Exploitation of Mangrove Endophytic Fungi for Infectious Disease Drug Discovery
Abstract
1. Introduction
2. Results
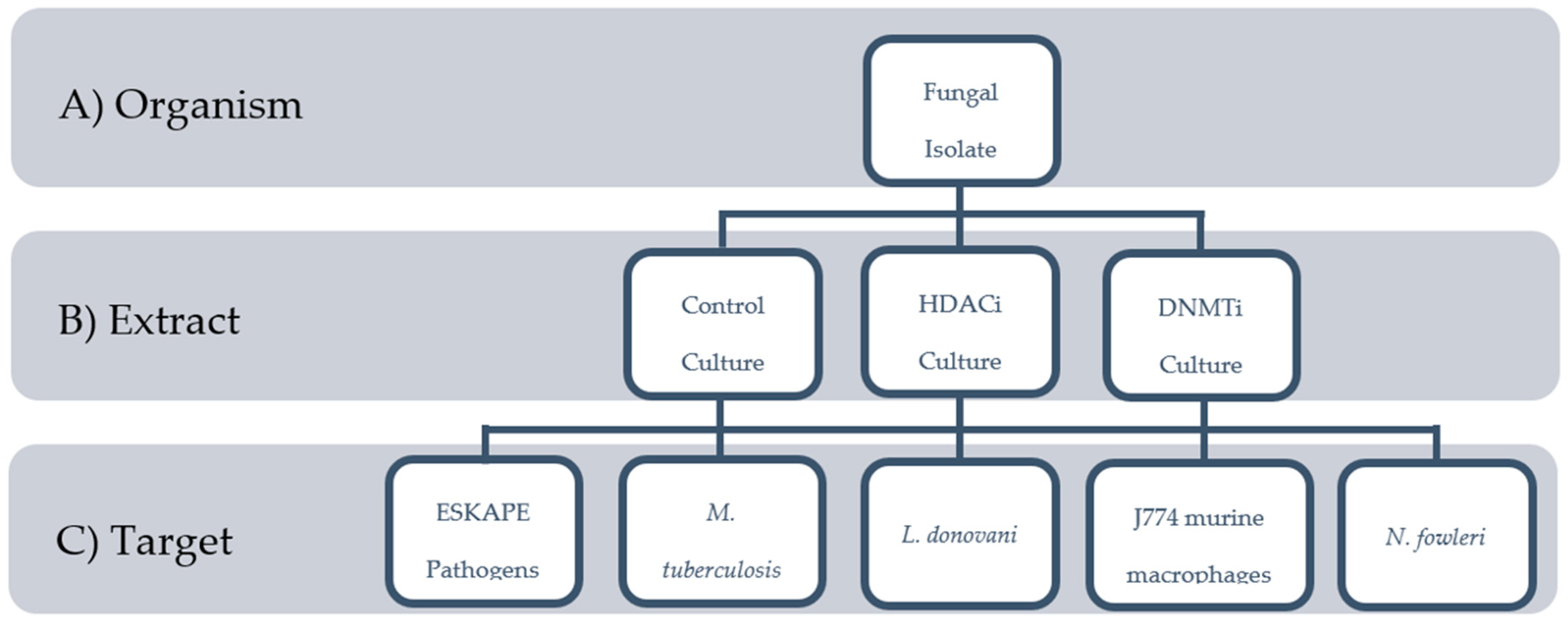
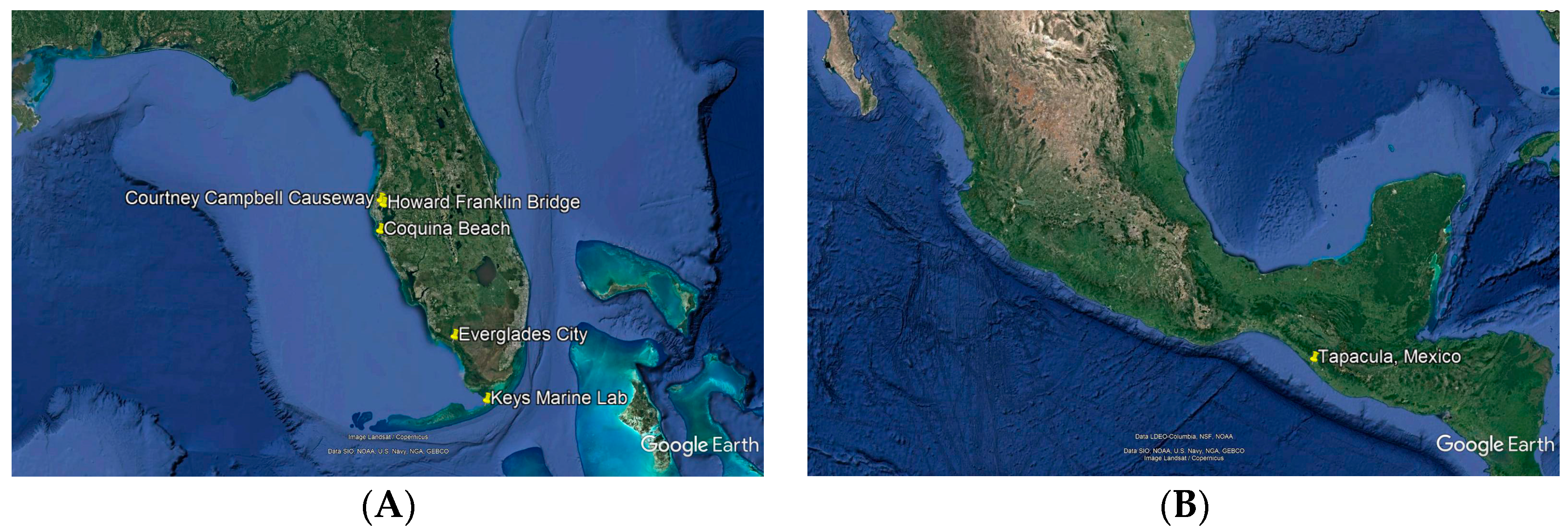
2.1. Biological Materials
2.2. Extract Library
2.3. Screening
2.3.1. ESKAPE Pathogens
2.3.2. Mycobacterium tuberculosis
2.3.3. Leishmania donovani
2.3.4. Naegleria fowleri
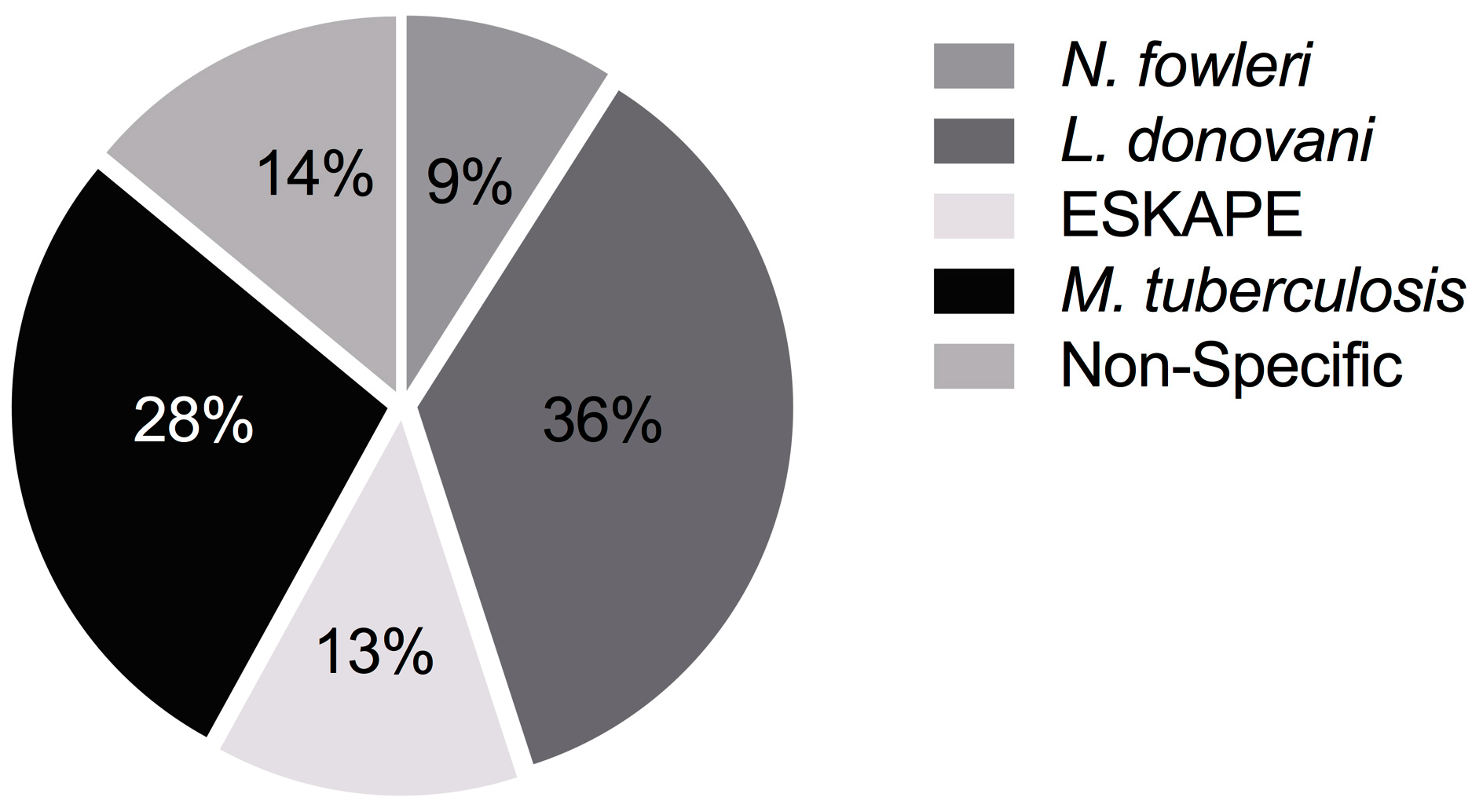
2.4. Overall Data Analysis
3. Discussion
4. Materials and Methods
4.1. Fungal Isolation
4.2. Miniaturized Culture Conditions and Extraction Procedures
4.3. ESKAPE Bacterial Strains, Growth Conditions, and Microtiter MIC Determination Assays
4.4. Analysis of Antimicrobial Activity against Replicating Mycobacterium tuberculosis
4.5. Leishmania donovani-Infected Macrophage (IM) Assay
4.6. Cytotoxicity of Mammalian Cell Line
4.7. Naegleria fowleri Culture and Cell Viability Assay
4.8. Data Analysis
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References and Notes
- Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. Available online: https://www.who.int/medicines/publications/global-priority-list-antibiotic-resistant-bacteria/en/ (accessed on 3 March 2017).
- Mdluli, K.; Kaneko, T.; Upton, A. The tuberculosis drug discovery and development pipeline and emerging drug targets. Cold Spring Harb. Perspect. Med. 2015, 5, a021154. [Google Scholar] [CrossRef] [PubMed]
- Pogue, J.M.; Kaye, K.S.; Cohen, D.A.; Marchaim, D. Appropriate antimicrobial therapy in the era of multidrug-resistant human pathogens. Clin. Microbiol. Infect. 2015, 21, 302–312. [Google Scholar] [CrossRef] [PubMed]
- Lechartier, B.; Rybniker, J.; Zumla, A.; Cole, S.T. Tuberculosis drug discovery in the post-post-genomic era. EMBO Mol. Med. 2014, 6, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Yoder, J.S.; Eddy, B.A.; Visvesvara, G.S.; Capewell, L.; Beach, M.J. The epidemiology of primary amoebic meningoencephalitis in the USA, 1962–2008. Epidemiol. Infect. 2010, 138, 968–975. [Google Scholar] [CrossRef] [PubMed]
