are extracted by acetonitrile aqueous solution with 1% TFA. The crude extract is fractionated by Size Exclusion Chromatography (SEC), and then purified by C18 reverse-phase chromatography with gradient acetonitrile/water solution with 0.1% TFA as mobile phase. In order to gain enough target conopeptides for the subsequent characterization, enough crude venom and rigorous purification skills are required. The purified conotoxins are subjected to de novo sequencing through Edman degradation [58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87] or MS sequencing [81,88,89,90] after sequential disulfide bond reduction, hydrosulphonyl alkylation, and enzymolysis, which make sequencing process much easier. PTMs are assigned with the aid of MS techniques [59,65,66,69,70,71,75,76,77,78,80,83,89,90,91,92]. The targeted conotoxins are then chemically synthesized through SPPS, following the subsequent oxidative folding. HPLC co-elution of the synthesized peptides and the purified native conotoxins could validate the sequencing results [69,73,75,76,81,83,84,89]. For gene superfamily identification of the native peptides, their precursor genomic genes or cDNAs could be cloned by various PCR methods or identified by venom transcriptome sequencing. According to the signal peptide homology of the precursors, their gene superfamily could be determined and classified [31,62,65,68,74,75,79,82,86].
Conotoxins purified from Conus venom during the last ten years are summarized in Table 2. Fifty conotoxins in total were discovered during the last 10 years and only five in average were found and characterized per year, indicating that conotoxin discovery from crude venom isolation was stagnant. More efficient omic study is developing in full swing in recent years. It is difficult to isolate a novel conotoxin from limited amount of crude venom that consists of more than 1000 venom peptides. Blind search policy always makes the native peptide isolation process time-consuming and laborious. Therefore, only limited random conotoxins were discovered, which belonged to a few gene superfamilies (Table 2). Hence, more effective bioassay-guided and MS-sequence-tag guided fractionation methods are under development to facilitate the rapid discovery of novel native conotoxins from different crude venoms [62,86,87].

Table 2.
Conotoxins isolated from cone snail venom during recent ten years.
4. Gene Cloning to Discover New Conotoxins
To overcome the limitations of crude venom purification strategy, gene cloning for novel conotoxins discovery has emerged in 1990s [94]. Since a conotoxin is expressed by a specific gene, it can be amplified by PCR technique with specific primers [53,95,96,97,98].
Generally, genomic DNA is extracted from snail tissue of an individual specimen, or cDNA is prepared by reversed transcription of mRNA extracted from dissected venom duct. The resulting total DNA or cDNA is served as a template for PCR amplification with forward and reverse primers to perform 3′- and 5′-RACE. The primers are designed and synthesized on the basis of the conserved sequence in signal region (Figure 3, primer 1) or untranslated region of 3′- or 5′-UTRs (Figure 3, primer 2 and 3) of specific known conotoxin precursor, or its relatively conserved introns (Figure 3, primer 4). The PCR products are purified by electrophoresis on agarose gel and are ligated into a plasmid vector for sequencing. The annotation of possible conotoxin-encoding genes is conducted based on homologous searching. The resulting conotoxin sequences are analyzed and assigned by CLUSTALX [99]. The signal region sequences can be predicted by SignalP 3.0 server (http://www.cbs. dtu.dk/services/SignalP/) [99,100,101].
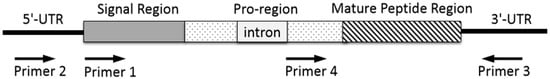
Figure 3.
PCR amplification strategy to clone conotoxin precursor genes from genomic DNA.
Primers make it possible for specific conotoxin-encoding genes to be amplified from the total genomic DNA or RNA of a cone snail. Thus the PCR primer design is a key factor for conotoxin discovery by gene cloning. Generally, the resulting PCR sequence of a conotoxin precursor gene generated by primer 1 or primer 2 pairing with primer 3, contains a complete open reading frame (ORF) sequence, which includes a signal region, a pro-region, and a mature peptide region (Figure 3). When primer 4 pairing with primer 3 is used to clone a conotoxin precursor gene, it starts with partial pro-region without signal peptide. Representative α-family (α*-) conotoxins discovered by gene cloning during the last ten years are shown in Table 3. The resulting sequences of Pu14.1 and GeXIVA consist of complete precursor sequences including signal regions which facilitate to assign gene superfamily category. Previous study showed that the sequences of the α-conotoxin intron in pro-region is highly conserved [97]. Many new α-conotoxins have been discovered by PCR technique using its conserved intron and 3′-UTR primers in our lab, such as α-conotoxin TxIB, TxID, LvIA, etc., which do not contain signal regions (Table 3). A forward primer and its paired reverse primer could be designed according to the conserved intron of a known gene superfamily and its 3′-UTR, such as A-, O-, or other superfamily, to clone novel conotoxin precursor genes. Random cDNA sequencing can also obtain the complete precursor sequence, e.g., VxXXIVA, but this method is not as targeted as the strategy with delicately designed primers.

Table 3.
Representative α*-conotoxins discovered by gene cloning during the last ten years.
When compared with crude venom purification, the gene cloning strategy is more resource-saving. Generally, several or even one specimen is enough for conotoxin gene cloning. However, the mature peptide sequences are speculated from their precursor genes, so no PTMs identification is involved. On the other hand, gene cloning strategy is relatively low-throughput, when compared with the transcriptomic approach that arose in 2010s. In addition, the primers for gene cloning are designed according to the conserved sequences of known family or superfamily, so new family or superfamily conotoxins are difficult to be discovered by this way.
5. Cone Snail Multi-Omics
Although big efforts have been made for novel conotoxin discovery from natural crude venom and gene cloning, most of the total estimated conotoxins have not been characterized yet [108]. More efficient, resource-saving, and high-throughput methodology urgently needs to be exploited. “Omics” such as transcriptomics and proteomics, and “Multi-omics” by integrating them together, have opened a new era for conotoxin discovery and rapidly accelerate the rate of conotoxin discovery [108,109,110].
5.1. Transcriptomics—A Useful Pathway to Identify Putative Conotoxins
Transcriptomics aims to identify and profile the holistic gene (including the conotoxin-encoding genes) transcription and expression at RNA level. Venom duct is an ideal material for transcriptomic analysis, because the number and level of conotoxin-encoding transcripts from venom duct are much larger than those from other tissues [50,111]. Conus venom duct transcriptomics is able to describe the conotoxin expression and it has presented a useful method to rapidly identify putative conopeptide sequences. In addition, transcriptomics using next generation sequencing (NGS) technology [112] makes large scale sequencing time- and cost-effective.
The transcriptomic pipeline (Figure 4) starts from the total RNA extraction of dissected venom duct. Then, mRNAs are served as reverse-transcriptional templates for cDNA library construction. PCR amplification is conducted while using cDNA as template and specific sequences as primers. The resulting cDNA or the raw RNA sequences are sequencing using NGS platforms, such as 454 (Roche, Branford, CT, USA), Illumina (Illumina, San Diego, CA, USA), Ion Torrent Personal Genome Machine (Thermo Fisher, Waltham, MA, USA), Nanopore (Oxford, UK), ABI 3730 Series (Applied Biosystems, Foster City, CA, USA), and PacBio (Pacific Biosciences, Menlo Park, CA, USA) [108,113]. Illumina and Roche 454 are the most widely-used NGS platform at present (Table 4). The raw reads generated from NGS platforms require data assembly to remove artifacts, poor quality raw reads, as well as redundant and aberrant sequences [114]. The trimmed sequences are then deciphered into peptide primary sequences according to opening reading frames (ORFs) [112] by ConoPrec [1,53,54,111,115,116] or SignalP4.0 [115,116,117,118], which may locate the signal peptides and predict their cleavage sites. Profile Hidden Markov Models (pHMMs) [1,111,118,119] is a useful tool of ConoSorter [1,110,118,119], which could identify the putative precursors of conopeptides and categorize their superfamilies. Homology search and analysis by running BLAST against the combined searchable online databases, like ConoServer (The university of Queensland, Brisbane, Australia) (http://research1t.imb.uq.edu.au/conoserver/) [120,121], UniProtKB/Swiss-Prot (http://www.uniprot.org/downloads) [122,123], and NCBI (http://www.ncbi.nlm.nih.gov/), may enable the rapid identification of known and novel conotoxins. ConoSorter also facilitates to illustrating relative sequence frequency, length, number of cysteines, N-terminal hydrophobicity, and sequence similarity score [118]. Thus, a unique transcriptomic dataset for an individual specimen from a specific Conus specie might be established.
