Loss of Function in Zeaxanthin Epoxidase of Dunaliella tertiolecta Caused by a Single Amino Acid Mutation within the Substrate-Binding Site
Abstract
1. Introduction
2. Results and Discussion
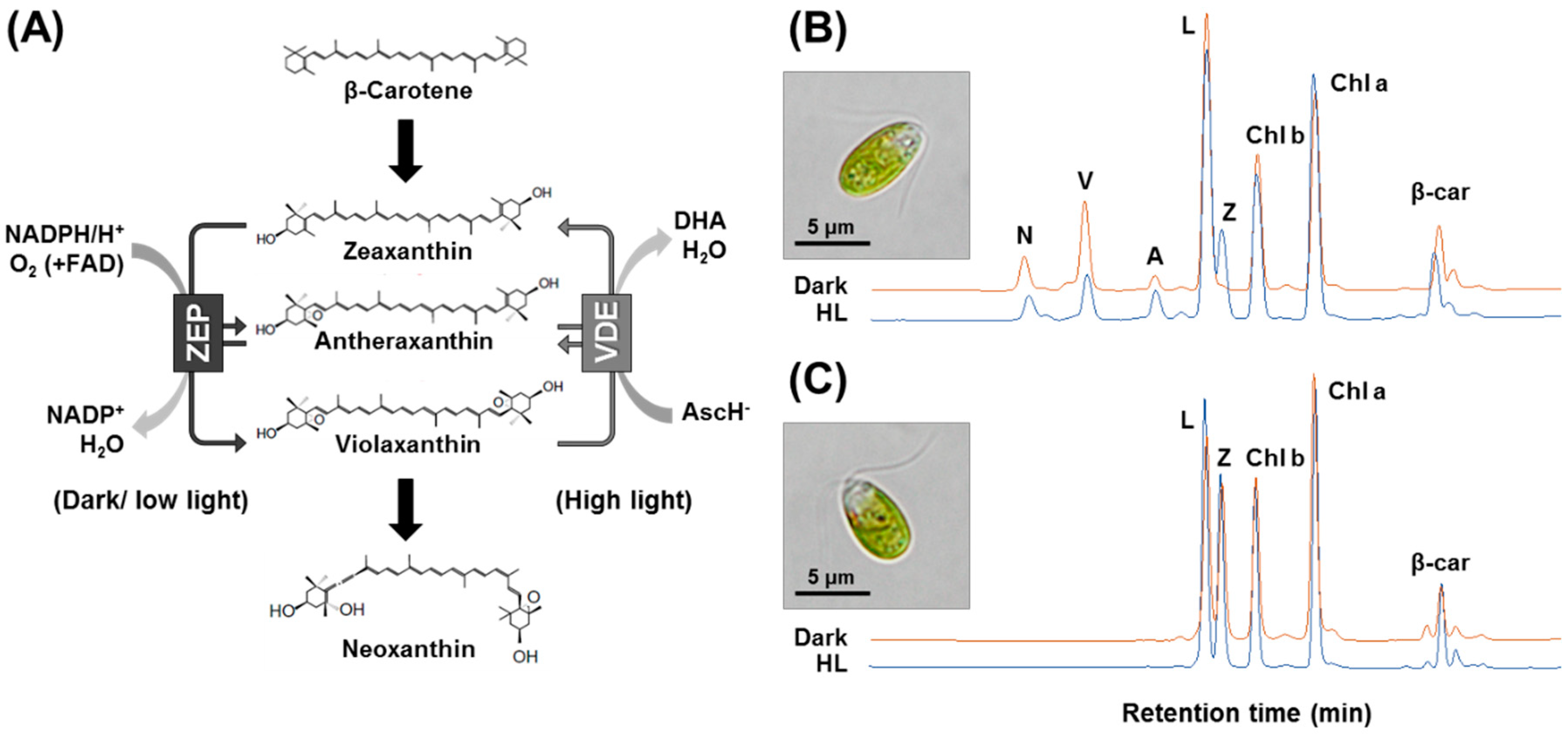
2.1. Comparison of Pigment Profile between Wild-Type and zea1
2.2. Isolation of DtZEP and Sequence Analysis
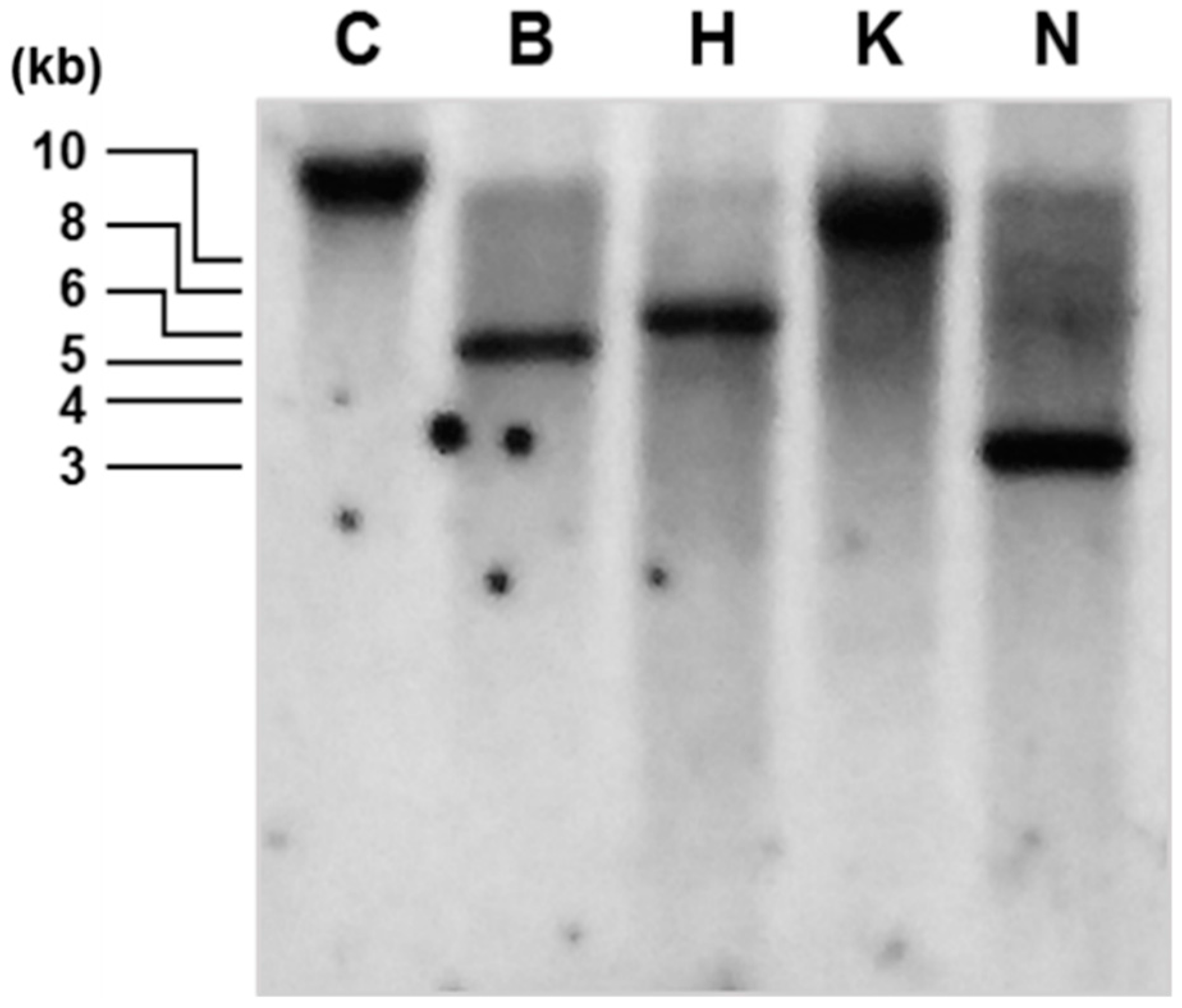
2.3. Comparative Genotype Analysis of Wild-Type and zea1
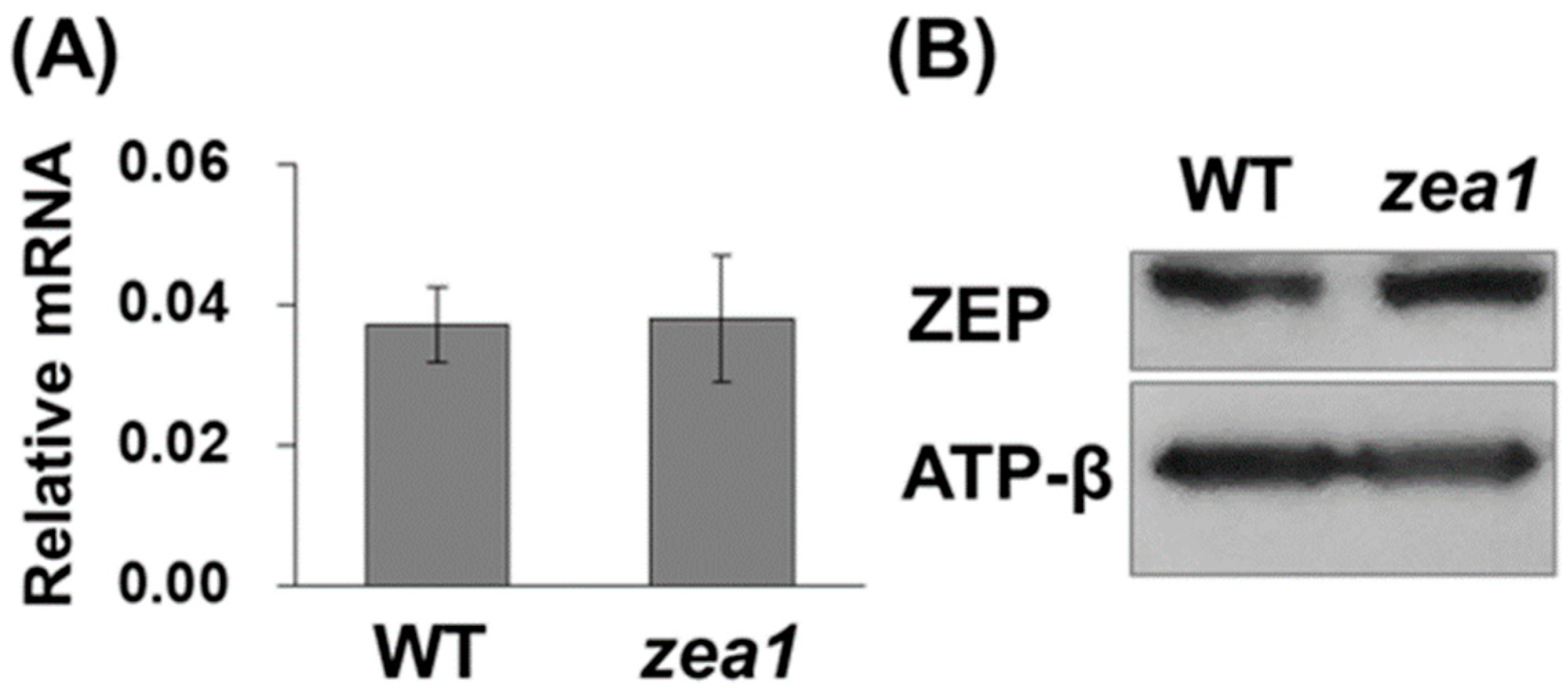
2.4. A Single Amino Acid Change Does Not Affect the Levels of ZEP mRNA and Protein in zea1
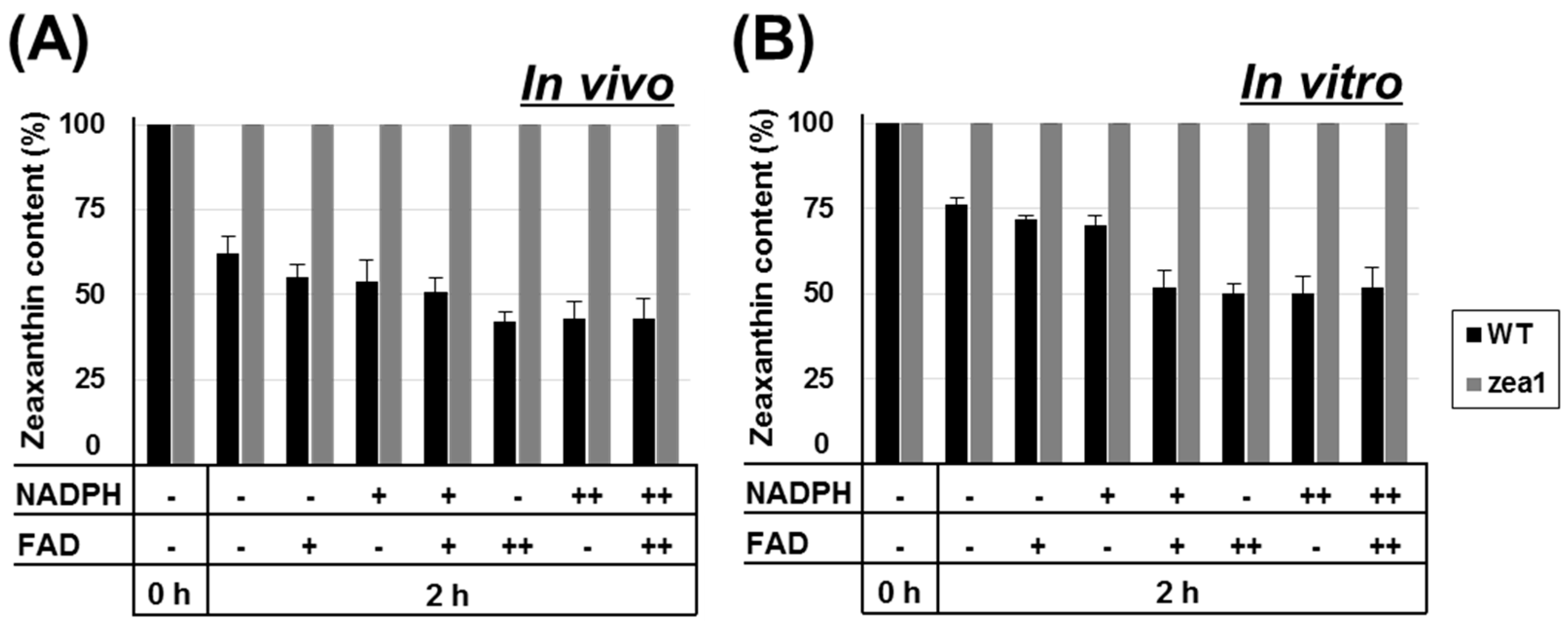
2.5. Additional Cofactor Supply Does Not Affect the Enzymatic Activity of ZEP in zea1
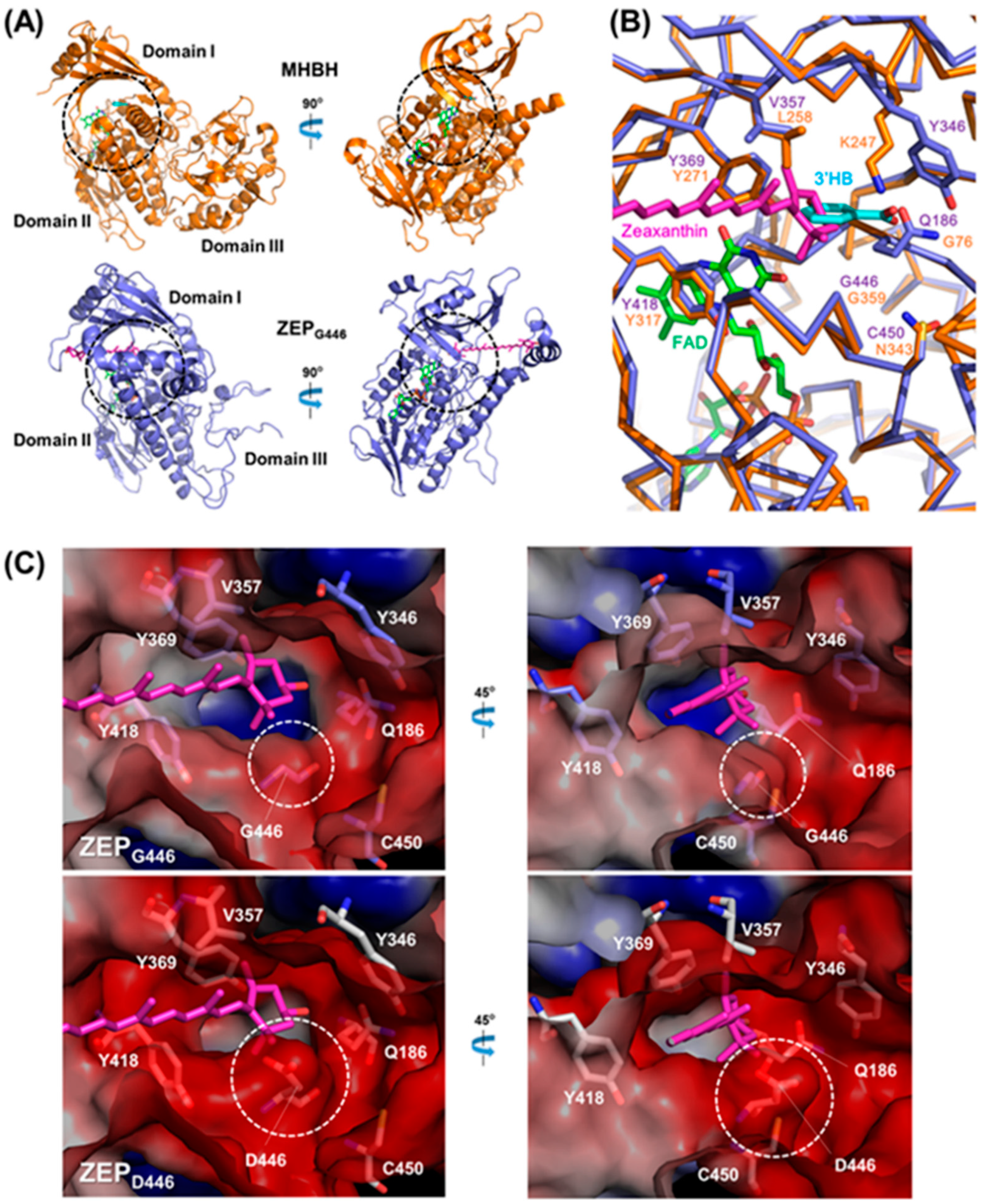
2.6. Comparison of the Three-Dimensional ZEP Structures of Wild-Type and zea1
3. Materials and Methods
3.1. Strains and Culture Conditions
3.2. Cloning of Zeaxanthin Epoxidase Gene
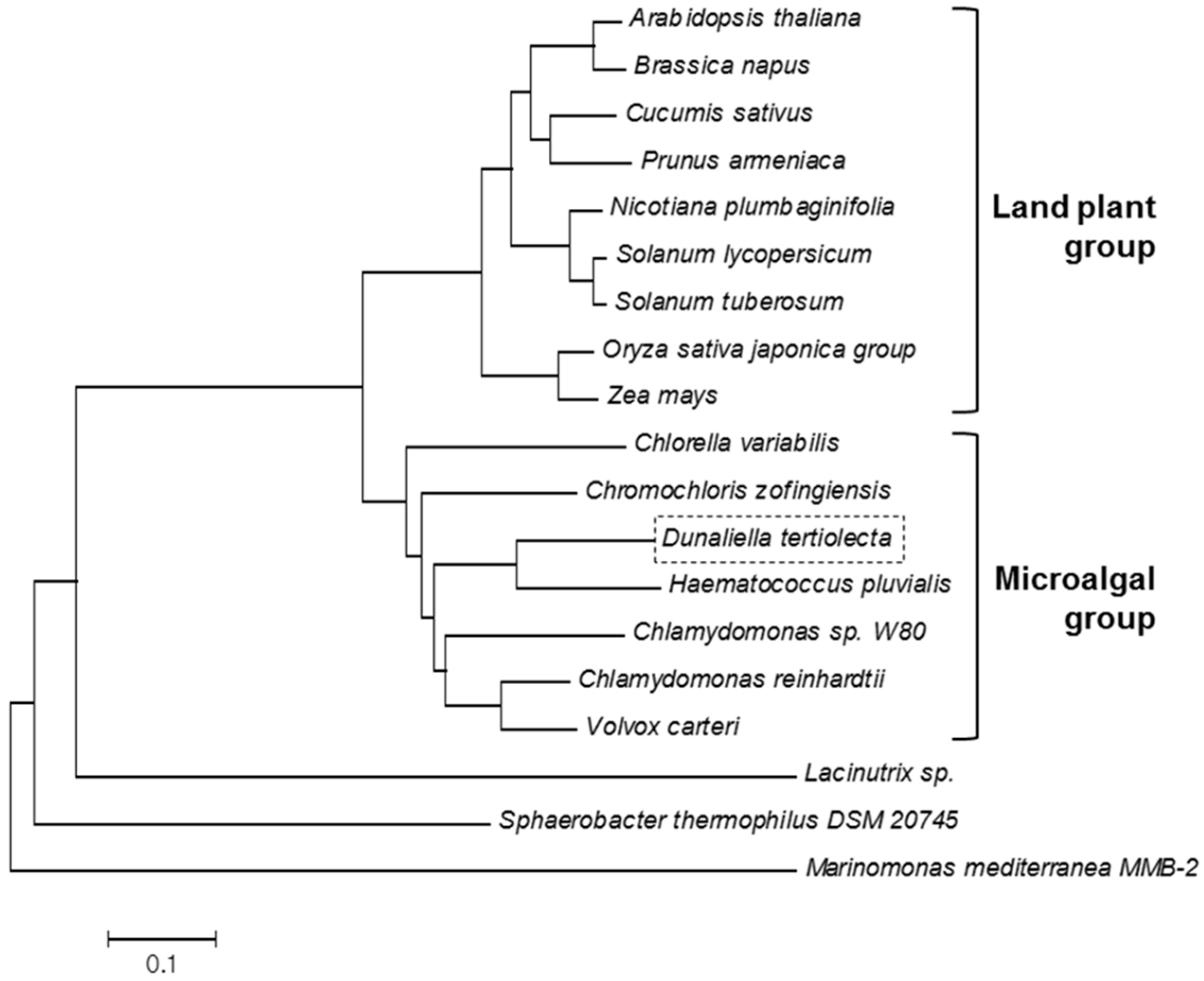
3.3. Sequence and Phylogenetic Analysis
3.4. Isolation of Genomic DNA and Southern Blot Analysis
3.5. Quantitative Real-Time PCR
3.6. Western Blot Analysis
3.7. Treatment with ZEP Cofactors
3.8. Xanthophyll Composition Analysis
3.9. Prediction of Protein Structure by Comparative Modeling
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Munro, M.H.; Blunt, J.W.; Dumdei, E.J.; Hickford, S.J.; Lill, R.E.; Li, S.; Battershill, C.N.; Duckworth, A.R. The discovery and development of marine compounds with pharmaceutical potential. J. Biotechnol. 1999, 70, 15–25. [Google Scholar] [CrossRef]
- Apt, K.E.; Behrens, P.W. Commercial developments in microalgal biotechnology. J. Phycol. 1999, 35, 215–226. [Google Scholar] [CrossRef]
- Borowitzka, M.A. Commercial production of microalgae: Ponds, tanks, tubes and fermenters. J. Biotechnol. 1999, 70, 313–321. [Google Scholar] [CrossRef]