- Debroy, S.; Prosper, O.; Mishoe, A.; Mubayi, A. Challenges in modeling complexity of neglected tropical diseases: A review of dynamics of visceral leishmaniasis in resource limited settings. Emerg. Themes Epidemiol. 2017, 14, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Akhoundi, M.; Kuhls, K.; Cannet, A.; Votýpka, J.; Marty, P.; Delaunay, P.; Sereno, D. A historical overview of the classification, evolution, and dispersion of Leishmania parasites and sandflies. PLoS Negl. Trop. Dis. 2016, 10, 1–40. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Kumar, M.; Singh, R.K. Leishmaniasis: Current status of available drugs and new potential drug targets. Asian Pac. J. Trop. Med. 2012, 5, 485–497. [Google Scholar] [CrossRef]
- No, J.H. Visceral leishmaniasis: Revisiting current treatments and approaches for future discoveries. Acta Trop. 2016, 155, 113–123. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.; Cragg, G. Drugs and drug candidates from marine sources: An assessment of the current “state of play”. Planta Med. 2016, 82, 775–789. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 2016, 79, 629–661. [Google Scholar] [CrossRef] [PubMed]
- Baker, B.J. (Ed.) Marine Biomedicine; CRC Press: Boca Raton, FL, USA, 2015; 594p, ISBN 978-1-4665-8212-5. [Google Scholar]
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Munro, M.H.G.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2017, 34, 235–294. [Google Scholar] [CrossRef] [PubMed]
- Wright, G.D. Opportunities for natural products in 21st century antibiotic discovery. Nat. Prod. Rep. 2017, 34, 694–701. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues Felix, C.; Gupta, R.; Geden, S.; Roberts, J.; Winder, P.; Pomponi, S.A.; Diaz, M.C.; Reed, J.K.; Wright, A.E.; Rohde, K.H. Selective killing of dormant Mycobacterium tuberculosis by marine natural products. Antimicrob. Agents Chemother. 2017, 61, AAC.00743-17. [Google Scholar] [CrossRef] [PubMed]
- Cheuka, P.M.; Mayoka, G.; Mutai, P.; Chibale, K. The role of natural products in drug discovery and development against neglected tropical diseases. Molecules 2017, 22, 58. [Google Scholar] [CrossRef] [PubMed]
- Von Salm, J.L.; Wilson, N.G.; Vesely, B.A.; Kyle, D.E.; Cuce, J.; Baker, B.J. Shagenes A and B, new tricyclic sesquiterpenes produced by an undescribed antarctic octocoral. Org. Lett. 2014, 16, 2630–2633. [Google Scholar] [CrossRef] [PubMed]
- Von Salm, J.; Witowski, C.; Demers, D.; Young, R.; Calcul, L.; Baker, B. Screening marine microbial libraries. In Marine Biomedicine; Baker, B.J., Ed.; CRC Press: Boca Raton, FL, USA, 2015; pp. 105–134. ISBN 9781466582125. [Google Scholar]