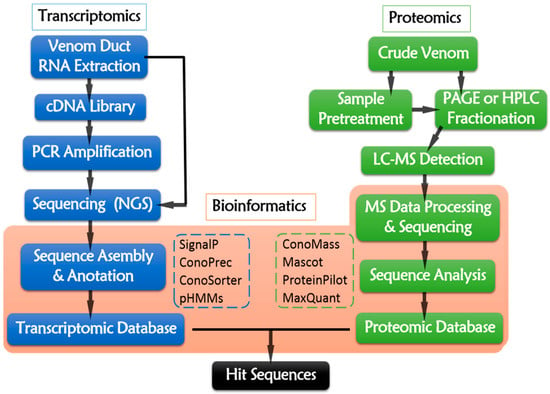
Figure 4.
Multi-omic pipeline of conotoxin discovery.

Table 4.
The reported transcriptomic and proteomic data from various cone snails during the past decade.
Compared with traditional isolation and gene cloning, venom duct transcriptomic approach is a rapid, efficient, resource-saving, and high-throughput way to identify massive conotoxins from different cone snails, which greatly extends our cognition of conotoxin resource (Table 4). During the last decade, many putative conopeptide precursors have been identified from transcriptome of different Conus species (Table 4). At least 30 conopeptides precursors were discovered from C. bullatus by transcriptome sequencing. Surprisingly, as many as 3305 novel conopeptide precursors were discovered from a single Conus episcopatus specimen by sequencing its transcriptome (Table 4).
Phylogeny-based conotoxin discovery utilizes the known conserved sequence to design specific primers for PCR amplification, which enables to find more conopeptides belonging to known superfamilies from different Conus species [124]. Additionally, specific PCR primers might be designed according to incomplete sequences that were obtained by MS-sequencing-tag or Edman degradation [124,125], which is also applied to clone new conotoxin precursors from venom transcriptome, cDNA, and its genomic DNA of various Conus species. It provides a feasible way to explore novel conotoxins belonging to new superfamilies [124]. cDNA library normalization is an effective and commonly-used method to equalize some specific cDNA, which facilitates to identify conotoxin genes with a relatively low level expression level [114,124]. Normalization suppresses highly abundant transcript reads and increases rare transcripts, so as to maximize the identified number of unique conotoxins [119].
Thanks to transcriptomic study, venom insulins, which target the heterospecific insulin receptors of prey, predators, and competitors, have been proven to be expressed in many worm- and snail-hunting cone snails [126]. Six insecticidal conotoxins have been validated and screened out from the transcriptomic dataset of 215 precursors by homologous search with α-conotoxin ImI [127]. These findings reveal that Conus transcriptomic database can promote the extension for new knowledge and find various new conopeptides.
5.2. Proteomics—An Effective Approach to Discovery Natural Conotoxins
Traditional proteomic identification depends on Edman degradation and amino acid composition analysis to assign the peptide sequences, but its sample-consuming and low-throughput characters make it difficult to be extensively applied. As the high-resolution MS instrument appears [135], venom proteomic study with the aid of modern MS technology has proven to be an effective and high-throughput approach for novel conotoxin discovery [108,109].
The general proteomic procedure is presented in Figure 4. Briefly, the venom sample is prepared by squeezing the venom from dissected venom duct (one-off operation), or collecting the secreted venom that is induced by pray from living cone snail individuals (reproducible operation) [1]. As the MS techniques develop, the required venom amount for experimental analysis of proteomics is decreased. Even about 7% or less of crude venom from one specimen is enough [136]. The proteomic data detected from different Conus species, especially for those cone snails hunting different preys, are quite different from each other, because different species and the food preference are the key factors for the evolution of venom diversity [137].
In traditional bottom-up proteomics, pretreatment of venom sample, such as reduction, alkylation, and enzymatic digestion, is carried out before HPLC-MS analysis in order to eliminate the influence of disulfide bonds, although it leads to partial loss of conopeptides during processing [50,53,111,115,138]. In top-down proteomic approach, intact disulfide-bridged venom peptides are remained, which makes it more applicable to analyze simple peptide mixtures, like highly purified venom subfractions, whereas bottom-up approach is more suitable for complex crude venom [50,53,139]. The combination of top-down and bottom-up approach enables the identification of several unexpected cleavage sites during conotoxin maturation [53].
The resulting vast MS data generated from LC-MS analysis are subjected to bioinformatic tools for further data processing and mining. Raw MS data are inputted into Mascot for Peptide mass fingerprint. ProteinPilot™ [1,49,54,115] is used for sequence identification and the annotation of precursor ions by searching the MS/MS mass list obtained at a relatively high precise level [54]. Parameters for enzymolysis and various types of PTMs are imported into ProteinPilot to identify PTMs and fragment splicing. ConoMass [49,115] and ProteinPilot are able to identify nearly all the PTMs except glycosylation, which requires assignment by de novo sequencing. The sequences are homologically searched and matched against databases, such as ConoServer, UniProtKB/Swiss-Prot, NCBI, and known transcriptomic dataset from its own, to identify the known and novel conopeptides as well as their gene superfamilies. The subsequent results are presented by various peptide sequences with a series of statistical data to profile the venom components.
Advanced mass analyzer, like TOF, especially Quadrupole-TOF (Q-TOF), shows rapid acquisition, high resolution, first-class sensitivity, and excellent mass accuracy. Ionization methods, such as ESI, MALDI, CID, ETD, EThcD, etc., provide options for obtaining alternative mass data for different purpose. ESI and MALDI are generally for proteomic study, whereas CID, ETD, EThcD are commonly for de novo MS sequencing by providing different dissociation patterns to acquire variable specific peptide fragments. More mature peptides can be detected by using superior MS instrument with advanced mass analyzer and efficient ionization technique. For instance, from venom proteomics of Conus marmoreus, there were 2710 peptide sequences revealed by MALDI-TOF; 3172 peptide sequences were detected by ESI-Q-TOF with regular electrospray; and 6254 peptide sequences were disclosed using ESI-Q-TOF, which is equipped with a DuoSpray ionization source [49]. ETD ionization strategy combined with targeted chemical derivatization has been applied to increase the charge state of conopeptides so as to maximize the detectable mass range, because the molecular masses of conotoxins usually exceed the optimum detective coverage [132]. Superior mass analyzer and various ionization methods are combined and applied to expand the boundary of accessible venom repertoire. Modern venom proteomics provides a methodology, not only for the rapid detection and characterization of specific conotoxins, but also for profiling an overview of the complex venom components.
5.3. Bioinformaics—An Efficient Tool for Massive Data Processing and Integrating
Bioinformatics is an efficient tool for massive data processing and integrating, which has been deeply penetrative during the raw data processing, sequence identification, and superfamily classification by exquisite analytical softwares and algorithms with the introduction of integrated databases [108,140]. Venom duct transcriptomics and venom proteomics both benefit from the emergence and development of bioinformatics, especially the improvements on bioinformatic softwares, algorithms and expansion of searchable databases. The functions of the frequently-used tools for transcriptomics and proteomics are presented in Table 5. Transcriptomic and proteomic studies are quite reliant on the foundation database, which provides templates for sequence searching, matching, and annotating. The databases for sequence identification and BLAST should be the latest updated version, which should be composed of complete or partial natural precursor and mature toxin sequences generated either from conotoxin genes, transcripts, or proteins, as well as artificially synthesized conotoxins. Discovery of novel sequences using different approaches, in return, expands the capacity of the databases.