- Fraser, P.D.; Bramley, P.M. The biosynthesis and nutritional uses of carotenoids. Prog. Lipid Res. 2004, 43, 228–265. [Google Scholar] [CrossRef] [PubMed]
- Demmig-Adams, B.; Adams, W.W. Antioxidants in photosynthesis and human nutrition. Science 2002, 298, 2149–2153. [Google Scholar] [CrossRef] [PubMed]
- Johnson, E.J.; Krinsky, N.I. Carotenoids and coronary heart disease. In Carotenoids; Birkhäuser: Basel, Switzerland, 2009; pp. 287–300. [Google Scholar]
- Fiedor, J.; Burda, K. Potential role of carotenoids as antioxidants in human health and disease. Nutrients 2014, 6, 466–488. [Google Scholar] [CrossRef] [PubMed]
- Nwachukwu, I.D.; Udenigwe, C.C.; Aluko, R.E. Lutein and zeaxanthin: Production technology, bioavailability, mechanisms of action, visual function, and health claim status. Trends Food Sci. Technol. 2016, 49, 74–84. [Google Scholar] [CrossRef]
- Bernstein, P.S.; Li, B.; Vachali, P.P.; Gorusupudi, A.; Shyam, R.; Henriksen, B.S.; Nolan, J.M. Lutein, zeaxanthin, and meso-zeaxanthin: The basic and clinical science underlying carotenoid-based nutritional interventions against ocular disease. Prog. Retin. Eye Res. 2016, 50, 34–66. [Google Scholar] [CrossRef] [PubMed]
- Niyogi, K.K. Photoprotection revisited: Genetic and molecular approaches. Annu. Rev. Plant Boil. 1999, 50, 333–359. [Google Scholar] [CrossRef] [PubMed]
- Vass, I.; Styring, S.; Hundal, T.; Koivuniemi, A.; Aro, E.; Andersson, B. Reversible and irreversible intermediates during photoinhibition of photosystem II: Stable reduced QA species promote chlorophyll triplet formation. Proc. Natl. Acad. Sci. USA 1992, 89, 1408–1412. [Google Scholar] [CrossRef] [PubMed]