- Moore, B.S.; Gerwick, W.H. Lessons from the past and charting the future of marine natural products drug discovery and chemical biology. ACS Chem. Biol. 2012, 19, 85–98. [Google Scholar] [CrossRef]
- Kjer, J.; Debbab, A.; Aly, A.H.; Proksch, P. Methods for isolation of marine-derived endophytic fungi and their bioactive secondary products. Nat. Protoc. 2010, 5, 479–490. [Google Scholar] [CrossRef] [PubMed]
- Pettit, R.K. Culturability and secondary metabolite diversity of extreme microbes: Expanding contribution of deep sea and deep-sea vent microbes to natural product discovery. Mar. Biotechnol. 2011, 13, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Van der Lee, T.A.J.; Medema, M.H. Computational strategies for genome-based natural product discovery and engineering in fungi. Fungal Genet. Biol. 2016, 89, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Brakhage, A.A.; Schroeckh, V. Fungal secondary metabolites–strategies to activate silent gene clusters. Fungal Genet. Biol. 2011, 48, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Brakhage, A.A. Regulation of fungal secondary metabolism. Nat. Rev. Microbiol. 2013, 11, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Jensen, P.R.; Chavarria, K.L.; Fenical, W.; Moore, B.S.; Ziemert, N. Challenges and triumphs to genomics-based natural product discovery. J. Ind. Microbiol. Biotechnol. 2014, 41, 203–209. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.F.; Tsai, K.J.S.; Harvey, C.J.B.; Li, J.J.; Ary, B.E.; Berlew, E.E.; Boehman, B.L.; Findley, D.M.; Friant, A.G.; Gardner, C.A.; et al. Comprehensive curation and analysis of fungal biosynthetic gene clusters of published natural products. Fungal Genet. Biol. 2016, 89, 18–28. [Google Scholar] [CrossRef] [PubMed]
- Thatoi, H.; Behera, B.C.; Mishra, R.R.; Dutta, S.K. Biodiversity and biotechnological potential of microorganisms from mangrove ecosystems: A review. Ann. Microbiol. 2013, 63, 1–19. [Google Scholar] [CrossRef]
- Pang, K.L.; Vrijmoed, L.L.P.; Khiang Goh, T.; Plaingam, N.; Jones, E.B.G. Fungal endophytes associated with Kandelia candel (Rhizophoraceae) in Mai Po Nature Reserve, Hong Kong. Bot. Mar. 2008, 51, 171–178. [Google Scholar] [CrossRef]