Table 5.
Frequently-used bioinformatic tools for cone snail venom transcriptomics and proteomics.
5.4. Multi-Omics Integration
A comprehensive strategy, named “multi-omics” or “venomics” [108,140], by integrating transcriptomics with proteomics through bioinformatics, is popular in the field of conotoxin research [110]. Although venom duct transcriptomics and venom proteomics both have proven to be effective and high-throughput methods to identify massive conopeptide sequences, the conotoxin sequences that are generated from transcriptomics are putative precursor peptides that need to be further confirmed for their real existence at protein level. Furthermore, no PTMs could be predicted from the precursor sequences. Luckily, venom proteomics is able to validate the putative peptides at protein level (Table 4) and identify nearly all the PTMs [49,128]. The validated sequencing and PTMs data can help to illustrate the processing mechanisms (transcript variation, VPP, PTMs) from precursor peptides (transcriptomic data) to the corresponding mature peptides (proteomic data).
Just as not every putative precursor can be validated by proteomic data, not all peptide sequences from proteomic data can find their corresponding precursors (Table 4). In fact, they were overlapped and matched with a very small percentage of 9.98% for Conus episcopatus [50]. The significant variations between the datasets of transcriptomics and proteomics actually exist, and the overlapped data (hit sequences) are not big enough. How to extend the datasets for making access to the completed repertoire of conotoxins? How to decode the variation so as to expand the overlapped or matched precursors with their corresponding mature peptides? Since one precursor can generate various mature conopeptides by the different PTMs. Theoretically, more mature peptides should be detected by proteomics. In practical use, the detected number always varies greatly with different MS instrument, bioinformatic analytical methods, fractionation, and sample pretreatment processes, etc. Rare transcripts at a low translational level are difficult to be recognized, which also contribute to the disparity. Thus, standardized processing protocols, reliable detection methods, dedicated integrated databases, and robust data analysis tools are needed.
6. Conclusions and Prospects
In this review, we introduced the discovery methodology of novel conotoxins from various Conus species. It focused on obtaining full N- to C-terminal sequences, regardless of disulfide connectivity through crude venom purification, conotoxin precursor gene cloning, venom duct transcriptomics, venom proteomics, and multi-omic methods. The protocols, advantages, disadvantages, and developments of different approaches during the last decade and the promising prospects are summarized and discussed. To overcome the limitations of crude venom purification strategy, gene cloning technique have been developed and it temporarily slows down the deprivation of the native cone snail resource.
In order to improve efficiency, high-throughput omic and multi-omic strategies have opened a new era for conotoxin discovery. Transcriptomics and proteomics are now acknowledged to be effective, resource-saving, and high-throughput approaches for novel conotoxin discovery. Multi-omic strategy is more efficient than using transcriptomics or proteomics alone. Efforts should be made to decode and reduce the variations between transcriptomic and proteomic data in order to expand the accessible repertoire of known conotoxins. The precursor processing mechanisms need to be illustrated as well. Thus, standardized processing protocols, reliable detection methods, dedicated integrated databases, and robust data analysis tools for transcriptomic, proteomic, and multi-omic studies are required to speed up novel conotoxin discovery.
Author Contributions
Y.F. and S.L. conceived and designed the article; C.L., Y.W. and S.D. collected the data; Y.F. and C.L. wrote the article, while S.L., D.Z. and S.D. revised the article.
Funding
This work was supported by Natural Science Foundation of Hainan Province (No. 817001), Changjiang Scholars and Innovative Research Team in University Grant (IRT_15R15), and National Natural Science Foundation of China (No. 81660585, No.31760249).
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| BLAST | Basic local alignment search tool |
| CID | Collision induced dissociation |
| ESI | Electrospray ionization |
| ETD | Electron transfer dissociation |
| EThcD | Electron transfer higher energy collision dissociation |
| GPCRs | G protein-coupled receptors |
| HPLC | High performance liquid chromatography |
| LTQ-Orbitrap | Linear Trap Quatropole-Orbitrap |
| MS | Mass spectrometry |
| MALDI | Matrix-assisted laser desorption ionization |
| nAChRs | Nicotinic acetylcholine receptors |
| NCBI | National center of biotechnology information |
| NMR | Nuclear magnetic resonance |
| NMDA | N-methyl-d-aspartic acid receptor |
| PAGE | Poly acrylamide gel electrophoresis |
| PCR | Polymerase chain reaction |
| RACE | Rapid amplification of cDNA ends |
| SILAC | Stable isotope labeling by amino acids in cell culture |
| SPPS | Solid phase peptide synthesis |
| TOF | Time of flight |
References
- Prashanth, J.R.; Dutertre, S.; Jin, A.H.; Lavergne, V.; Hamilton, B.; Cardoso, F.C.; Griffin, J.; Venter, D.J.; Alewood, P.F.; Lewis, R.J. The role of defensive ecological interactions in the evolution of conotoxins. Mol. Ecol. 2016, 25, 598–615. [Google Scholar] [CrossRef] [PubMed]
- Endean, R.; Duchemin, C. The venom apparatus of Conus magus. Toxicon 1967, 4, 275–284. [Google Scholar] [CrossRef]
- Dutertre, S.; Jin, A.H.; Vetter, I.; Hamilton, B.; Sunagar, K.; Lavergne, V.; Dutertre, V.; Fry, B.G.; Antunes, A.; Venter, D.J. Evolution of separate predation- and defence-evoked venoms in carnivorous cone snails. Nat. Commun. 2014, 5, 3521. [Google Scholar] [CrossRef] [PubMed]
- Marshall, J.; Kelley, W.P.; Rubakhin, S.S.; Bingham, J.P.; Sweedler, J.V.; Gilly, W.F. Anatomical correlates of venom production in Conus californicus. Biol. Bull. 2002, 203, 27–41. [Google Scholar] [CrossRef] [PubMed]
- Salisbury, S.M.; Martin, G.G.; Kier, W.M.; Schulz, J.R. Venom kinematics during prey capture in Conus: The biomechanics of a rapid injection system. J. Exp. Biol. 2010, 213, 673–682. [Google Scholar] [CrossRef] [PubMed]
