Biological and Chemical Diversity of Ascidian-Associated Microorganisms
Abstract
1. Introduction
2. Microorganisms Associated with Ascidians
2.1. Geographical Distribution of Microorganisms Associated with Ascidians
2.2. Diversity of Culturable Microorganisms Associated with Ascidians
2.2.1. Bacteria
2.2.2. Actinobacteria
2.2.3. Cyanobacteria
2.2.4. Fungi
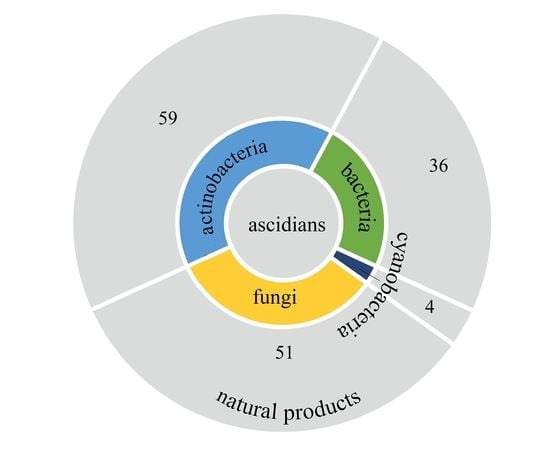
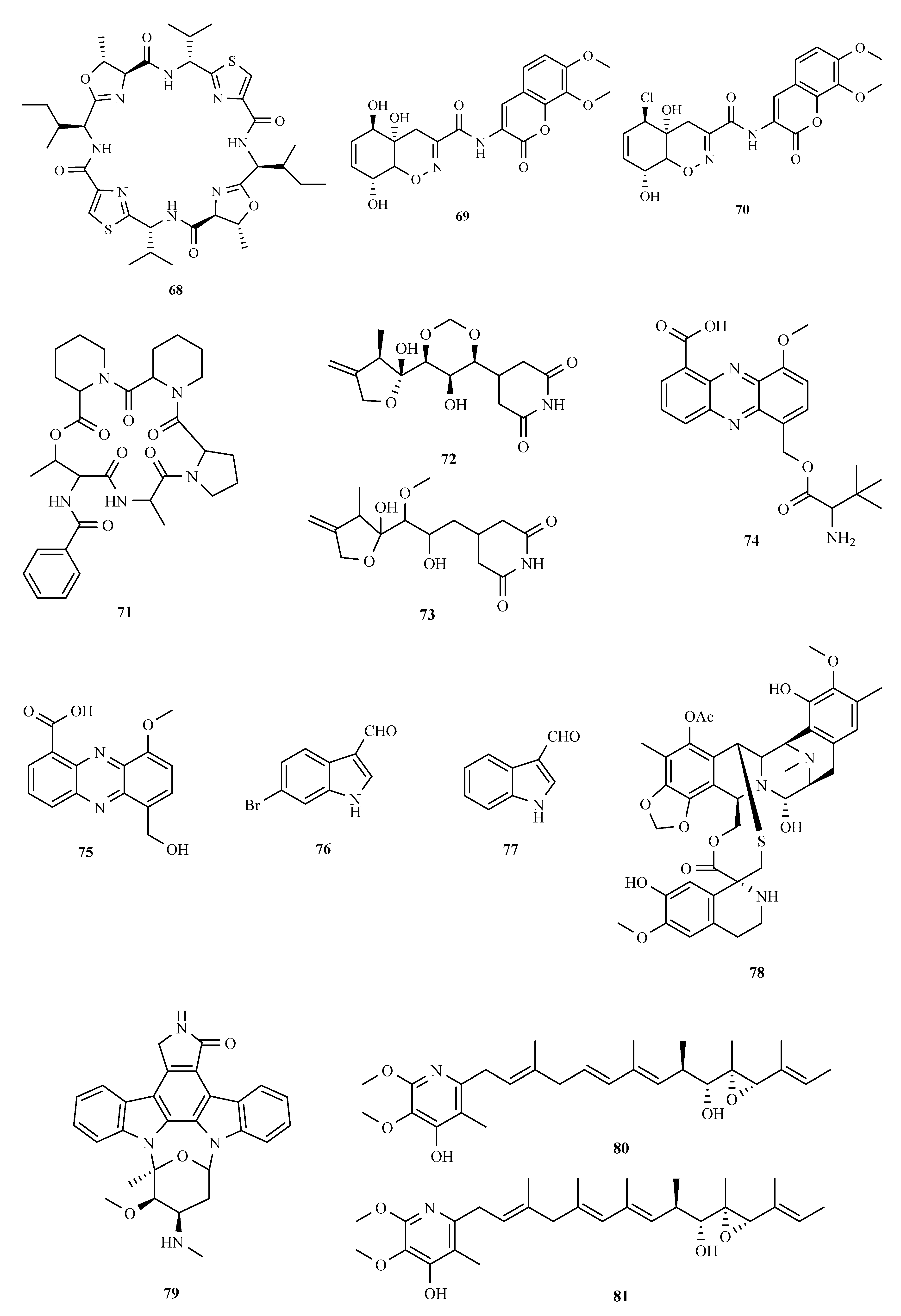
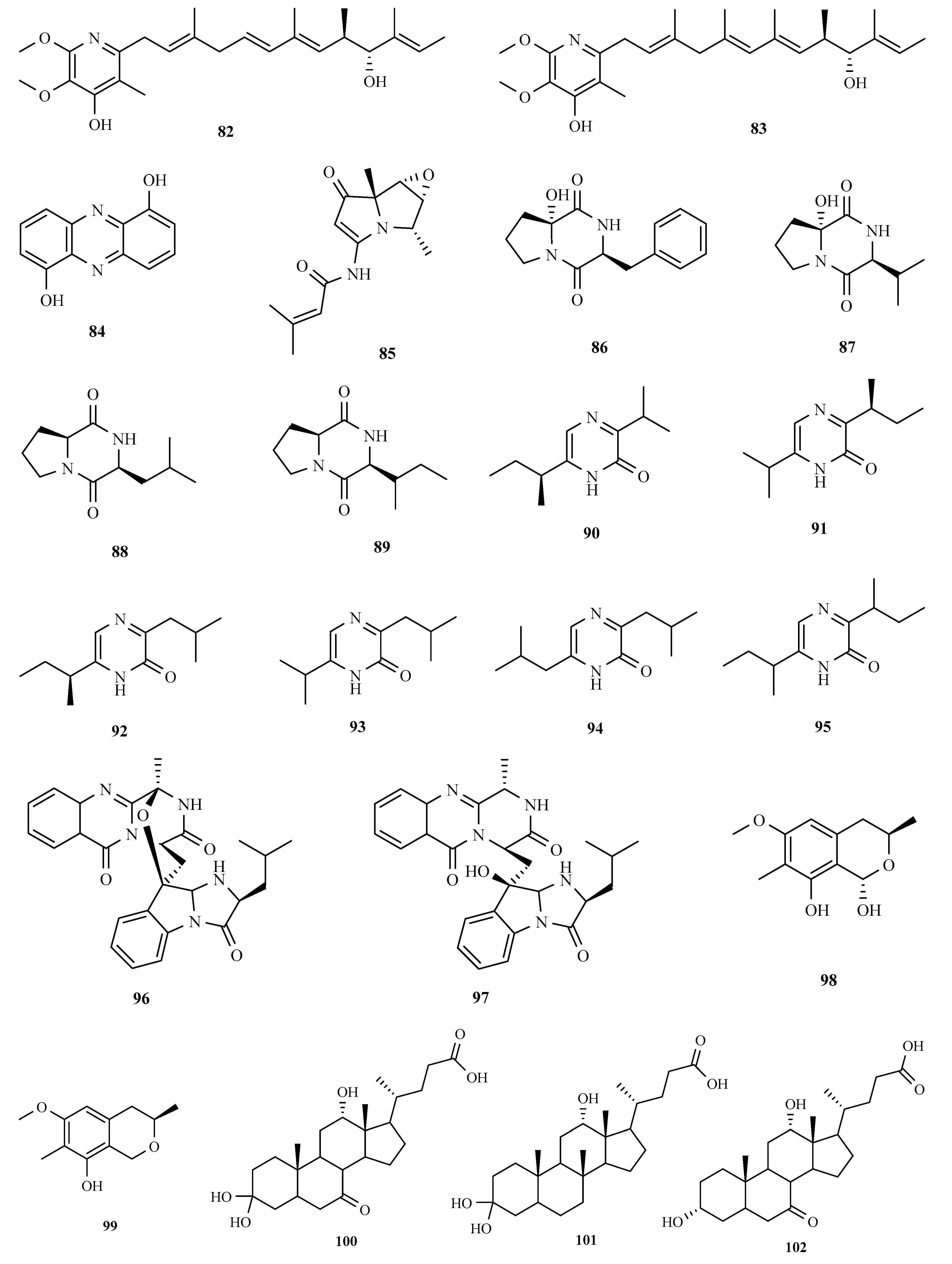
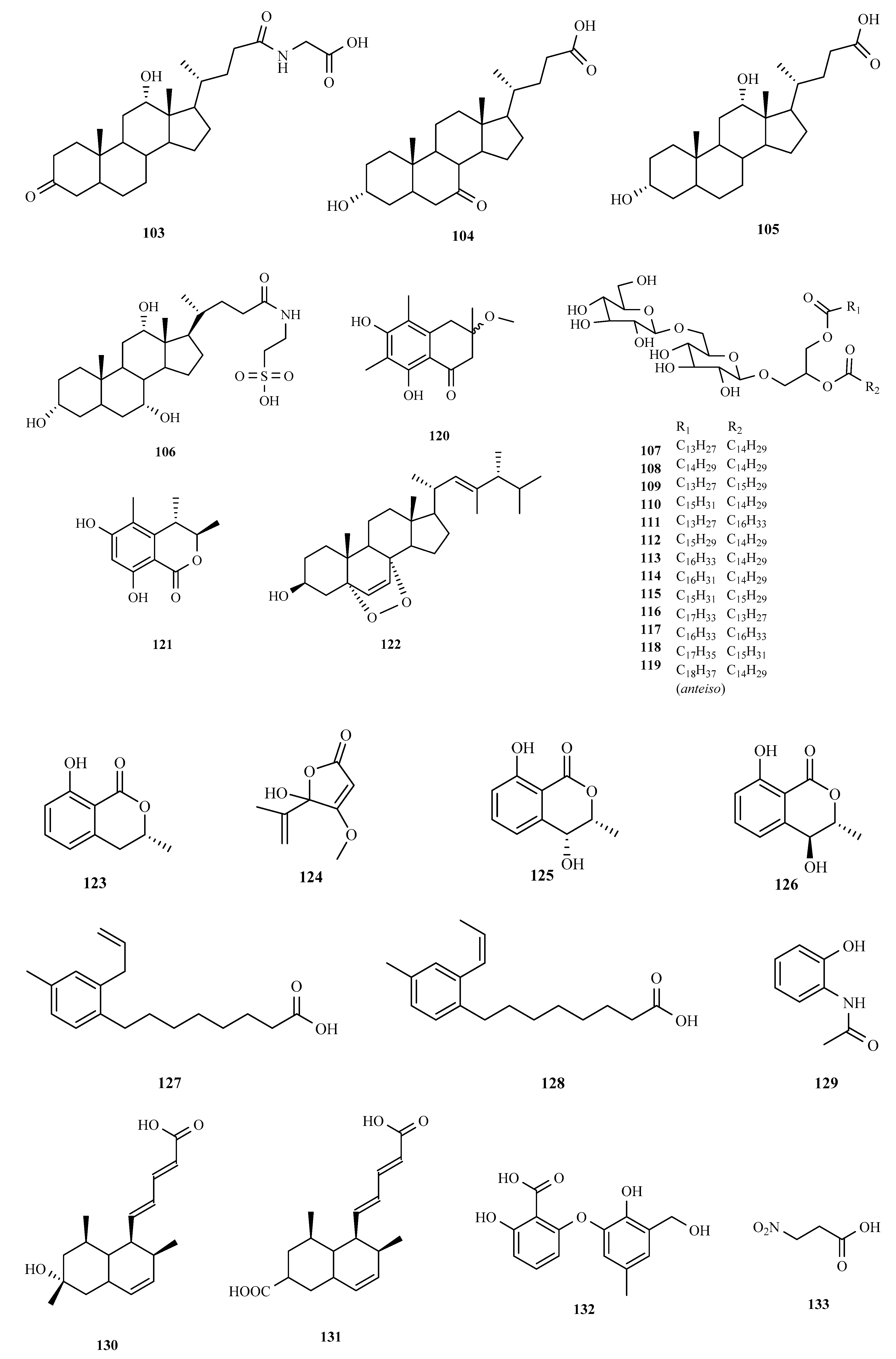