- Schaller, S.; Wilhelm, C.; Strzałka, K.; Goss, R. Investigating the interaction between the violaxanthin cycle enzyme zeaxanthin epoxidase and the thylakoid membrane. J. Photochem. Photobiol. B Boil. 2012, 114, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Hieber, A.D.; Bugos, R.C.; Yamamoto, H.Y. Plant lipocalins: Violaxanthin de-epoxidase and zeaxanthin epoxidase. Biochim. Biophys. Acta (BBA) Protein Struct. Mol. Enzym. 2000, 1482, 84–91. [Google Scholar] [CrossRef]
- Müller, P.; Li, X.-P.; Niyogi, K.K. Non-photochemical quenching. A response to excess light energy. Plant Physiol. 2001, 125, 1558–1566. [Google Scholar] [CrossRef] [PubMed]
- Jahns, P.; Holzwarth, A.R. The role of the xanthophyll cycle and of lutein in photoprotection of photosystem II. Biochim. Biophys. Acta (BBA) Bioenerg. 2012, 1817, 182–193. [Google Scholar] [CrossRef] [PubMed]
- Davison, P.; Hunter, C.; Horton, P. Overexpression of β-carotene hydroxylase enhances stress tolerance in Arabidopsis. Nature 2002, 418, 203–206. [Google Scholar] [CrossRef] [PubMed]
- Niyogi, K.K.; Grossman, A.R.; Björkman, O. Arabidopsis mutants define a central role for the xanthophyll cycle in the regulation of photosynthetic energy conversion. Plant Cell 1998, 10, 1121–1134. [Google Scholar] [CrossRef] [PubMed]
- Niyogi, K.K.; Bjorkman, O.; Grossman, A.R. Chlamydomonas xanthophyll cycle mutants identified by video imaging of chlorophyll fluorescence quenching. Plant Cell Online 1997, 9, 1369–1380. [Google Scholar] [CrossRef] [PubMed]
- Krieg, D.R. Ethyl methanesulfonate-induced reversion of bacteriophage T4rII mutants. Genetics 1963, 48, 561–580. [Google Scholar] [PubMed]
- Jin, E.; Feth, B.; Melis, A. A mutant of the green alga Dunaliella salina constitutively accumulates zeaxanthin under all growth conditions. Biotechnol. Bioeng. 2003, 81, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.-W.; Jiang, J.-G.; Wu, G.-H. Biosynthesis and regulation of carotenoids in Dunaliella: Progresses and prospects. Biotechnol. Adv. 2008, 26, 352–360. [Google Scholar] [CrossRef] [PubMed]