- Kohn, A.J. Cone Shell Stings. Recent Cases of Human Injury due to Venomous Marine Snails of the Genus Conus. Hawaii Med. J. 1958, 17, 528. [Google Scholar] [PubMed]
- Terlau, H.; Olivera, B.M. Conus venoms: A rich source of novel ion channel-targeted peptides. Physiol. Rev. 2004, 84, 41–68. [Google Scholar] [CrossRef] [PubMed]
- Tosti, E.; Boni, R.; Gallo, A. µ-Conotoxins Modulating Sodium Currents in Pain Perception and Transmission: A Therapeutic Potential. Mar. Drugs 2017, 15, 295. [Google Scholar] [CrossRef] [PubMed]
- Oliver, K.; Mcarthur, J.R.; Adams, D.J. Conotoxins Targeting Neuronal Voltage-Gated Sodium Channel Subtypes: Potential Analgesics? Toxins 2012, 4, 1236–1260. [Google Scholar] [CrossRef]
- Leipold, E.; Ullrich, F.; Thiele, M.; Tietze, A.A.; Terlau, H.; Imhof, D.; Heinemann, S.H. Subtype-specific block of voltage-gated K+ channels by μ-conopeptides. Biochem. Biophys. Res. Commun. 2017, 482, 1135–1140. [Google Scholar] [CrossRef] [PubMed]
- Ramírez, D.; Gonzalez, W.; Fissore, R.A.; Carvacho, I. Conotoxins as Tools to Understand the Physiological Function of Voltage-Gated Calcium (CaV) Channels. Mar. Drugs 2017, 15, 313. [Google Scholar] [CrossRef] [PubMed]
- Bourinet, E.; Zamponi, G.W. Block of voltage-gated calcium channels by peptide toxins. Neuropharmacology 2017, 127, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Giribaldi, J.; Dutertre, S. α-Conotoxins to explore the molecular, physiological and pathophysiological functions of neuronal nicotinic acetylcholine receptors. Neurosci. Lett. 2018, 679, 24–34. [Google Scholar] [CrossRef] [PubMed]
- Dutertre, S.; Nicke, A.; Tsetlin, V.I. Nicotinic acetylcholine receptor inhibitors derived from snake and snail venoms. Neuropharmacology 2017, 127, 196–223. [Google Scholar] [CrossRef] [PubMed]
- Azam, L.; Mcintosh, J.M. Alpha-conotoxins as pharmacological probes of nicotinic acetylcholine receptors. Acta Pharmacol. Sin. 2009, 30, 771. [Google Scholar] [CrossRef] [PubMed]
- England, L.J.; Imperial, J.; Jacobsen, R.; Craig, A.G.; Gulyas, J.; Akhtar, M.; Rivier, J.; Julius, D.; Olivera, B.M. Inactivation of a serotonin-gated ion channel by a polypeptide toxin from marine snails. Science 1998, 281, 575–578. [Google Scholar] [CrossRef] [PubMed]
- Barton, M.E.; White, H.S.; Wilcox, K.S. The effect of CGX-1007 and CI-1041, novel NMDA receptor antagonists, on NMDA receptor-mediated EPSCs. Epilepsy Res. 2004, 59, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Castro, J.; Harrington, A.M.; Garciacaraballo, S.; Maddern, J.; Grundy, L.; Zhang, J.; Page, G.; Miller, P.E.; Craik, D.J.; Adams, D.J. α-Conotoxin Vc1.1 inhibits human dorsal root ganglion neuroexcitability and mouse colonic nociception via GABAB receptors. Gut 2017, 66, 1083–1094. [Google Scholar] [CrossRef] [PubMed]
- Daniel, J.T.; Clark, R.J. G-Protein Coupled Receptors Targeted by Analgesic Venom Peptides. Toxins 2017, 9, 372. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Rogge, G.; Hague, C.; Alewood, D.; Colless, B.; Lewis, R.J.; Minneman, K.P. Subtype-selective noncompetitive or competitive inhibition of human alpha1-adrenergic receptors by rho-TIA. J. Biol. Chem. 2004, 279, 35326–35333. [Google Scholar] [CrossRef] [PubMed]
- Sharpe, I.A.; Gehrmann, J.; Loughnan, M.L.; Thomas, L.; Adams, D.A.; Atkins, A.; Palant, E.; Craik, D.J.; Adams, D.J.; Alewood, P.F. Two new classes of conopeptides inhibit the alpha1-adrenoceptor and noradrenaline transporter. Nat. Neurosci. 2001, 4, 902–907. [Google Scholar] [CrossRef] [PubMed]
- Möller, C.; Marí, F. A vasopressin/oxytocin-related conopeptide with gamma-carboxyglutamate at position 8. Biochem. J. 2007, 404, 413–419. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.K.; Zhang, L.; Smith, M.D.; Walewska, A.; Vellore, N.A.; Baron, R.; Mcintosh, J.M.; White, H.S.; Olivera, B.M.; Bulaj, G. A marine analgesic peptide, Contulakin-G, and neurotensin are distinct agonists for neurotensin receptors: Uncovering structural determinants of desensitization properties. Front. Pharmacol. 2015, 6, 11. [Google Scholar] [CrossRef] [PubMed]
- Paczkowski, F.A.; Sharpe, I.A.; Dutertre, S.; Lewis, R.J. chi-Conotoxin and tricyclic antidepressant interactions at the norepinephrine transporter define a new transporter model. J. Biol. Chem. 2007, 282, 17837. [Google Scholar] [CrossRef] [PubMed]
- Romero, H.K.; Christensen, S.B.; Di Cesare Mannelli, L.; Gajewiak, J.; Ramachandra, R.; Elmslie, K.S.; Vetter, D.E.; Ghelardini, C.; Iadonato, S.P.; Mercado, J.L.; et al. Inhibition of alpha9alpha10 nicotinic acetylcholine receptors prevents chemotherapy-induced neuropathic pain. Proc. Natl. Acad. Sci. USA 2017, 114, E1825–E1832. [Google Scholar] [CrossRef] [PubMed]
- Hannon, H.E.; Atchison, W.D. Omega-Conotoxins as Experimental Tools and Therapeutics in Pain Management. Mar. Drugs 2013, 11, 680. [Google Scholar] [CrossRef] [PubMed]
- Crooks, P.A.; Bardo, M.T.; Dwoskin, L.P. Nicotinic receptor antagonists as treatments for nicotine abuse. Adv. Pharmacol. 2014, 69, 513–551. [Google Scholar] [CrossRef] [PubMed]
- Gandini, M.A.; Sandoval, A.; Felix, R. Toxins targeting voltage-activated Ca2+ channels and their potential biomedical applications. Curr. Top. Med. Chem. 2015, 15, 604–616. [Google Scholar] [CrossRef] [PubMed]
- Irasema, O.P.; Mario, N.; Cervantes-Luevano, K.E.; Carolina, Á.-D.; Guy, S.; Sanchez-Campos, L.N.; Licea-Navarro, A.F. Apoptosis Activation in Human Lung Cancer Cell Lines by a Novel Synthetic Peptide Derived from Conus californicus Venom. Toxins 2016, 8, 38. [Google Scholar] [CrossRef]
- Yang, R.; Liu, Y.; Hou, X.; Fan, Y.; Li, J.; Chen, M.; Wang, Y.; Zhang, X.; Zhang, M. MAPKs-mediated modulation of the myocyte voltage-gated K+ channels is involved in ethanol-induced rat coronary arterial contraction. Eur. J. Pharmacol. 2018, 834, 274–280. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Dendorfer, A.; Finolurdaneta, R.K.; Terlau, H.; Olivera, B.M. Biochemical Characterization of κM-RIIIJ, a Kv1.2 Channel Blocker. J. Biol. Chem. 2010, 285, 14882–14889. [Google Scholar] [CrossRef] [PubMed]
- Vetter, I.; Lewis, R.J. Therapeutic potential of cone snail venom peptides (conopeptides). Curr. Top. Med. Chem. 2012, 12, 1546–1552. [Google Scholar] [CrossRef] [PubMed]
- Lewis, R.J.; Dutertre, S.; Vetter, I.; Christie, M.J. Conus venom peptide pharmacology. Pharmacol. Rev. 2012, 64, 259. [Google Scholar] [CrossRef] [PubMed]