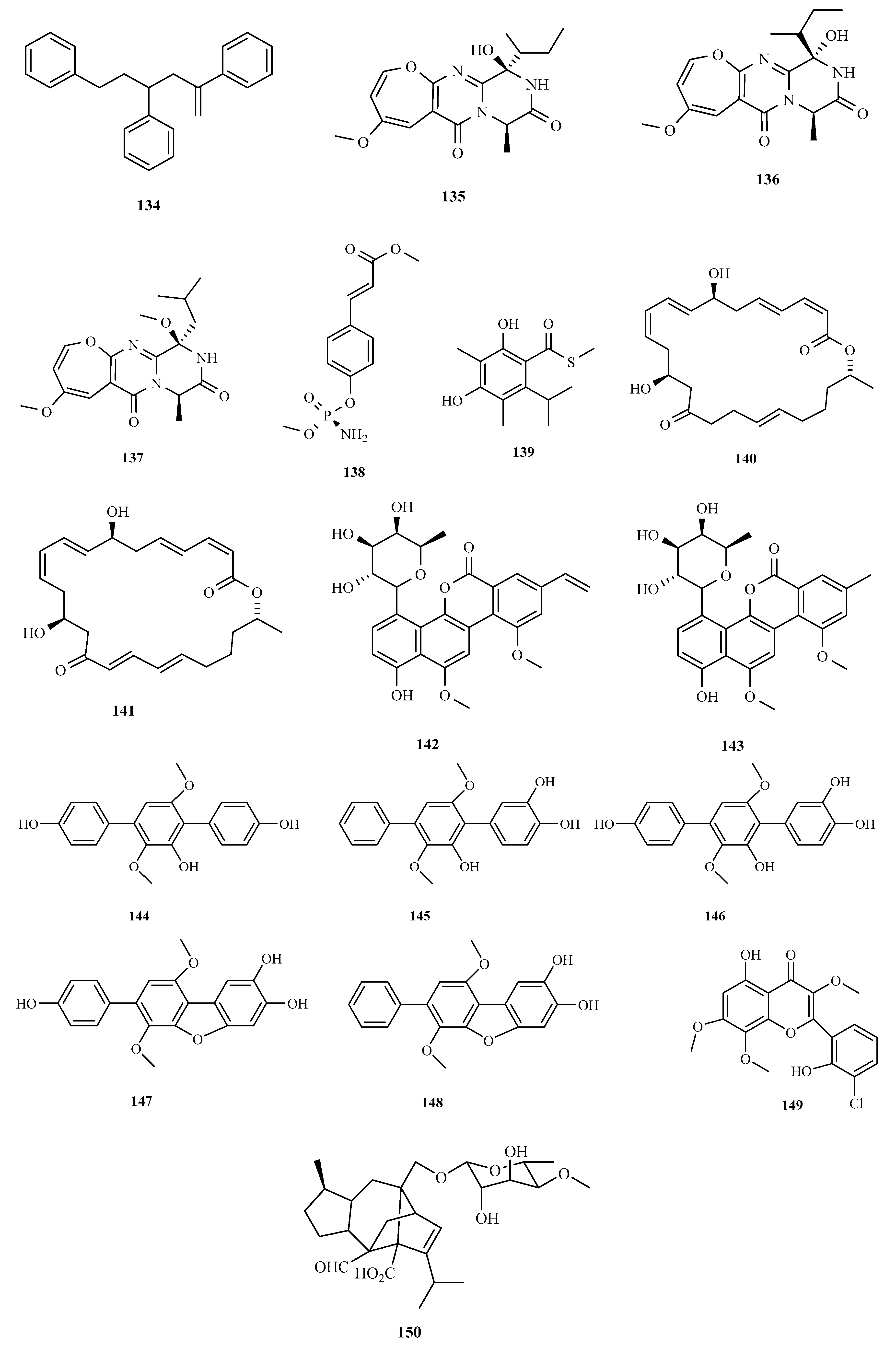
3. Structure and Bioactivities of Natural Products
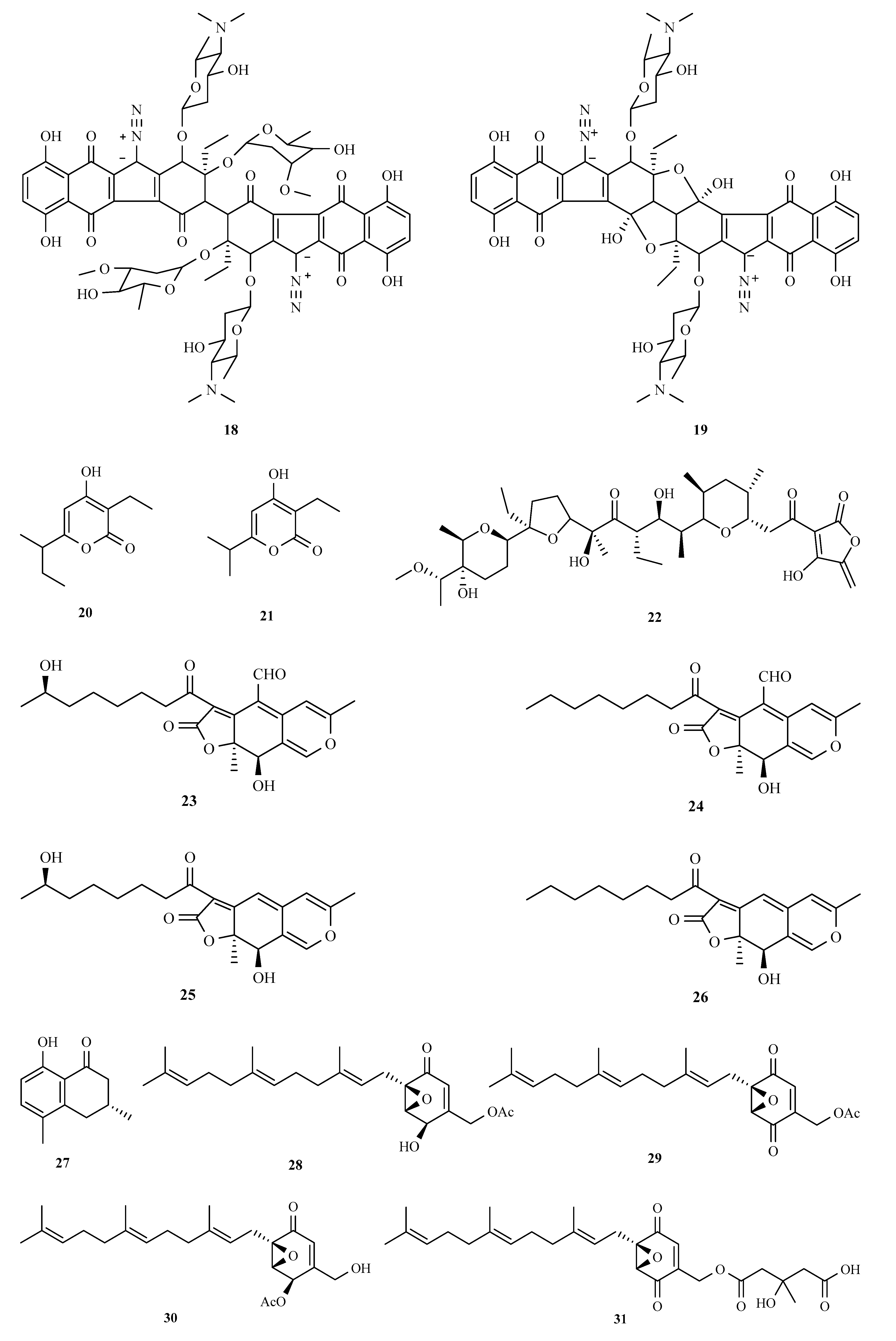
3.1. Polyketides
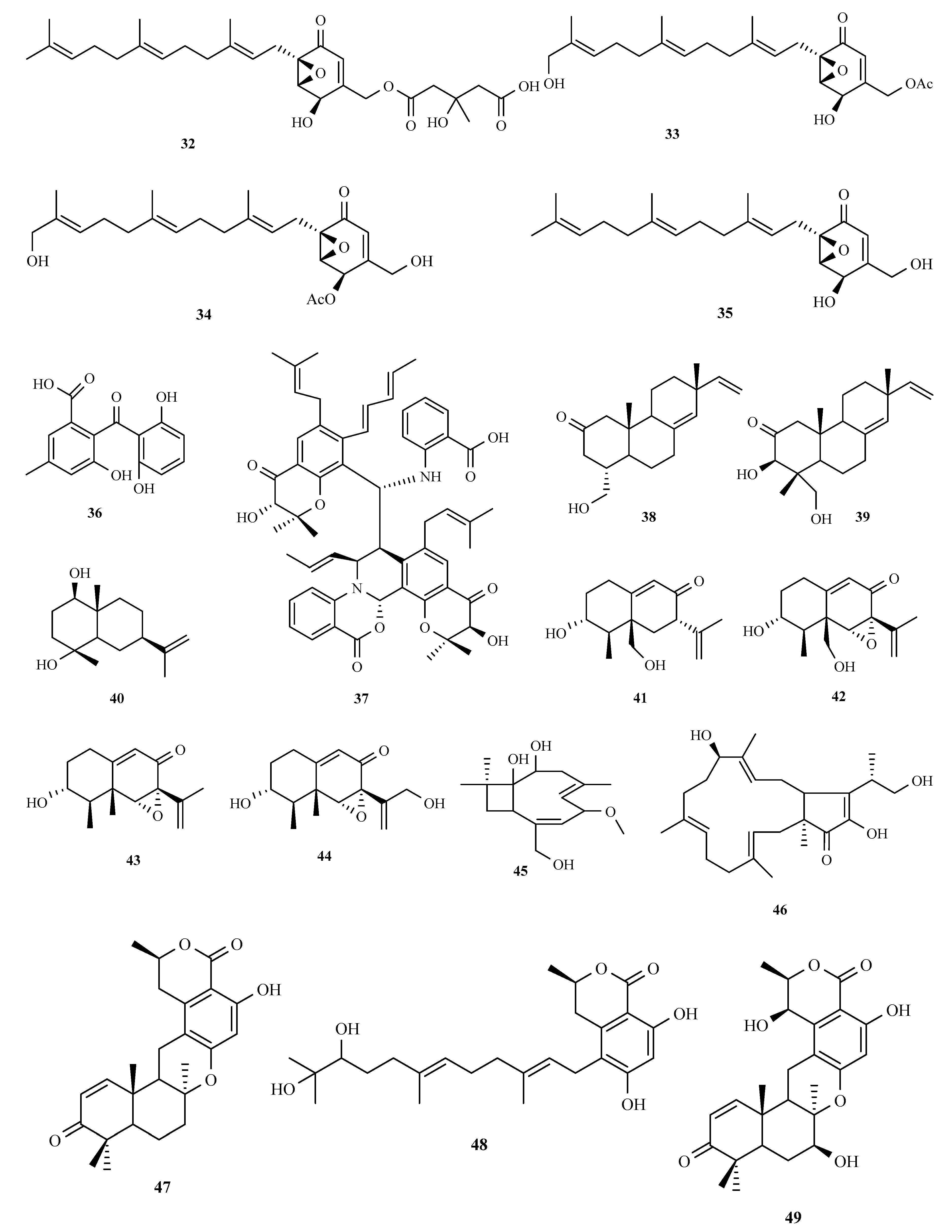
3.2. Terpenoids and Meroterpenoids