- Jin, E.; Yokthongwattana, K.; Polle, J.E.; Melis, A. Role of the reversible xanthophyll cycle in the photosystem II damage and repair cycle in Dunaliella salina. Plant Physiol. 2003, 132, 352–364. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Ahn, J.; Jeon, H.; Jin, E. Development of a Dunaliella tertiolecta Strain with Increased Zeaxanthin Content Using Random Mutagenesis. Mar. Drugs 2017, 15, 189. [Google Scholar] [CrossRef] [PubMed]
- North, H.M.; Frey, A.; Boutin, J.-P.; Sotta, B.; Marion-Poll, A. Analysis of xanthophyll cycle gene expression during the adaptation of Arabidopsis to excess light and drought stress: Changes in RNA steady-state levels do not contribute to short-term responses. Plant Sci. 2005, 169, 115–124. [Google Scholar] [CrossRef]
- Thompson, A.J.; Jackson, A.C.; Parker, R.A.; Morpeth, D.R.; Burbidge, A.; Taylor, I.B. Abscisic acid biosynthesis in tomato: Regulation of zeaxanthin epoxidase and 9-cis-epoxycarotenoid dioxygenase mRNAs by light/dark cycles, water stress and abscisic acid. Plant Mol. Boil. 2000, 42, 833–845. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.E.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook; Humana Press: New York, NY, USA, 2005; pp. 571–607. [Google Scholar]
- Couso, I.; Cordero, B.F.; Vargas, M.Á.; Rodríguez, H. Efficient heterologous transformation of Chlamydomonas reinhardtii npq2 mutant with the zeaxanthin epoxidase gene isolated and characterized from Chlorella Zofingiensis. Mar. Drugs 2012, 10, 1955–1976. [Google Scholar] [CrossRef] [PubMed]
- Blanc, G.; Duncan, G.; Agarkova, I.; Borodovsky, M.; Gurnon, J.; Kuo, A.; Lindquist, E.; Lucas, S.; Pangilinan, J.; Polle, J. The Chlorella variabilis NC64A genome reveals adaptation to photosymbiosis, coevolution with viruses, and cryptic sex. Plant Cell 2010, 22, 2943–2955. [Google Scholar] [CrossRef] [PubMed]