- Layer, R.T.; Mcintosh, J.M. Conotoxins: Therapeutic Potential and Application. Mar. Drugs 2006, 4, 119–142. [Google Scholar] [CrossRef]
- Akondi, K.B.; Muttenthaler, M.; Dutertre, S.; Kaas, Q.; Craik, D.J.; Lewis, R.J.; Alewood, P.F. Discovery, synthesis, and structure: Activity relationships of conotoxins. Chem. Rev. 2014, 114, 5815–5847. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.; Peng, C.; Yang, J.; Yi, Y.; Zhang, J.; Shi, Q. Cone Snails: A Big Store of Conotoxins for Novel Drug Discovery. Toxins 2017, 9, 397. [Google Scholar] [CrossRef] [PubMed]
- Prashanth, J.R.; Brust, A.; Jin, A.H.; Alewood, P.F.; Dutertre, S.; Lewis, R.J. Cone snail venomics: From novel biology to novel therapeutics. Future Med. Chem. 2014, 6, 1659–1675. [Google Scholar] [CrossRef] [PubMed]
- Halai, R.; Craik, D.J. Conotoxins: Natural product drug leads. Nat. Prod. Rep. 2009, 26, 526–536. [Google Scholar] [CrossRef] [PubMed]
- Miljanich, G.P. Ziconotide: Neuronal calcium channel blocker for treating severe chronic pain. Curr. Med. Chem. 2004, 11, 3029–3040. [Google Scholar] [CrossRef] [PubMed]
- Pope, J.E.; Deer, T.R. Ziconotide: A clinical update and pharmacologic review. Expert Opin. Pharmacother. 2013, 14, 957–966. [Google Scholar] [CrossRef] [PubMed]
- Obata, H.; Conklin, D.; Eisenach, J.C. Spinal noradrenaline transporter inhibition by reboxetine and Xen2174 reduces tactile hypersensitivity after surgery in rats. Pain 2005, 113, 271–276. [Google Scholar] [CrossRef] [PubMed]
- Lubbers, N.L.; Campbell, T.J.; Polakowski, J.S.; Bulaj, G.; Layer, R.T.; Moore, J.; Gross, G.J.; Cox, B.F. Postischemic administration of CGX-1051, a peptide from cone snail venom, reduces infarct size in both rat and dog models of myocardial ischemia and reperfusion. J. Cardiovasc. Pharm. 2005, 46, 141–146. [Google Scholar] [CrossRef]
- Clark, R.J.; Fischer, H.; Nevin, S.T.; Adams, D.J.; Craik, D.J. The synthesis, structural characterization, and receptor specificity of the alpha-conotoxin Vc1.1. J. Biol. Chem. 2006, 281, 23254–23263. [Google Scholar] [CrossRef] [PubMed]
- Kern, S.; Allen, J.; Wagstaff, J.S.; Yaksh, T. The pharmacokinetics of the conopeptide contulakin-G (CGX-1160) after intrathecal administration: An analysis of data from studies in beagles. Anesth. Analg. 2007, 104, 1514–1520. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.J.; Zhang, M.M.; Azam, L.; Olivera, B.M.; Bulaj, G.; Yoshikami, D. Navβ subunits modulate the inhibition of Nav1.8 by the analgesic gating modifier μO-conotoxin MrVIB. J. Pharmacol. Exp. Ther. 2011, 338, 687–693. [Google Scholar] [CrossRef] [PubMed]
- Hieble, J.P.; Robert, R.R., Jr. The use of alpha-adrenoceptor antagonists in the pharmacological management of benign prostatic hypertrophy: An overview. Pharmacol. Res. 1996, 33, 145–160. [Google Scholar] [CrossRef] [PubMed]
- Brust, A.; Palant, E.; Croker, D.E.; Colless, B.; Drinkwater, R.; Patterson, B.; Schroeder, C.I.; Wilson, D.; Nielsen, C.K.; Smith, M.T. chi-Conopeptide pharmacophore development: Toward a novel class of norepinephrine transporter inhibitor (Xen2174) for pain. J. Med. Chem. 2009, 52, 6991–7002. [Google Scholar] [CrossRef] [PubMed]
- Puillandre1, N.; Duda, T.F.; Meyer, C.; Olivera, B.M.; Bouchet, P. One, four or 100 genera? A new classification of the cone snails. J. Molluscan Stud. 2015, 81, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Dutertre, S.; Jin, A.H.; Kaas, Q.; Jones, A.; Alewood, P.F.; Lewis, R.J. Deep venomics reveals the mechanism for expanded peptide diversity in cone snail venom. Mol. Cell. Proteom. 2013, 12, 312–329. [Google Scholar] [CrossRef] [PubMed]
- Lavergne, V.; Harliwong, I.; Jones, A.; Miller, D.; Taft, R.J.; Alewood, P.F. Optimized deep-targeted proteotranscriptomic profiling reveals unexplored Conus toxin diversity and novel cysteine frameworks. Proc. Natl. Acad. Sci. USA 2015, 112, E3782. [Google Scholar] [CrossRef] [PubMed]
- Davis, J.; Jones, A.; Lewis, R.J. Remarkable inter- and intra-species complexity of conotoxins revealed by LC/MS. Peptides 2009, 30, 1222–1227. [Google Scholar] [CrossRef] [PubMed]
- Woodward, S.R.; Cruz, L.J.; Olivera, B.M.; Hillyard, D.R. Constant and hypervariable regions in conotoxin propeptides. EMBO J. 1990, 9, 1015–1020. [Google Scholar] [CrossRef] [PubMed]
- Lu, A.; Yang, L.; Xu, S.; Wang, C. Various Conotoxin Diversifications Revealed by a Venomic Study of Conus flavidus. Mol. Cell. Proteom. 2014, 13, 105–118. [Google Scholar] [CrossRef] [PubMed]
- Jin, A.H.; Dutertre, S.; Kaas, Q.; Lavergne, V.; Kubala, P.; Lewis, R.J.; Alewood, P.F. Transcriptomic messiness in the venom duct of Conus miles contributes to conotoxin diversity. Mol. Cell. Proteom. 2013, 12, 3824–3833. [Google Scholar] [CrossRef] [PubMed]
- Jakubowski, J.A.; Kelley, W.P.; Sweedler, J.V. Screening for post-translational modifications in conotoxins using liquid chromatography/mass spectrometry: An important component of conotoxin discovery. Toxicon 2006, 47, 688–699. [Google Scholar] [CrossRef] [PubMed]
- Riveraortiz, J.A.; Cano, H.; Marí, F. Intraspecies variability and conopeptide profiling of the injected venom of Conus ermineus. Peptides 2011, 32, 306–316. [Google Scholar] [CrossRef] [PubMed]
- Cruz, L.J.; Gray, W.R.; Olivera, B.M. Purification and properties of a myotoxin from Conus geographus venom. Arch. Biochem. Biophys. 1978, 190, 539–548. [Google Scholar] [CrossRef]
- Edman, P.; Begg, G. A Protein Sequenator. FEBS J. 1967, 1, 80–91. [Google Scholar] [CrossRef]
- Wang, L.; Liu, J.; Pi, C.; Zeng, X.; Zhou, M.; Jiang, X.; Chen, S.; Ren, Z.; Xu, A. Identification of a novel M-superfamily conotoxin with the ability to enhance tetrodotoxin sensitive sodium currents. Arch. Toxicol. 2009, 83, 925–932. [Google Scholar] [CrossRef] [PubMed]
- Van, D.H.A.; Peigneur, S.; Dyubankova, N.; Möller, C.; Marí, F.; Diego-García, E.; Naudé, R.; Lescrinier, E.; Herdewijn, P.; Tytgat, J. Pc16a, the first characterized peptide from Conus pictus venom, shows a novel disulfide connectivity. Peptides 2012, 34, 106–113. [Google Scholar] [CrossRef]
- Bernáldez, J.; Romángonzález, S.A.; Martínez, O.; Jiménez, S.; Vivas, O.; Arenas, I.; Corzo, G.; Arreguín, R.; García, D.E.; Possani, L.D. A Conus regularis Conotoxin with a Novel Eight-Cysteine Framework Inhibits CaV2.2 Channels and Displays an Anti-Nociceptive Activity. Mar. Drugs 2013, 11, 1188–1202. [Google Scholar] [CrossRef] [PubMed]