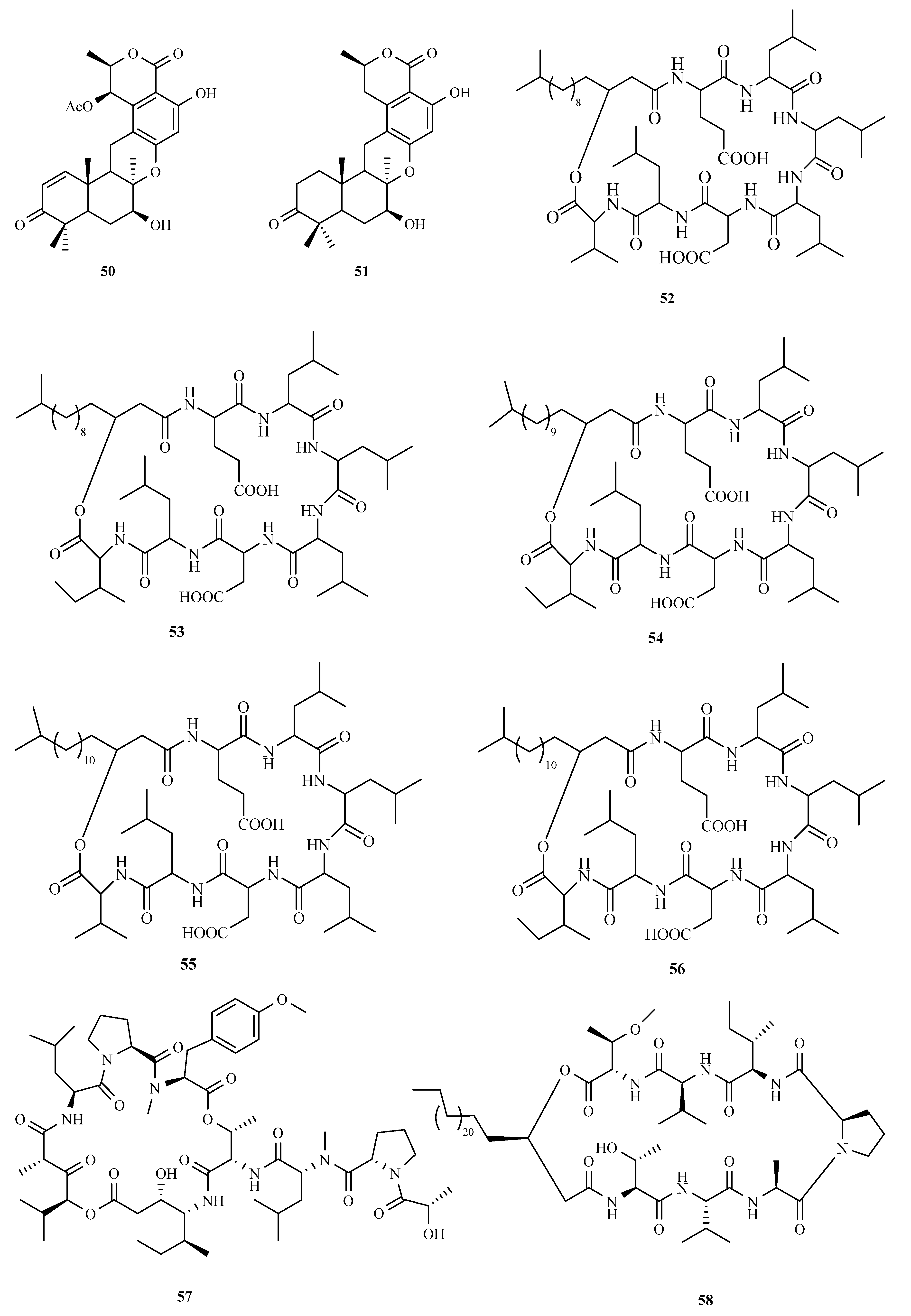
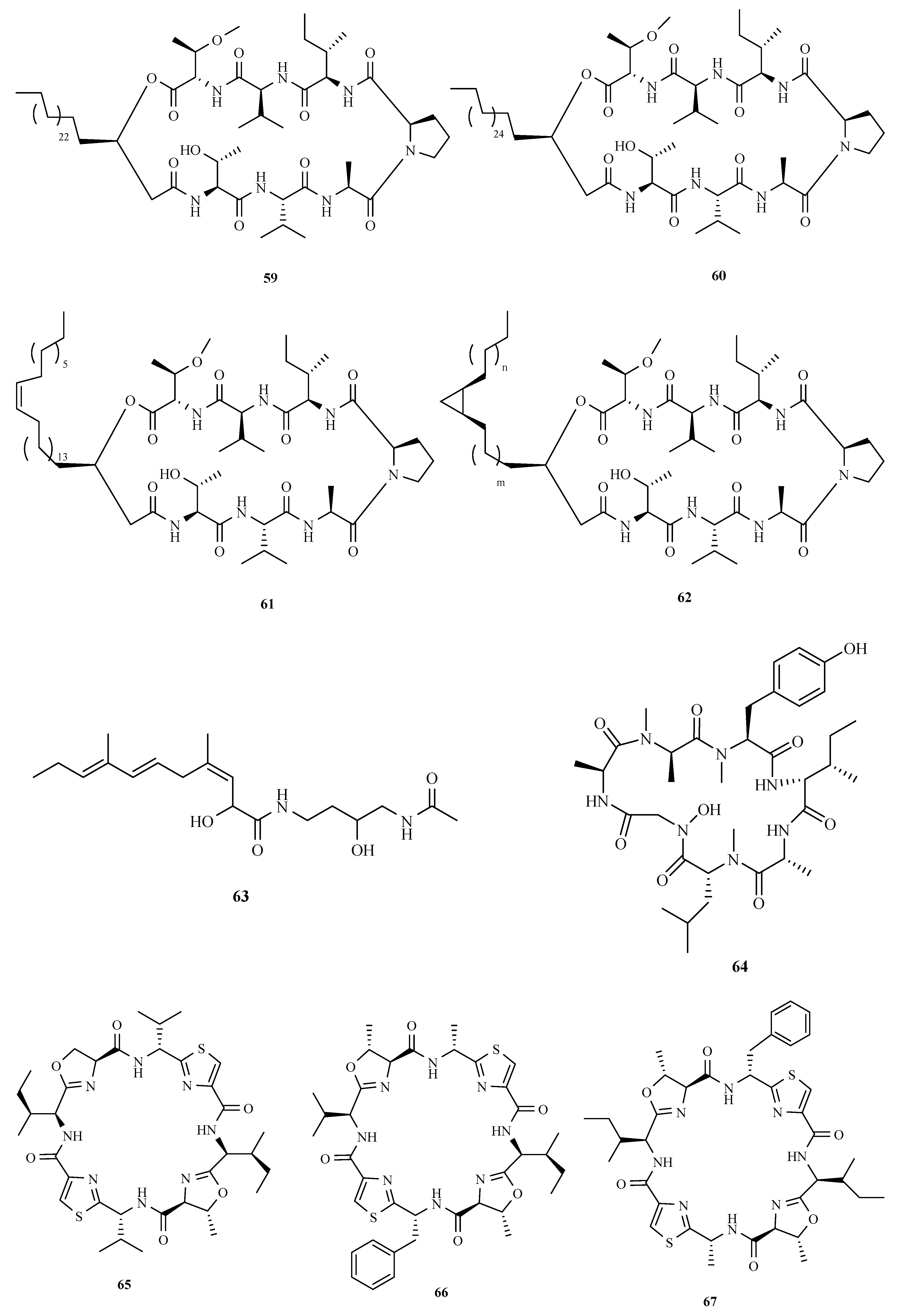
3.3. Peptides
3.4. Alkaloids
3.5. Other Types of Compounds Isolated from Ascidian-Associated Microorganisms
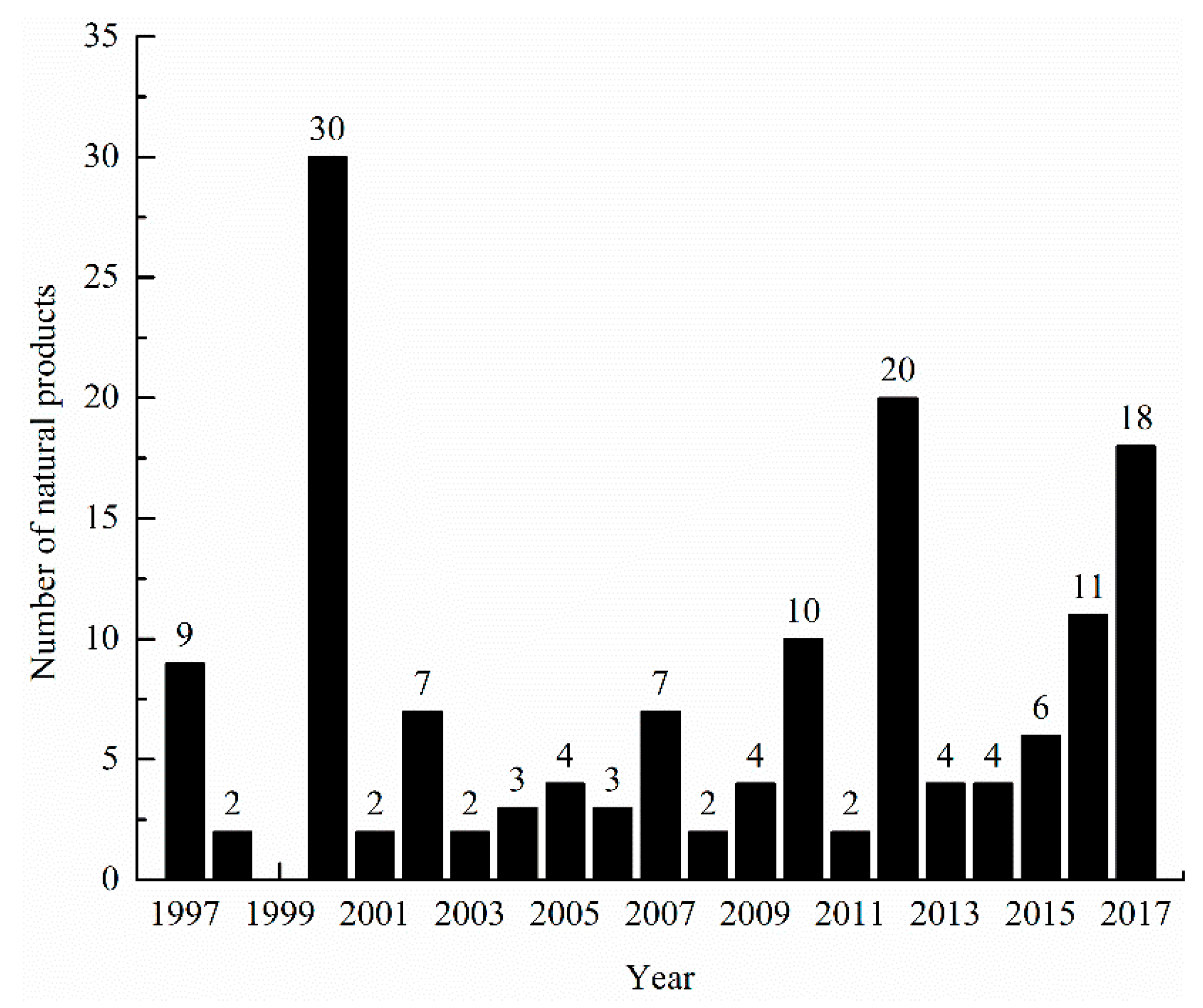
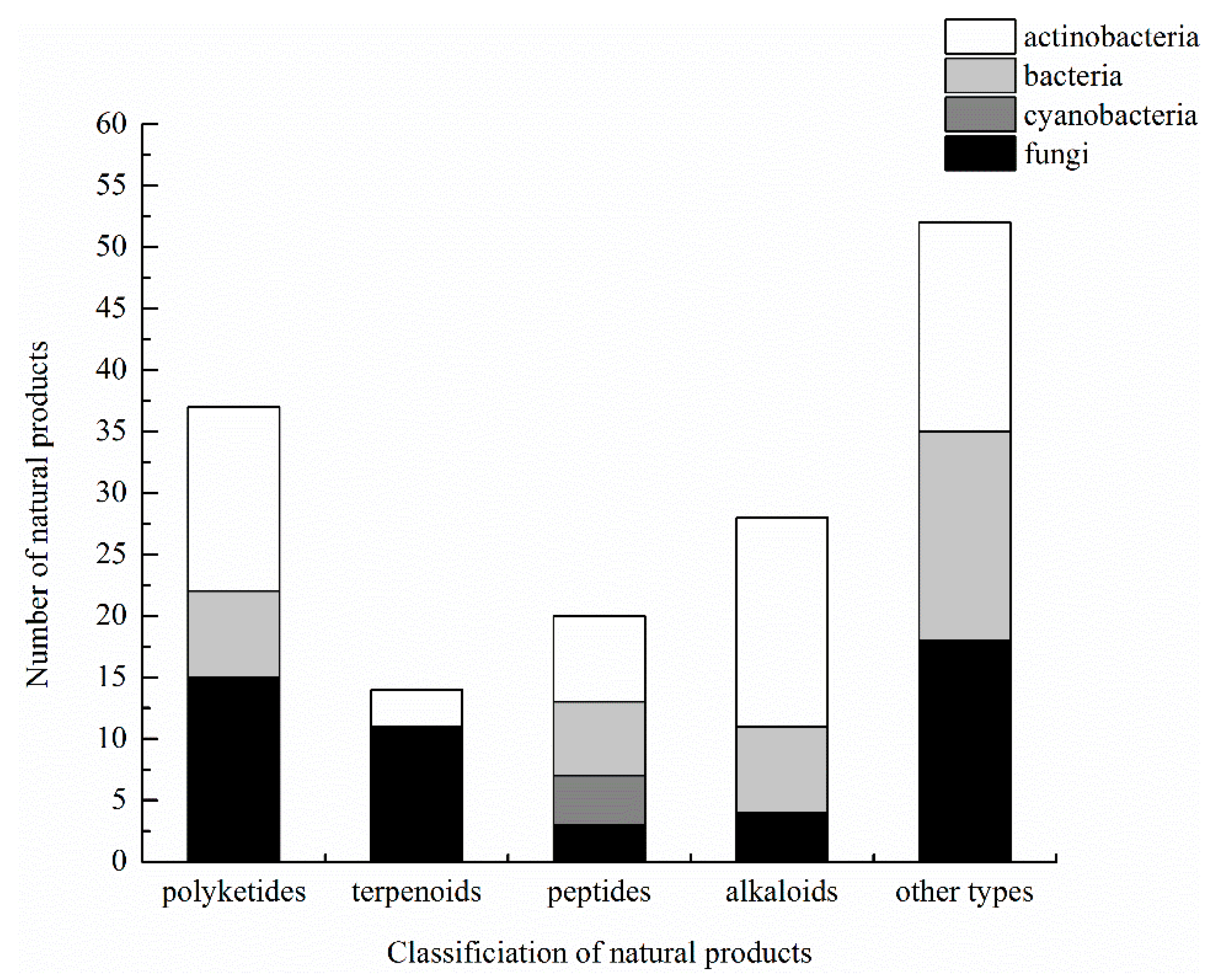