- Wang, K.-W.; Wang, S.-W.; Wu, B.; Wei, J.-G. Bioactive natural compounds from the magrove endophytic fungi. Mini Rev. Med. Chem. 2014, 14, 370–391. [Google Scholar] [CrossRef] [PubMed]
- Calcul, L.; Waterman, C.; Ma, W.S.; Lebar, M.D.; Harter, C.; Mutka, T.; Morton, L.; Maignan, P.; Van Olphen, A.; Kyle, D.E.; et al. Screening mangrove endophytic fungi for antimalarial natural products. Mar. Drugs 2013, 11, 5036–5050. [Google Scholar] [CrossRef] [PubMed]
- Waterman, C.; Calcul, L.; Beau, J.; Ma, W.S.; Lebar, M.D.; von Salm, J.L.; Harter, C.; Mutka, T.; Morton, L.C.; Maignan, P.; et al. Miniaturized cultivation of microbiota for antimalarial drug discovery. Med. Res. Rev. 2016, 36, 144–168. [Google Scholar] [CrossRef] [PubMed]
- Bok, J.W.; Keller, N.P. LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot. Cell 2004, 3, 527–535. [Google Scholar] [CrossRef] [PubMed]
- Hoffmeister, D.; Keller, N.P. Natural products of filamentous fungi: Enzymes, genes, and their regulation. Nat. Prod. Rep. 2007, 24, 393–416. [Google Scholar] [CrossRef] [PubMed]
- Shwab, E.K.; Jin, W.B.; Tribus, M.; Galehr, J.; Graessle, S.; Keller, N.P. Histone deacetylase activity regulates chemical diversity in Aspergillus. Eukaryot. Cell 2007, 6, 1656–1664. [Google Scholar] [CrossRef] [PubMed]
- Bode, H.B.; Bethe, B.; Höfs, R.; Zeeck, A. Big effects from small changes: Possible ways to explore nature’s chemical diversity. ChemBioChem 2002, 3, 619–627. [Google Scholar] [CrossRef]
- Van Lanen, S.G.; Shen, B. Microbial genomics for the improvement of natural product discovery. Curr. Opin. Microbiol. 2006, 9, 252–260. [Google Scholar] [CrossRef] [PubMed]
- Williams, R.B.; Henrikson, J.C.; Hoover, A.R.; Lee, A.E.; Cichewicz, R.H. Epigenetic remodeling of the fungal secondary metabolome. Org. Biomol. Chem. 2008, 6, 1895–1897. [Google Scholar] [CrossRef] [PubMed]
- Challis, G.L. Mining microbial genomes for new natural products and biosynthetic pathways. Microbiology 2008, 154, 1555–1569. [Google Scholar] [CrossRef] [PubMed]
- Scherlach, K.; Hertweck, C. Triggering cryptic natural product biosynthesis in microorganisms. Org. Biomol. Chem. 2009, 7, 1753–1760. [Google Scholar] [CrossRef] [PubMed]