- Lebbe, E.K.; Peigneur, S.; Maiti, M.; Mille, B.G.; Devi, P.; Ravichandran, S.; Lescrinier, E.; Waelkens, E.; D’Souza, L.; Herdewijn, P. Discovery of a new subclass of α-conotoxins in the venom of Conus australis. Toxicon 2014, 91, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Lebbe, E.K.M.; Peigneur, S.; Maiti, M.; Devi, P.; Ravichandran, S.; Lescrinier, E.; Ulens, C.; Waelkens, E.; D’Souza, L.; Herdewijn, P. Structure-Function Elucidation of a New α-Conotoxin, Lo1a, from Conus longurionis. J. Biol. Chem. 2014, 91, 170–171. [Google Scholar] [CrossRef]
- Nguyen, B.; Le, C.J.; Aráoz, R.; Thai, R.; Lamthanh, H.; Benoit, E.; Molgó, J. Isolation, purification and functional characterization of alpha-BnIA from Conus bandanus venom. Toxicon 2014, 91, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Zhang, T.; Kompella, S.N.; Yan, M.; Lu, A.; Wang, Y.; Shao, X.; Chi, C.; Adams, D.J.; Ding, J.; et al. Conotoxin alphaD-GeXXA utilizes a novel strategy to antagonize nicotinic acetylcholine receptors. Sci. Rep. 2015, 5, 14261. [Google Scholar] [CrossRef] [PubMed]
- Lei, W.; Liu, J.; Ren, Z.; Yu, C.; Xu, A. Discovery of two P-superfamily conotoxins, lt9a and lt9b, with different modifications on voltage-sensitive sodium channels. Toxicon 2017, 134, 6–13. [Google Scholar] [CrossRef]
- Jiang, S.; Tae, H.S.; Xu, S.; Shao, X.; Adams, D.J.; Wang, C. Identification of a Novel O-Conotoxin Reveals an Unusual and Potent Inhibitor of the Human α9α10 Nicotinic Acetylcholine Receptor. Mar. Drugs 2017, 15, 170. [Google Scholar] [CrossRef] [PubMed]
- Yuan, D.D.; Liu, L.; Shao, X.X.; Peng, C.; Chi, C.W.; Guo, Z.Y. New conotoxins define the novel I3-superfamily. Peptides 2009, 30, 861–865. [Google Scholar] [CrossRef] [PubMed]
- Ni, H.; Chen, F.; Cai, H.; Xiao, A.; You, Q.; Lu, Y. Isolation and Characterization of Conotoxin bt5a from Conus betulinus. Chin. J. Nat. Med. 2010, 8, 132–136. [Google Scholar] [CrossRef]
- Möller, C.; Marí, F. 9.3 KDa components of the injected venom of Conus purpurascens define a new five-disulfide conotoxin framework. Biopolymers 2011, 96, 158–165. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, M.B.; Zugasti-Cruz, A.; Falcón, A.; Batista, C.V.F.; Olivera, B.M.; Cotera, E.P.H.D.L. A novel arrangement of Cys residues in a paralytic peptide of Conus cancellatus (jr. syn.: Conus austini), a worm-hunting snail from the Gulf of Mexico. Peptides 2013, 41, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Heghinian, M.D.; Mejia, M.; Adams, D.J.; Godenschwege, T.A.; Marí, F. Inhibition of cholinergic pathways in Drosophila melanogaster by α-conotoxins. FASEB J. 2015, 29, 1011–1018. [Google Scholar] [CrossRef] [PubMed]
- Peng, C.; Ye, M.; Wang, Y.; Shao, X.; Yuan, D.; Liu, J.; Hawrot, E.; Wang, C.; Chi, C. A new subfamily of conotoxins belonging to the A-superfamily. Peptides 2010, 31, 2009–2016. [Google Scholar] [CrossRef] [PubMed]
- Christensen, S.B.; Bandyopadhyay, P.K.; Olivera, B.M.; McIntosh, J.M. αS-conotoxin GVIIIB potently and selectively blocks α9α10 nicotinic acetylcholine receptors. Biochem. Pharmacol. 2015, 96, 349–356. [Google Scholar] [CrossRef] [PubMed]
- Espino, S.S.; Dilanyan, T.; Imperial, J.S.; Aguilar, M.B.; Teichert, R.W.; Bandyopadhyay, P.; Olivera, B.M. Glycine-rich Conotoxins from the Virgiconus clade. Toxicon 2016, 113, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Lebbe, E.K.; Ghequire, M.G.; Peigneur, S.; Mille, B.G.; Devi, P.; Ravichandran, S.; Waelkens, E.; D’Souza, L.; De, M.R.; Tytgat, J. Novel Conopeptides of Largely Unexplored Indo Pacific Conus sp. Mar. Drugs 2016, 14, 199. [Google Scholar] [CrossRef] [PubMed]
- Echterbille, J.; Gilles, N.; Araóz, R.; Mourier, G.; Amar, M.; Servent, D.; Pauw, E.D.; Quinton, L. Discovery and characterization of EII B, a new α-conotoxin from Conus ermineus venom by nAChRs affinity capture monitored by MALDI-TOF/TOF mass spectrometry. Toxicon 2017, 130, 1. [Google Scholar] [CrossRef] [PubMed]
- Hoggard, M.F.; Rodriguez, A.M.; Cano, H.; Clark, E.; Tae, H.S.; Adams, D.J.; Godenschwege, T.A.; Marí, F. In vivo and in vitro testing of native α-conotoxins from the injected venom of Conus purpurascens. Neuropharmacology 2017, 127, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Kauferstein, S.; Kendel, Y.; Nicke, A.; Coronas, F.I.V.; Possani, L.D.; Favreau, P.; Krizaj, I.; Wunder, C.; Kauert, G.; Mebs, D. New conopeptides of the D-superfamily selectively inhibiting neuronal nicotinic acetylcholine receptors. Toxicon 2009, 54, 295–301. [Google Scholar] [CrossRef] [PubMed]
- Jimenez, E.C.; Olivera, B.M. Divergent M- and O-superfamily peptides from venom of fish-hunting Conus parius. Peptides 2010, 31, 1678–1683. [Google Scholar] [CrossRef] [PubMed]
- Ye, M.; Hong, J.; Zhou, M.; Huang, L.; Shao, X.; Yang, Y.; Sigworth, F.J.; Chi, C.; Lin, D.; Wang, C. A novel conotoxin, qc16a, with a unique cysteine framework and folding. Peptides 2011, 32, 1159–1165. [Google Scholar] [CrossRef] [PubMed]
- Ye, M.; Khoo, K.K.; Xu, S.; Zhou, M.; Boonyalai, N.; Perugini, M.A.; Shao, X.; Chi, C.; Galea, C.A.; Wang, C. A helical conotoxin from Conus imperialis has a novel cysteine framework and defines a new superfamily. J. Biol. Chem. 2012, 287, 14973–14983. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Shao, X.; Yan, M.; Chi, C.; Lu, A.; Wang, C. Identification of Two Novel O2-Conotoxins from Conus generalis. Int. J. Pept. Res. Ther. 2015, 21, 81–89. [Google Scholar] [CrossRef]
- Vetter, I.; Dekan, Z.; Knapp, O.; Adams, D.J.; Alewood, P.F.; Lewis, R.J. Isolation, characterization and total regioselective synthesis of the novel μO-conotoxin MfVIA from Conus magnificus that targets voltage-gated sodium channels. Biochem. Pharmacol. 2012, 84, 540–548. [Google Scholar] [CrossRef] [PubMed]
- Inserra, M.C.; Kompella, S.N.; Vetter, I.; Brust, A.; Daly, N.L.; Cuny, H.; Craik, D.J.; Alewood, P.F.; Adams, D.J.; Lewis, R.J. Isolation and characterization of α-conotoxin LsIA with potent activity at nicotinic acetylcholine receptors. Biochem. Pharmacol. 2013, 86, 791–799. [Google Scholar] [CrossRef] [PubMed]
- Franco, A.; Kompella, S.N.; Akondi, K.B.; Melaun, C.; Daly, N.L.; Luetje, C.W.; Alewood, P.F.; Craik, D.J.; Adams, D.J.; Mari, F. RegIIA: An alpha 4/7-conotoxin from the venom of Conus regius that potently blocks alpha 3 beta 4 nAChRs. Biochem. Pharmacol. 2012, 83, 419–426. [Google Scholar] [CrossRef] [PubMed]