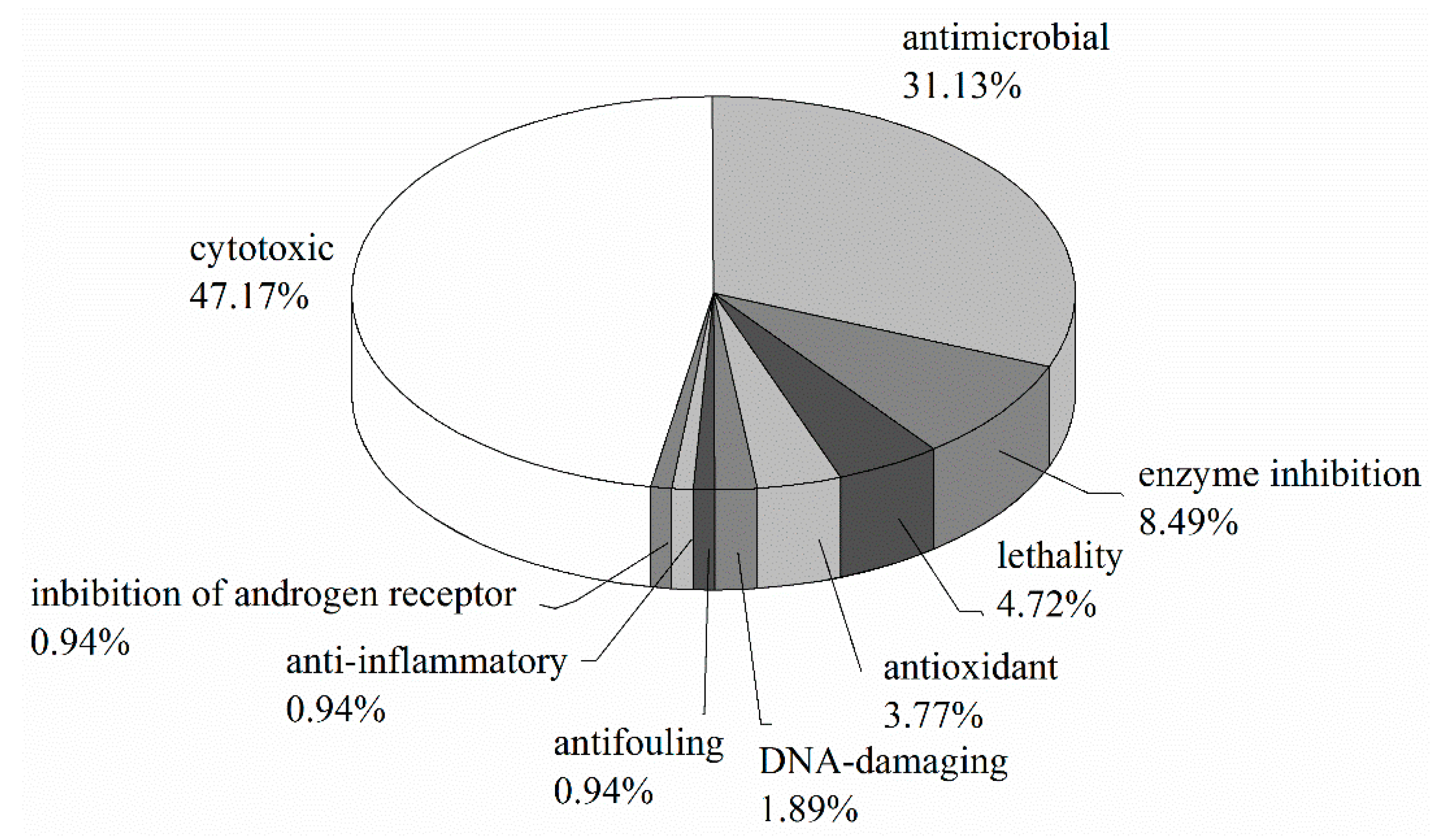
3.6. Summary of Natural Products
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Ascidiacea World Database. Available online: www.marinespecies.org/ascidiacea (accessed on 11 February 2017).