- Merchant, S.S.; Prochnik, S.E.; Vallon, O.; Harris, E.H.; Karpowicz, S.J.; Witman, G.B.; Terry, A.; Salamov, A.; Fritz-Laylin, L.K.; Maréchal-Drouard, L. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 2007, 318, 245–250. [Google Scholar] [CrossRef] [PubMed]
- Marin, E.; Nussaume, L.; Quesada, A.; Gonneau, M.; Sotta, B.; Hugueney, P.; Frey, A.; Marion-Poll, A. Molecular identification of zeaxanthin epoxidase of Nicotiana plumbaginifolia, a gene involved in abscisic acid biosynthesis and corresponding to the ABA locus of Arabidopsis thaliana. EMBO J. 1996, 15, 2331–2342. [Google Scholar] [CrossRef] [PubMed]
- Burbidge, A.; Grieve, T.; Terry, C.; Corlett, J.; Thompson, A.; Taylor, I. Structure and expression of a cDNA encoding zeaxanthin epoxidase, isolated from a wilt-related tomato (Lycopersicon esculentum Mill.) library. J. Exp. Bot. 1997, 48, 1749–1750. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, Y.; Chang, L.; Zhang, T.; An, J.; Liu, Y.; Cao, Y.; Zhao, X.; Sha, X.; Hu, T. MsZEP, a novel zeaxanthin epoxidase gene from alfalfa (Medicago sativa), confers drought and salt tolerance in transgenic tobacco. Plant Cell Rep. 2016, 35, 439–453. [Google Scholar] [CrossRef] [PubMed]
- Coesel, S.N.; Baumgartner, A.C.; Teles, L.M.; Ramos, A.A.; Henriques, N.M.; Cancela, L.; Varela, J.C.S. Nutrient limitation is the main regulatory factor for carotenoid accumulation and for Psy and Pds steady state transcript levels in Dunaliella salina (Chlorophyta) exposed to high light and salt stress. Mar. Biotechnol. 2008, 10, 602. [Google Scholar] [CrossRef] [PubMed]
- Eilers, U.; Dietzel, L.; Breitenbach, J.; Büchel, C.; Sandmann, G. Identification of genes coding for functional zeaxanthin epoxidases in the diatom Phaeodactylum Tricornutum. J. Plant Physiol. 2016, 192, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Büch, K.; Stransky, H.; Hager, A. FAD is a further essential cofactor of the NAD (P) H and O2-dependent zeaxanthin-epoxidase. FEBS Lett. 1995, 376, 45–48. [Google Scholar] [CrossRef]