- Braga, M.C.; Nery, A.A.; Ulrich, H.; Konno, K.; Sciani, J.M.; Pimenta, D.C. α-RgIB: A Novel Antagonist Peptide of Neuronal Acetylcholine Receptor Isolated from Conus regius Venom. Int. J. Pept. 2013, 2013, 543028. [Google Scholar] [CrossRef] [PubMed]
- Favreau, P.; Benoit, E.; Hocking, H.G.; Carlier, L.; Hoedt, D.D.; Leipold, E.; Markgraf, R.; Schlumberger, S.; Córdova, M.A.; Gaertner, H. A novel µ-conopeptide, CnIIIC, exerts potent and preferential inhibition of NaV1.2/1.4 channels and blocks neuronal nicotinic acetylcholine receptors. Br. J. Pharmacol. 2012, 166, 1654–1668. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, B.; Caer, J.P.; Mourier, G.; Thai, R.; Lamthanh, H.; Servent, D.; Benoit, E.; Molgó, J. Characterization of a Novel Conus bandanus Conopeptide Belonging to the M-Superfamily Containing Bromotryptophan. Mar. Drugs 2014, 12, 3449–3465. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Wahab, M.; Miyashita, M.; Kitanaka, A.; Juichi, H.; Sarhan, M.; Fouda, M.; Abdel-Rahman, M.; Saber, S.; Nakagawa, Y. Characterization of the venom of the vermivorous cone snail Conus fulgetrum. Biosci. Biotechnol. Biochem. 2016, 80, 1879–1882. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Du, T.; Liu, Z.; Wu, Q.; Feng, G.; Dong, M.; Zhou, X.; Jiang, L.; Dai, Q. Im10A, a short conopeptide isolated from Conus imperialis and possesses two highly concentrated disulfide bridges and analgesic activity. Peptides 2016, 81, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Johanna, B.; Samanta, J.; Javier, G.L.; Noda, F.J.; Enrique, S.; Emilio, S.; Daniela, C.; Aguilar, M.B.; Alexei, L.N. A New Member of Gamma-Conotoxin Family Isolated from Conus princeps Displays a Novel Molecular Target. Toxins 2016, 8, 39. [Google Scholar] [CrossRef]
- Han, T.S.; Teichert, R.W.; Olivera, B.M.; Bulaj, G. Conus venoms—A rich source of peptide-based therapeutics. Curr. Pharm. Des. 2008, 14, 2462–2479. [Google Scholar] [CrossRef] [PubMed]
- Shon, K.J.; Grilley, M.M.; Marsh, M.; Yoshikami, D.; Hall, A.R.; Kurz, B.; Gray, W.R.; Imperial, J.S.; Hillyard, D.R.; Olivera, B.M. Purification, characterization, synthesis, and cloning of the lockjaw peptide from Conus purpurascens venom. Biochemistry 1995, 34, 4913–4918. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.D.; Mcintosh, J.M.; Hillyard, D.R.; Cruz, L.J.; Olivera, B.M. The A-superfamily of conotoxins: Structural and functional divergence. J. Biol. Chem. 2004, 279, 17596–17606. [Google Scholar] [CrossRef] [PubMed]
- Mcintosh, J.M.; Plazas, P.V.; Watkins, M.; Gomezcasati, M.E.; Olivera, B.M.; Elgoyhen, A.B. A Novel α-Conotoxin, PeIA, Cloned from Conus pergrandis, Discriminates between Rat α9α10 and α7 Nicotinic Cholinergic Receptors. J. Biol. Chem. 2005, 280, 30107–30112. [Google Scholar] [CrossRef] [PubMed]
- Yuan, D.D.; Han, Y.H.; Wang, C.G.; Chi, C.W. From the identification of gene organization of conotoxins to the cloning of novel toxins. Toxicon 2007, 49, 1135–1149. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Jiang, H.; Han, Y.H.; Yuan, D.D.; Chi, C.W. Two different groups of signal sequence in M-superfamily conotoxins. Toxicon 2008, 51, 813–822. [Google Scholar] [CrossRef] [PubMed]
- Peng, C.; Liu, L.; Shao, X.; Chi, C.; Wang, C. Identification of a novel class of conotoxins defined as V-conotoxins with a unique cysteine pattern and signal peptide sequence. Peptides 2008, 29, 985–991. [Google Scholar] [CrossRef] [PubMed]
- Yuan, D.D.; Liu, L.; Shao, X.X.; Peng, C.; Chi, C.W.; Guo, Z.Y. Isolation and cloning of a conotoxin with a novel cysteine pattern from Conus caracteristicus. Peptides 2008, 29, 1521–1525. [Google Scholar] [CrossRef] [PubMed]
- Bendtsen, J.D.; Nielsen, H.; Von, H.G.; Brunak, S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004, 340, 783–795. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.; Zhangsun, D.; Harvey, P.J.; Kass, Q.; Wu, Y.; Zhu, X.; Hu, Y.; Li, X.; Tsetlin, V.I.; Christensen, S. Cloning, synthesis, and characterization of αO-conotoxin GeXIVA, a potent α9α10 nicotinic acetylcholine receptor antagonist. Proc. Natl. Acad. Sci. USA 2015, 112, E4026. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.; Zhangsun, D.; Wu, Y.; Zhu, X.; Hu, Y.; Mcintyre, M.; Christensen, S.; Akcan, M.; Craik, D.J.; Mcintosh, J.M. Characterization of a Novel α-Conotoxin from Conus textile that Selectively Targets α6/α3β2β3 Nicotinic Acetylcholine Receptors. J. Biol. Chem. 2013, 288, 894–902. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.; Zhangsun, D.; Zhu, X.; Wu, Y.; Hu, Y.; Christensen, S.; Harvey, P.J.; Akcan, M.; Craik, D.J.; Mcintosh, J.M. Characterization of a Novel Alpha-Conotoxin TxID from Conus textile that Potently Blocks rat Alpha3beta4 Nicotinic Acetylcholine Receptors. J. Med. Chem. 2013, 56, 9655–9663. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.; Zhangsun, D.; Schroeder, C.I.; Zhu, X.; Hu, Y.; Wu, Y.; Weltzin, M.M.; Eberhard, S.; Kaas, Q.; Craik, D.J.; et al. A novel α4/7-conotoxin LvIA from Conus lividus that selectively blocks α3β2 vs. α6/α3β2β3 nicotinic acetylcholine receptors. FASEB J. 2014, 28, 1842–1853. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Liang, L.; Ning, H.; Cai, F.; Liu, Z.; Zhang, L.; Zhou, L.; Dai, Q. Cloning, Synthesis and Functional Characterization of a Novel α-Conotoxin Lt1.3. Mar. Drugs 2018, 16, 112. [Google Scholar] [CrossRef] [PubMed]
- Zhangsun, D.; Luo, S.; Wu, Y.; Zhu, X.; Hu, Y.; Xie, L. Novel O-superfamily conotoxins identified by cDNA cloning from three vermivorous Conus species. Chem. Biol. Drug Des. 2010, 68, 256–265. [Google Scholar] [CrossRef] [PubMed]
- Swa, H.; Lewis, R.J. Venomics-Accelerated Cone Snail Venom Peptide Discovery. Int. J. Mol. Sci. 2018, 19, 788. [Google Scholar] [CrossRef]
- Utkin, Y.N. Modern trends in animal venom research—Omics and nanomaterials. World J. Biol. Chem. 2017, 8, 4–12. [Google Scholar] [CrossRef] [PubMed]
- Utkin, Y.N. Animal venom studies: Current benefits and future developments. World J. Biol. Chem. 2015, 6, 28–33. [Google Scholar] [CrossRef] [PubMed]
- Peng, C.; Yao, G.; Gao, B.M.; Fan, C.X.; Bian, C.; Wang, J.; Cao, Y.; Wen, B.; Zhu, Y.; Ruan, Z. High-throughput identification of novel conotoxins from the Chinese tubular cone snail (Conus betulinus) by multi-transcriptome sequencing. Gigascience 2016, 5, 17. [Google Scholar] [CrossRef] [PubMed]
- Mardis, E.R. Next-generation sequencing platforms. Annu. Rev. Anal. Chem. 2013, 6, 287–303. [Google Scholar] [CrossRef] [PubMed]