- Davidson, B.S. Ascidians: Producers of amino acid-derived metabolites. Chem. Rev. 1993, 93, 1771–1791. [Google Scholar] [CrossRef]
- Chen, L.; Fu, C.M.; Wang, G.Y. Microbial diversity associated with ascidians: A review of research methods and application. Symbiosis 2017, 71, 19–26. [Google Scholar] [CrossRef]
- Fermé, C.; Mateos, M.V.; Szyldergemajn, S.; Corrado, S.C.; Zucca, E.; Extremera, S.; Gianni, A.M.; Vandermeeren, A.; Ribrag, V. Aplidin® (Plitidepsin) activity in peripheral T-cell lymphoma (PTCL): Final results. Blood 2010, 116, 1767. [Google Scholar]
- Stone, R.M.; Mandrekar, S.; Sanford, B.L.; Geyer, S.; Bloomfield, C.D.; Dohner, K.; Thiede, C.; Marcucci, G.; Lo-Coco, F.; Klisovic, R.B.; et al. The multi-kinase inhibitor midostaurin (M) prolongs survival compared with placebo (P) in combination with daunorubicin (D)/cytarabine (C) induction (ind), high-dose C consolidation (consol), and as maintenance (maint) therapy in newly diagnosed acute myeloid leukemia (AML) patients (pts) age 18–60 with FLT3 mutations (muts): An international prospective randomized (rand) P-controlled double-blind trial (CALGB 10603/RATIFY [Alliance]). In Proceedings of the American Society of Hematology (ASH) 57th Annual Meeting, Orlando, FL, USA, 5–8 December 2005. [Google Scholar]
- Levis, M.; Ravandi, F.; Wang, E.S.; Baer, M.R.; Perl, A.; Coutre, S.; Erba, H.; Stuart, R.K.; Baccarani, M.; Cripe, L.D.; et al. Results from a randomized trial of salvage chemotherapy followed by lestaurtinib for patients with FLT3 mutant AML in first relapse. Blood 2011, 117, 3294–3301. [Google Scholar] [CrossRef] [PubMed]
- Saif, M.W.; Diasio, R.B. Edotecarin: A novel topoisomerase I inhibitor. Clin. Colorectal Cancer 2005, 5, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.Y. Advances in marine microbial symbionts in the China Sea and related pharmaceutical metabolites. Mar. Drugs 2009, 7, 113–129. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, E.W.; Donia, M.S. Life in cellulose houses: Symbiotic bacterial biosynthesis of ascidian drugs and drug leads. Curr. Opin. Biotechnol. 2010, 21, 827–833. [Google Scholar] [CrossRef] [PubMed]
- Lars, S.; Kjeldsen, K.U.; Funch, P.; Jensen, J.; Obst, M.; López-Legentil, S.; Schramm, A. Endozoicomonas are specific, facultative symbionts of sea squirts. Front. Microbiol. 2016, 7, 1042. [Google Scholar] [CrossRef]
- Das, S.; Lyla, P.S.; Khan, S.A. Marine microbial diversity and ecology: Importance and future perspectives. Curr. Sci. 2006, 90, 1325–1335. [Google Scholar]
- Olguin-Uribe, G.; Abou-Mansour, E.; Boulander, A.; Débard, H.; Francisco, C.; Combaut, G. 6-bromoindole-3-carbaldehyde, from an Acinetobacter sp. bacterium associated with the ascidian Stomozoa murrayi. J. Chem. Ecol. 1997, 23, 2507–2521. [Google Scholar] [CrossRef]
- Socha, A.M.; Garcia, D.; Sheffer, R.; Rowley, D.C. Antibiotic bisanthraquinones produced by a Streptomycete isolated from a cyanobacterium associated with Ecteinascidia turbinata. J. Nat. Prod. 2006, 69, 1070–1073. [Google Scholar] [CrossRef] [PubMed]
- Asolkar, R.N.; Kirkland, T.N.; Jensen, P.R.; Fenical, W. Arenimycin, an antibiotic effective against rifampin- and methicillin-resistant Staphylococcus aureus from the marine actinomycete Salinispora arenicola. J. Antibiot. (Tokyo) 2010, 63, 37–39. [Google Scholar] [CrossRef] [PubMed]
- Rinehart, K.L.; Holt, T.G.; Fregeau, N.L.; Stroh, J.G.; Keifer, P.A.; Sun, F.; Li, L.H.; Martin, D.G. Ecteinascidins 729, 743, 745, 759A, 759B, and 770: Potent antitumor agents from the Caribbean tunicate Ecteinascidia turbinata. J. Org. Chem. 1990, 55, 4512–4515. [Google Scholar] [CrossRef]
- Rath, C.M.; Janto, B.; Earl, J.; Ahmed, A.; Hu, F.Z.; Hiller, L.; Dahlgren, M.; Kreft, R.; Yu, F.; Wolff, J.J.; et al. Metaomic characterization of the marine invertebrate microbial consortium that produces the chemotherapeutic natural product ET-743. ACS Chem. Biol. 2011, 6, 1244–1256. [Google Scholar] [CrossRef] [PubMed]
- Rinehart, K.L.; Gloer, J.B.; Hughes, R.G.; Renis, H.E.; Mcgovern, J.P.; Swynenberg, E.B.; Stringfellow, D.A.; Kuentzel, S.L.; Li, L.H. Didemnins: Antiviral and antitumor depsipeptides from a Caribbean tunicate. Science 1981, 212, 933–935. [Google Scholar] [CrossRef] [PubMed]
- Tsukimoto, M.; Nagaoka, N.; Shishido, Y.; Fujimoto, J.; Nishisaka, F.; Matsumoto, S.; Harunari, E.; Imada, C.; Matsuzaki, T. Bacterial production of the tunicate-derived antitumor cyclic depsipeptide Didemnin B. J. Nat. Prod. 2011, 74, 2329–2331. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Kersten, R.D.; Nam, S.G.; Lu, L.; Al-Suwailem, A.M.; Zheng, H.; Fenical, W.; Dorrestein, P.C.; Moore, B.S.; Qian, P.Y. Bacterial biosynthesis and maturation of the didemnin anti-cancer agents. J. Am. Chem. Soc. 2012, 134, 8625–8632. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, E.W. The secret to a successful relationship: Lasting chemistry between ascidians and their symbiotic bacteria. Invertebr. Biol. 2014, 134, 88–102. [Google Scholar] [CrossRef] [PubMed]
- Tianero, M.D.; Kwan, J.C.; Wyche, T.P.; Presson, A.P.; Koch, M.; Barrows, L.R.; Bugni, T.S.; Schmidt, E.W. Species specificity of symbiosis and secondary metabolism in ascidians. ISME J. 2015, 9, 615–628. [Google Scholar] [CrossRef] [PubMed]
- Tait, E.; Sievert, C.S.M. Phylogenetic diversity of bacteria associated with ascidians in Eel Pond (Woods Hole, Massachusetts, USA). J. Exp. Mar. Biol. Ecol. 2007, 342, 138–146. [Google Scholar] [CrossRef]
- Abdelmohsen, U.R.; Bayer, K.; Hentschel, U. Diversity, abundance and natural products of marine sponge-associated actinomycetes. Nat. Prod. Rep. 2014, 31, 381–399. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Zhang, L.M.; Guo, X.X.; Dai, X.; Liu, L.; Wang, J.; Song, L.; Wang, Y.Z.; Zhu, Y.X.; Huang, L.; et al. Diversity, biogeography, and biodegradation potential of actinobacteria in the deep-sea sediments along the Southwest Indian Ridge. Front. Microbiol. 2016, 7, 1340. [Google Scholar] [CrossRef] [PubMed]
- Sarmientovizcaíno, A.; González, V.; Braña, A.F.; Palacios, J.J.; Otero, L.; Fernández, J.; Molina, A.; Kulik, A.; Vázquez, F.; Acuña, J.L.; et al. Pharmacological potential of phylogenetically diverse actinobacteria isolated from deep-sea coral ecosystems of the submarine aviles canyon in the Cantabrian Sea. Microbiol. Ecol. 2016, 73, 338–352. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.; Kato, C.; Runko, G.M.; Nogi, Y.; Hori, T.; Li, J.T.; Morono, Y.; Inagaki, F. Predominance of viable spore-forming piezophilic bacteria in high-pressure enrichment cultures from ~1.5 to 2.4 km-deep coal-bearing sediments below theocean floor. Front. Microbiol. 2017, 8, 137. [Google Scholar] [CrossRef] [PubMed]