- Baek, K.; Kim, D.H.; Jeong, J.; Sim, S.J.; Melis, A.; Kim, J.-S.; Jin, E.; Bae, S. DNA-free two-gene knockout in Chlamydomonas reinhardtii via CRISPR-Cas9 ribonucleoproteins. Sci. Rep. 2016, 6, 30620. [Google Scholar] [CrossRef] [PubMed]
- Karplus, P.A.; Daniels, M.J. Atomic structure of ferredoxin-NADP+ reductase: Prototype for a structurally novel flavoenzyme family. Science 1991, 251, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Lambers, H.; Chapin, F.S.; Pons, T.L. Photosynthesis. In Plant Physiological Ecology; Springer: New York, NY, USA, 2008; pp. 11–99. [Google Scholar]
- Thaipratum, R.; Melis, A.; Svasti, J.; Yokthongwattana, K. Analysis of non-photochemical energy dissipating processes in wild type Dunaliella salina (green algae) and in zea1, a mutant constitutively accumulating zeaxanthin. J. Plant Res. 2009, 122, 465–476. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Ortiz-Maldonado, M.; Entsch, B.; Massey, V.; Ballou, D.; Gatti, D.L. Protein and ligand dynamics in 4-hydroxybenzoate hydroxylase. Proc. Natl. Acad. Sci. USA 2002, 99, 608–613. [Google Scholar] [CrossRef] [PubMed]
- Hiromoto, T.; Fujiwara, S.; Hosokawa, K.; Yamaguchi, H. Crystal structure of 3-hydroxybenzoate hydroxylase from Comamonas testosteroni has a large tunnel for substrate and oxygen access to the active site. J. Mol. Boil. 2006, 364, 878–896. [Google Scholar] [CrossRef] [PubMed]
- Ukaegbu, U.E.; Kantz, A.; Beaton, M.; Gassner, G.T.; Rosenzweig, A.C. Structure and ligand binding properties of the epoxidase component of styrene monooxygenase. Biochemistry 2010, 49, 1678–1688. [Google Scholar] [CrossRef] [PubMed]
- Honig, B.; Nicholls, A. Classical electrostatics in biology and chemistry. Science 1995, 268, 1144–1149. [Google Scholar] [CrossRef] [PubMed]