- Quail, M.A.; Miriam, S.; Paul, C.; Otto, T.D.; Harris, S.R.; Connor, T.R.; Anna, B.; Swerdlow, H.P.; Yong, G. A tale of three next generation sequencing platforms: Comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genom. 2012, 13, 341. [Google Scholar] [CrossRef] [PubMed]
- Safavi-Hemami, H.; Young, N.D.; Williamson, N.A.; Purcell, A.W. Proteomic interrogation of venom delivery in marine cone snails: Novel insights into the role of the venom bulb. J. Proteome Res. 2010, 9, 5610–5619. [Google Scholar] [CrossRef] [PubMed]
- Himaya, S.W.; Jin, A.H.; Dutertre, S.; Giacomotto, J.; Mohialdeen, H.; Vetter, I.; Alewood, P.F.; Lewis, R.J. Comparative Venomics Reveals the Complex Prey Capture Strategy of the Piscivorous Cone Snail Conus catus. J. Proteome Res. 2015, 14, 4372–4381. [Google Scholar] [CrossRef] [PubMed]
- Barghi, N.; Concepcion, G.P.; Olivera, B.M.; Lluisma, A.O. Comparison of the Venom Peptides and Their Expression in Closely Related Conus Species: Insights into Adaptive Post-speciation Evolution of Conus Exogenomes. Genome Biol. Evol. 2015, 7, 1797–1814. [Google Scholar] [CrossRef] [PubMed]
- Petersen, T.N.; Brunak, S.; Von, G.H.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef] [PubMed]
- Lavergne, V.; Dutertre, S.; Jin, A.H.; Lewis, R.J.; Taft, R.J.; Alewood, P.F. Systematic interrogation of the Conus marmoreus venom duct transcriptome with ConoSorter reveals 158 novel conotoxins and 13 new gene superfamilies. BMC Genom. 2013, 14, 708. [Google Scholar] [CrossRef] [PubMed]
- Robinson, S.D.; Safavihemami, H.; Mcintosh, L.D.; Purcell, A.W.; Norton, R.S.; Papenfuss, A.T. Diversity of Conotoxin Gene Superfamilies in the Venomous Snail, Conus victoriae. PLoS ONE 2014, 9, e87648. [Google Scholar] [CrossRef] [PubMed]
- Kaas, Q.; Westermann, J.C.; Halai, R.; Wang, C.K.L.; Craik, D.J. ConoServer, a database for conopeptide sequences and structures. Bioinformatics 2008, 24, 445–446. [Google Scholar] [CrossRef] [PubMed]
- Kaas, Q.; Yu, R.; Jin, A.H.; Dutertre, S.; Craik, D.J. ConoServer: Updated content, knowledge, and discovery tools in the conopeptide database. Nucleic Acids Res. 2012, 40, D325–D330. [Google Scholar] [CrossRef] [PubMed]
- Magrane, M.; Martin, M.J.; O’Donovan, C.; Apweiler, R. Protein Sequence Databases. Curr. Opin. Chem. Biol. 2004, 8, 76–80. [Google Scholar] [CrossRef]
- Consortium, U.P. UniProt: A hub for protein information. Nucleic Acids Res. 2015, 43, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Holford, M.; Zhang, M.M.; Gowd, K.H.; Azam, L.; Green, B.R.; Watkins, M.; Ownby, J.P.; Yoshikami, D.; Bulaj, G.; Olivera, B.M. Pruning nature: Biodiversity-derived discovery of novel sodium channel blocking conotoxins from Conus bullatus. Toxicon 2009, 53, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Gilly, W.F.; Richmond, T.A.; Duda, T.F., Jr.; Elliger, C.; Lebaric, Z.; Schulz, J.; Bingham, J.P.; Sweedler, J.V. A diverse family of novel peptide toxins from an unusual cone snail, Conus californicus. J. Exp. Biol. 2011, 214, 147–161. [Google Scholar] [CrossRef] [PubMed]
- Helena, S.H.; Lu, A.; Li, Q.; Fedosov, A.E.; Jason, B.; Patrice, S.C.; Jon, S.; Mark, Y.; Olivera, B.M. Venom Insulins of Cone Snails Diversify Rapidly and Track Prey Taxa. Mol. Biol. Evol. 2016, 33, 2924–2934. [Google Scholar] [CrossRef]
- Gao, B.; Peng, C.; Lin, B.; Chen, Q.; Zhang, J.; Shi, Q. Screening and Validation of Highly-Efficient Insecticidal Conotoxins from a Transcriptome-Based Dataset of Chinese Tubular Cone Snail. Toxins 2017, 9, 214. [Google Scholar] [CrossRef] [PubMed]
- Tayo, L.L.; Lu, B.; Cruz, L.J.; Rd, Y.J. Proteomic analysis provides insights on venom processing in Conus textile. J. Proteome Res. 2010, 9, 2292. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Bandyopadhyay, P.K.; Olivera, B.M.; Yandell, M. Characterization of the Conus bullatus genome and its venom-duct transcriptome. BMC Genom. 2011, 12, 60. [Google Scholar] [CrossRef] [PubMed]
- Terrat, Y.; Biass, D.; Dutertre, S.; Favreau, P.; Remm, M.; Stöcklin, R.; Piquemal, D.; Ducancel, F. High-resolution picture of a venom gland transcriptome: Case study with the marine snail Conus consors. Toxicon 2012, 59, 34–46. [Google Scholar] [CrossRef] [PubMed]
- Lluisma, A.O.; Milash, B.A.; Moore, B.; Olivera, B.M.; Bandyopadhyay, P.K. Novel venom peptides from the cone snail Conus pulicarius discovered through next-generation sequencing of its venom duct transcriptome. Mar. Genom. 2012, 5, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Barghi, N.; Concepcion, G.P.; Olivera, B.M.; Lluisma, A.O. High Conopeptide Diversity in Conus tribblei Revealed Through Analysis of Venom Duct Transcriptome Using Two High-Throughput Sequencing Platforms. Mar. Biotechnol. 2015, 17, 81. [Google Scholar] [CrossRef] [PubMed]
- Jin, A.H.; Vetter, I.; Himaya, S.W.A.; Alewood, P.F.; Lewis, R.J.; Dutertre, S. Transcriptome and proteome of Conus planorbis identify the nicotinic receptors as primary target for the defensive venom. Proteomics 2015, 15, 4030–4040. [Google Scholar] [CrossRef] [PubMed]
- Robinson, S.D.; Li, Q.; Lu, A.; Bandyopadhyay, P.K.; Yandell, M.; Olivera, B.M.; Safavihemami, H. The Venom Repertoire of Conus gloriamaris (Chemnitz, 1777), the Glory of the Sea. Mar. Drugs 2017, 15, 145. [Google Scholar] [CrossRef] [PubMed]
- Domon, B.; Aebersold, R. Mass spectrometry and protein analysis. Science 2006, 312, 212–217. [Google Scholar] [CrossRef] [PubMed]
- Ueberheide, B.M.; Fenyö, D.; Alewood, P.F.; Chait, B.T. Rapid sensitive analysis of cysteine rich peptide venom components. Proc. Natl. Acad. Sci. USA 2009, 106, 6910–6915. [Google Scholar] [CrossRef] [PubMed]
- Phuong, M.A.; Mahardika, G.N.; Alfaro, M.E. Dietary breadth is positively correlated with venom complexity in cone snails. BMC Genom. 2016, 17, 401. [Google Scholar] [CrossRef] [PubMed]
- Aebersold, R.; Mann, M. Mass-spectrometric exploration of proteome structure and function. Nature 2016, 537, 347. [Google Scholar] [CrossRef] [PubMed]
- Petras, D.; Heiss, P.; Süssmuth, R.D.; Calvete, J.J. Venom proteomics of Indonesian king cobra, Ophiophagus hannah: Integrating top-down and bottom-up approaches. J. Proteome Res. 2015, 14, 2539–2556. [Google Scholar] [CrossRef] [PubMed]
- Kaas, Q.; Craik, D.J. Bioinformatics-Aided Venomics. Toxins 2015, 7, 2159–2187. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).