- Olson, R.R. Photoadaptations of the Caribbean colonial ascidian-cyanophyte symbiosis Trididemnum solidum. Biol. Bull. 1986, 170, 62–74. [Google Scholar] [CrossRef]
- Qu, W.Y.; Chen, L.; Wang, G.Y.; Yan, P.S. Symbiosis between ascidian and cyanobacteria. Microbiol. China 2017, 44, 458–464. (In Chinese) [Google Scholar]
- López-Legentil, S.; Erwin, P.M.; Turon, M.; Yarden, O. Diversity of fungi isolated from three temperate ascidians. Symbiosis 2015, 66, 99–106. [Google Scholar] [CrossRef]
- Menezes, C.B.; Bonugli-Santo, R.C.; Miqueletto, P.B.; Passarini, M.R.Z.; Sliva, C.H.D.; Justo, M.R.; Leal, R.R.; Fantinatti-Garbonggini, F.; Oliveira, V.M.; Berlinck, R.G.S.; et al. Microbial diversity associated with algae, ascidians and sponges from the north coast of São Paulo state, Brazil. Microbiol. Res. 2010, 165, 466–482. [Google Scholar] [CrossRef] [PubMed]
- Acebal, C.; Alcazar, R.; Cañedo, L.M.; De, L.C.F.; Rodriguez, P.; Romero, F.; Puentes, J.L.F. Two marine Agrobacterium producers of sesbanimide antibiotics. J. Antibiot. 1998, 51, 64–67. [Google Scholar] [CrossRef] [PubMed]
- Kalinovskaya, N.I.; Kuznetsova, T.A.; Kalinovsky, A.I.; Denisenka, V.A.; Svetashev, V.I.; Romanenko, L.A. Structural characterization of gentiobiosyl diglycerides from Bacillus pumilus assoicated with ascidian Haloccynthia aurantium. Russ. Chem. Bull. 2000, 49, 169–173. [Google Scholar] [CrossRef]
- Kalinovskaya, N.I.; Kuznetsova, T.A.; Ivanova, E.P.; Romanenko, L.A.; Voinov, V.G.; Huth, F.; Laatsch, H. Characterization of surfactin-like cyclic depsipeptides Bacillus pumilus from ascidian Halocynthia aurantium. Mar. Biotechnol. 2002, 4, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Kwan, J.C.; Donia, M.S.; Han, A.W.; Hirose, E.; Haygood, M.G.; Schmidt, E.W. Genome streamlining and chemical defense in a coral reef symbiosis. Proc. Natl. Acad. Sci. USA 2012, 109, 20655–20660. [Google Scholar] [CrossRef] [PubMed]
- Romanenko, L.A.; Schumann, P.; Rohde, M.; Mikhailov, V.V.; Stackebrandt, E. Halomonas halocynthiae sp. nov., isolated from the marine ascidian Halocynthia aurantium. Int. J. Syst. Evol. Microbiol. 2002, 52, 1767–1772. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Shin, Y.K.; Sohn, Y.K.; Kwon, H.C. Two new cholic acid derivatives from the marine ascidian-associated bacterium Hasllibacter halocynthiae. Molecules 2012, 17, 12357–12364. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Yang, H.O.; Shin, Y.K.; Kwon, H.C. Hasllibacter halocynthiae gen. nov., sp. nov., a nutriacholic acid-producing bacterium isolated from the marine ascidian Halocynthia roretzi. Int. J. Syst. Evol. Microbiol. 2012, 62, 624–631. [Google Scholar] [CrossRef] [PubMed]
- Lu, D.C.; Zhao, J.X.; Wang, F.Q.; Xie, Z.H.; Du, Z.J. Labilibacter aurantiacus gen. nov., sp. nov., isolated from sea squirt (Styela clava) and reclassification of Saccharicrinis marinus as Labilibacter marinus comb. nov. Int. J. Syst. Evol. Microbiol. 2017, 67, 441–446. [Google Scholar] [CrossRef] [PubMed]
- Romanenko, L.A.; Uchino, M.; Falsen, E.; Lysenko, A.M.; Zhukova, N.V.; Mikhailov, V.V. Pseudomonas xanthomarina sp. nov., a novel bacterium isolated from marine ascidian. J. Gen. Appl. Microbiol. 2005, 51, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.; Matsuda, S.; Adachi, K.; Kasai, H.; Yokota, H. Rubritalea halochordaticola sp. nov., a carotenoid-producing verrucomicrobial species isolated from a marine chordate. Int. J. Syst. Evol. Microbiol. 2011, 61, 1515–1520. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.O.; Park, S.; Nam, B.H.; Kang, S.J.; Hur, Y.B.; Lee, S.J.; Oh, T.K.; Yoon, J.H. Ruegeria halocynthiae sp. nov., isolated from the sea squirt Halocynthia roretzi. Int. J. Syst. Evol. Microbiol. 2012, 62, 925–930. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.O.; Park, S.; Nam, B.H.; Jung, Y.T.; Kim, D.G.; Jee, Y.J.; Yoon, J.H. Tenacibaculum halocynthiae sp. nov., a member of the family Flavobacteriaceae isolated from sea squirt Halocynthia roretzi. Antonie Van Leeuwenhoek 2013, 103, 1321–1327. [Google Scholar] [CrossRef] [PubMed]
- Gnanakkan, A. Optimization of novel protease production through submerged fermetation by ascidian associated Vibrio. World J. Pharm. Pharm. Sci. 2014, 3, 765–770. [Google Scholar]
- Singh, M.P.; Menendez, A.T.; Petersen, P.J.; Ding, W.D.; Maiese, W.M.; Greenstein, M. Biological and mechanistic activities of phenazine antibiotics produced by culture LL-14I352. J. Antibiot. 1997, 50, 785–787. [Google Scholar] [CrossRef] [PubMed]
- Blasiak, L.C.; Zinder, S.H.; Buckley, D.H.; Hill, R.T. Bacterial diversity associated with the tunic of the model chordate Ciona intestinalis. ISME J. 2014, 8, 309–320. [Google Scholar] [CrossRef] [PubMed]
- Wyche, T.P.; Miranda, S.; Hou, Y.; Doug, B.; Johnson, D.A.; Johnson, J.A.; Bugni, T.S. Activation of the nuclear factor E2-related factor 2 pathway by novel natural products halomadurones A–D and a synthetic analogue. Mar. Drugs 2013, 11, 5089–5099. [Google Scholar] [CrossRef] [PubMed]
- Wyche, T.P.; Alvarenga, R.F.R.; Piotrowski, J.S.; Duster, M.N.; Warrack, S.R.; Cornilescu, G.; De Wolfe, T.J.; Hou, Y.; Braun, D.R.; Ellis, G.A.; et al. Chemical genomics, structure elucidation, and in vivo studies of the marine-derived Anticlostridial Ecteinamycin. ACS Chem. Biol. 2017, 12, 2287–2295. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Yang, H.O.; Sohn, Y.C.; Kwon, H.C. Aeromicrobium halocynthiae sp. nov., a taurocholic acid-producing bacterium isolated from the marine ascidian Halocynthia roretzi. Int. J. Syst. Evol. Microbiol. 2010, 60, 2793–2798. [Google Scholar] [CrossRef] [PubMed]
- De Menezes, C.B.A.; Afonso, R.S.; de Souza, W.R.; Parma, M.; de Melo, I.S.; Zucchi, T.D.; Garboggini, F.F. Gordonia didemni sp. nov. an actinomycete isolated from the marine ascidium Didemnum sp. Antonie Van Leeuwenhoek 2016, 109, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Jimenez, P.C.; Ferreira, E.G.; Araújo, L.A.; Guimarães, L.A.; Sousa, T.S.; Pessoa, O.D.L.; Lotufo, T.M.C.; Costa-Lotufo, L.V. Cytotoxicity of actinomycetes associated with the ascidian Eudistoma vannamei (Millar, 1977), endemic of northeastern coast of Brazil. Lat. Am. J. Aquat. Res. 2013, 41, 335–343. [Google Scholar] [CrossRef]
- Wyche, T.P.; Hou, Y.; Vazquez-Rivera, E.; Braun, D.; Bugni, T.S. Peptidolipins B–F, antibacterial lipopeptides from an ascidian-derived Nocardia sp. J. Nat. Prod. 2012, 75, 735–740. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsova, T.A.; Dmitrenok, A.S.; Sobolevskaya, M.P.; Shevchenko, L.S.; Mikhailov, V.V. Ubiquinone Q9 from a marine isolate of an actinobacterium Nocardia sp. Russ. Chem. Bull. 2002, 51, 1951–1953. [Google Scholar] [CrossRef]