- Fersht, A.R. Catalysis, binding and enzyme-substrate complementarity. Proc. R. Soc. Lond. B 1974, 187, 397–407. [Google Scholar] [CrossRef] [PubMed]
- Warshel, A.; Levitt, M. Theoretical studies of enzymic reactions: Dielectric, electrostatic and steric stabilization of the carbonium ion in the reaction of lysozyme. J. Mol. Boil. 1976, 103, 227–249. [Google Scholar] [CrossRef]
- Kahraman, A.; Morris, R.J.; Laskowski, R.A.; Thornton, J.M. Shape variation in protein binding pockets and their ligands. J. Mol. Boil. 2007, 368, 283–301. [Google Scholar] [CrossRef] [PubMed]
- Polle, J.E.; Barry, K.; Cushman, J.; Schmutz, J.; Tran, D.; Hathwaik, L.T.; Yim, W.C.; Jenkins, J.; McKie-Krisberg, Z.; Prochnik, S. Draft nuclear genome sequence of the halophilic and beta-carotene-accumulating green alga Dunaliella salina strain CCAP19/18. Genome Announc. 2017, 5, e01105-17. [Google Scholar] [CrossRef] [PubMed]
- Keeling, P.J.; Burki, F.; Wilcox, H.M.; Allam, B.; Allen, E.E.; Amaral-Zettler, L.A.; Armbrust, E.V.; Archibald, J.M.; Bharti, A.K.; Bell, C.J. The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): Illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol. 2014, 12, e1001889. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Boil. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Boil. Evol. 1987, 4, 406–425. [Google Scholar]
- Zuckerkandl, E.; Pauling, L. Evolutionary divergence and convergence in proteins. Evol. Genes Proteins 1965, 97, 97–166. [Google Scholar]
- Tanksley, S.; Ganal, M.; Prince, J.; De Vicente, M.; Bonierbale, M.; Broun, P.; Fulton, T.; Giovannoni, J.; Grandillo, S.; Martin, G. High density molecular linkage maps of the tomato and potato genomes. Genetics 1992, 132, 1141–1160. [Google