- Cichewicz, R.H. Epigenome manipulation as a pathway to new natural product scaffolds and their congeners. Nat. Prod. Rep. 2010, 27, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Chiang, Y.M.; Chang, S.L.; Oakley, B.R.; Wang, C.C.C. Recent advances in awakening silent biosynthetic gene clusters and linking orphan clusters to natural products in microorganisms. Curr. Opin. Chem. Biol. 2011, 15, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Pettit, R.K. Small-molecule elicitation of microbial secondary metabolites. Microb. Biotechnol. 2011, 4, 471–478. [Google Scholar] [CrossRef] [PubMed]
- Lim, F.Y.; Sanchez, J.F.; Wang, C.C.C.; Keller, N.P. Toward awakening cryptic secondary metabolite gene clusters in filamentous fungi. Methods Enzymol. 2012, 517, 303–324. [Google Scholar] [CrossRef] [PubMed]
- González-Menéndez, V.; Pérez-Bonilla, M.; Pérez-Victoria, I.; Martín, J.; Muñoz, F.; Reyes, F.; Tormo, J.; Genilloud, O. Multicomponent analysis of the differential induction of secondary metabolite profiles in fungal endophytes. Molecules 2016, 21, 234. [Google Scholar] [CrossRef] [PubMed]
- Beau, J.; Mahid, N.; Burda, W.N.; Harrington, L.; Shaw, L.N.; Mutka, T.; Kyle, D.E.; Barisic, B.; Van Olphen, A.; Baker, B.J. Epigenetic tailoring for the production of anti-infective cytosporones from the marine fungus Leucostoma persoonii. Mar. Drugs 2012, 10, 762–774. [Google Scholar] [CrossRef] [PubMed]
- Fleeman, R.; Lavoi, T.M.; Santos, R.G.; Morales, A.; Nefzi, A.; Welmaker, G.S.; Medina-Franco, J.L.; Giulianotti, M.A.; Houghten, R.A.; Shaw, L.N. Combinatorial libraries as a tool for the discovery of novel, broad-spectrum antibacterial agents targeting the ESKAPE pathogens. J. Med. Chem. 2015, 58, 3340–3355. [Google Scholar] [CrossRef] [PubMed]
- In our experience, chemotypic plasticity renders taxonomic identification irrelevant when targeting chemical diversity for drug discovery screening campaigns. Previously unpublished results from experiments in our lab have demonstrated that fungi identified as the same specimen by morphological taxonomic evaluation or even 16S sequencing can produce markedly different chemotypes in laboratory culture. With that in mind, fungi isolated for screening efforts, like those discussed here, are not identified or analyzed in any way based on taxonomy. Instead, the focus of this work is on accessing the greatest chemical potential of the greatest number of fungi via fermentation manipulation.