Scholar] [PubMed]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Masojídek, J.; Torzillo, G.; Koblížek, M.; Kopecký, J.; Bernardini, P.; Sacchi, A.; Komenda, J. Photoadaptation of two members of the Chlorophyta (Scenedesmus and Chlorella) in laboratory and outdoor cultures: Changes in chlorophyll fluorescence quenching and the xanthophyll cycle. Planta 1999, 209, 126–135. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Jung, G.; Hwang, Y.-S.; Jin, E. Dynamic response of the transcriptome of a psychrophilic diatom, Chaetoceros neogracile, to high irradiance. Planta 2010, 231, 349. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Voss, N.R.; Gerstein, M. 3V: Cavity, channel and cleft volume calculator and extractor. Nucleic Acids Res. 2010, 38 (Suppl. 2), W555–W562. [Google Scholar] [CrossRef] [PubMed]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, M.; Kang, J.; Kang, Y.; Kang, B.S.; Jin, E. Loss of Function in Zeaxanthin Epoxidase of Dunaliella tertiolecta Caused by a Single Amino Acid Mutation within the Substrate-Binding Site. Mar. Drugs 2018, 16, 418. https://doi.org/10.3390/md16110418
Kim M, Kang J, Kang Y, Kang BS, Jin E. Loss of Function in Zeaxanthin Epoxidase of Dunaliella tertiolecta Caused by a Single Amino Acid Mutation within the Substrate-Binding Site. Marine Drugs. 2018; 16(11):418. https://doi.org/10.3390/md16110418
Chicago/Turabian StyleKim, Minjae, Jisu Kang, Yongsoo Kang, Beom Sik Kang, and EonSeon Jin. 2018. "Loss of Function in Zeaxanthin Epoxidase of Dunaliella tertiolecta Caused by a Single Amino Acid Mutation within the Substrate-Binding Site" Marine Drugs 16, no. 11: 418. https://doi.org/10.3390/md16110418
APA StyleKim, M., Kang, J., Kang, Y., Kang, B. S., & Jin, E. (2018). Loss of Function in Zeaxanthin Epoxidase of Dunaliella tertiolecta Caused by a Single Amino Acid Mutation within the Substrate-Binding Site. Marine Drugs, 16(11), 418. https://doi.org/10.3390/md16110418