- Palomino, J.C.; Martin, A. Drug resistance mechanisms in Mycobacterium tuberculosis. Antibiotics 2014, 3, 317–340. [Google Scholar] [CrossRef] [PubMed]
- Rice, C.A.; Colon, B.L.; Alp, M.; Göker, H.; Boykin, D.W.; Kyle, D.E. Bis-benzimidazole hits against Naegleria fowleri discovered with new high-throughput screens. Antimicrob. Agents Chemother. 2015, 59, 2037–2044. [Google Scholar] [CrossRef] [PubMed]
- Alvar, J.; Velez, I.D.; Bern, C.; Herrero, M.; Desjeux, P.; Cano, J.; Jannin, J.; den Boer, M.; The WHO Leishmaniasis Control Team. Leishmaniasis worldwide and global estimates of its incidence. PLoS ONE 2012, 7, e35671. [Google Scholar] [CrossRef] [PubMed]
- Grace, E.; Asbill, S.; Virga, K. Naegleria fowleri: Pathogenesis, diagnosis, and treatment options. Antimicrob. Agents Chemother. 2015, 59, 6677–6681. [Google Scholar] [CrossRef] [PubMed]
- Okwor, I.; Uzonna, J. Social and economic burden of human leishmaniasis. Am. J. Trop. Med. Hyg. 2016, 94, 489–493. [Google Scholar] [CrossRef] [PubMed]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Demers, D.H.; Knestrick, M.A.; Fleeman, R.; Tawfik, R.; Azhari, A.; Souza, A.; Vesely, B.; Netherton, M.; Gupta, R.; Colon, B.L.; et al. Exploitation of Mangrove Endophytic Fungi for Infectious Disease Drug Discovery. Mar. Drugs 2018, 16, 376. https://doi.org/10.3390/md16100376
Demers DH, Knestrick MA, Fleeman R, Tawfik R, Azhari A, Souza A, Vesely B, Netherton M, Gupta R, Colon BL, et al. Exploitation of Mangrove Endophytic Fungi for Infectious Disease Drug Discovery. Marine Drugs. 2018; 16(10):376. https://doi.org/10.3390/md16100376
Chicago/Turabian StyleDemers, Danielle H., Matthew A. Knestrick, Renee Fleeman, Rahmy Tawfik, Ala Azhari, Ashley Souza, Brian Vesely, Mandy Netherton, Rashmi Gupta, Beatrice L. Colon, and et al. 2018. "Exploitation of Mangrove Endophytic Fungi for Infectious Disease Drug Discovery" Marine Drugs 16, no. 10: 376. https://doi.org/10.3390/md16100376
APA StyleDemers, D. H., Knestrick, M. A., Fleeman, R., Tawfik, R., Azhari, A., Souza, A., Vesely, B., Netherton, M., Gupta, R., Colon, B. L., Rice, C. A., Rodríguez-Pérez, M. A., Rohde, K. H., Kyle, D. E., Shaw, L. N., & Baker, B. J. (2018). Exploitation of Mangrove Endophytic Fungi for Infectious Disease Drug Discovery. Marine Drugs, 16(10), 376. https://doi.org/10.3390/md16100376