- Pei, Z.L.; Chen, L.; Xu, J.L.; Shao, C.L. Secondary metabiolities and their biological activities of two actinomycetes Streptomyces coelicoflavus and Nocardiopsis dassonvillei associated with ascidians Styela clava and Botryllus schlosseri. Chin. J. Mar. Drugs 2017, 36, 55–60. [Google Scholar]
- Takagi, M.; Motohashi, K.; Izumikawa, M.; Khan, S.T.; Hwang, J.H.; Shin-Ya, K. JBIR-66 a new metabolite isolated from tunicate-derived Saccharopolyspora sp. Biosci. Biotechnol. Biochem. 2010, 74, 2355–2357. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Ding, W.D.; Bernan, V.S.; Richardson, A.D.; Ireland, C.M.; Greenstein, M.; Ellestad, G.A.; Carter, G.T. Lomaiviticins A and B, potent antitumor antibiotics from Micromonospora lomaivitiensis. J. Am. Chem. Soc. 2001, 123, 5362–5363. [Google Scholar] [CrossRef] [PubMed]
- Janso, J.E.; Haltli, B.A.; Eustáquio, A.S.; Kulowski, K.; Waldman, A.J.; Zha, L.; Nakamura, H.; Bernan, V.S.; He, H.; Carter, G.T.; et al. Discovery of the lomaiviticin biosynthetic gene cluster in Salinispora pacifica. Tetrahedron 2014, 70, 4156–4164. [Google Scholar] [CrossRef] [PubMed]
- Steinert, G.; Taylor, M.W.; Schupp, P.J. Diversity of actinobacteria associated with the marine ascidian Eudistoma toealensis. Mar. Biotechnol. 2015, 17, 377–385. [Google Scholar] [CrossRef] [PubMed]
- Ellis, G.A.; Wyche, T.P.; Fry, C.G.; Braun, D.R.; Bugni, T.S. Solwaric acids A and B, antibacterial aromatic acids from a marine Solwaraspora sp. Mar. Drugs 2014, 12, 1013–1022. [Google Scholar] [CrossRef] [PubMed]
- Harunari, E.; Hamada, M.; Shibata, C.; Tamura, T.; Komaki, H.; Imada, C.; Igarashi, Y. Streptomyces hyaluromycini sp. nov., isolated from a tunicate (Molgula manhattensis). J. Antibiot. 2015, 69, 159–163. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Piel, J. A gene cluster from a marine Streptomyces encoding the biosynthesis of the aromatic spiroketal polyketide griseorhodin A. Chem. Biol. 2002, 9, 1017–1026. [Google Scholar] [CrossRef]
- Melanie, Q.; Tim, S.; Jörn, P.; Paultheo, V.Z.; Stephanie, G. The new metabolite (S)-cinnamoylphosphoramide from Streptomyces sp. and its total synthesis. Eur. J. Org. Chem. 2008, 2008, 5117–5124. [Google Scholar] [CrossRef]
- Shaala, L.A.; Youssef, D.T.A.; Badr, J.M.; Harakeh, S.M. Bioactive 2 (1H)-pyrazinones and diketopiperazine alkaloids from a tunicate-derived actinomycete Streptomyces sp. Molecules 2016, 21, 1116. [Google Scholar] [CrossRef] [PubMed]
- Sung, A.A.; Gromek, S.M.; Balunas, M.J. Upregulation and identification of antibiotic activity of a marine-derived Streptomyces sp. via co-cultures with human pathogens. Mar. Drugs 2017, 15, 250. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, E.W.; Nelson, J.T.; Rasko, D.A.; Sudek, S.; Eisen, J.A.; Haygood, M.G.; Ravel, J. Patellamide A and C biosynthesis by a microcin-like pathway in Prochloron didemni, the cyanobacterial symbiont of Lissoclinum patella. Proc. Natl. Acad. Sci. USA 2005, 102, 7315–7320. [Google Scholar] [CrossRef] [PubMed]
- Kott, P. Didemnid-algal symbiosis: Host species in the Western Pacific with notes on the symbiosis. Micronesica 1982, 18, 95–127. [Google Scholar]
- Long, P.F.; Dunlap, W.C.; Battershill, C.N.; Jaspars, M. Shotgun cloning and heterologous expression of the patellamide gene cluster as a strategy to achieving sustained metabolite production. Chembiochem 2005, 6, 1760–1765. [Google Scholar] [CrossRef] [PubMed]
- Hirose, E.; Turon, X.; López-Legentil, S.; Erwin, P.M.; Hirose, M. First records of didemnid ascidians harbouring Prochloron from Caribbean Panama: Genetic relationships between Caribbean and Pacific photosymbionts and host ascidians. Syst. Biodivers. 2012, 10, 435–445. [Google Scholar] [CrossRef]
- Schmidt, E.W.; Sudek, S.; Haygood, M.G. Genetic evidence supports secondary metabolic diversity in Prochloron spp. the cyanobacterial symbiont of a tropical ascidian. J. Nat. Prod. 2004, 67, 1341–1345. [Google Scholar] [CrossRef] [PubMed]
- Lewin, R.A. A marine Synechocystis (Cyanophyta, Chroococcales) epizoic on ascidians. Phycologia 1975, 14, 153–160. [Google Scholar] [CrossRef]
- Kott, P. Algal-bearing didemnid ascidians in the Indo-West Pacific. Mem. Qd Mus. 1980, 20, 1–47. [Google Scholar]
- Hirose, E.; Uchida, H.; Murakami, A. Ultrastructural and microspectrophotometric characterization of multiple species of cyanobacterial photosymbionts coexisting in the colonial ascidian Trididemnum clinides (Tunicata, Ascidiacea, Didemnidae). Eur. J. Phycol. 2009, 44, 365–375. [Google Scholar] [CrossRef]
- Belofsky, G.N.; Anguera, M.; Jensen, P.R.; Fenical, W.; Kock, M. Oxepinamides A–C and fumiquinazolines H–I: Bioactive metabolites from a marine isolate of a fungus of the genus Acremonium. Chem. Eur. J. 2000, 6, 1355–1360. [Google Scholar] [CrossRef]
- Yurchenko, A.N.; Ivanets, E.V.; Smetanina, O.F.; Pivkin, M.V.; Dyshlovoi, S.A.; Amsberg, G.V.; Afiyatullov, S.S. Metabolites of the marine fungus Aspergillus candidus KMM 4676 associated with a Kuril colonial ascidian. Chem. Nat. Compd. 2017, 53, 747–749. [Google Scholar] [CrossRef]
- Bugni, T.S.; Abbanat, D.; Bernan, V.S.; Maiese, W.M.; Greenstein, M.; Wagoner, R.M.A.; Ireland, C.M. Yanuthones: Novel metabolites from a marine isolate of Aspergillus niger. J. Org. Chem. 2000, 65, 7195–7200. [Google Scholar] [CrossRef] [PubMed]
- Montenegro, T.G.C.; Rodrigues, F.A.R.; Jimenez, P.C.; Angelim, A.L.; Melo, V.M.M.; Filho, E.R.; de Oliveira, M.C.F.; Costa-Lotufo, L.V. Cytotoxic activity of fungal strains isolated from the ascidian Eudistoma vannamei. Chem. Biodivers. 2012, 9, 2203–2209. [Google Scholar] [CrossRef] [PubMed]
- Smetanina, O.F.; Kuznetsova, T.A.; Gerasimenko, A.V.; Kalinovsky, A.I.; Pivkin, M.V.; Dmitrenok, P.C.; Elyakov, G.B. Metabolites of the marine fungus Humicola fuscoatra, KMM 4629. Russ. Chem. Bull. 2004, 36, 2643–2646. [Google Scholar] [CrossRef]
- Pei, Z.L. Secondary Metabiolities and Bioactivity of Three Microorganisms Associated with Ascidians. Master’s Thesis, Harbin Institute of Technology, Weihai, China, 2016. [Google Scholar]
- Malmstrùm, J.; Christophersen, C.; Frisvad, J.C. Secondary metabolites characteristic of Penicillium citrinum, Penicillium steckii and related species. Phytochemistry 2000, 54, 301–309. [Google Scholar] [CrossRef]
- Xin, Z.H.; Tian, L.; Zhu, T.J.; Wang, W.L.; Du, L.; Fang, Y.C.; Gu, Q.Q.; Zhu, W.M. Isocoumarin derivatives from the sea squirt-derived fungus Penicillium stoloniferum QY2-10 and the halotolerant fungus Penicillium notatum B-52. Arch. Pharm. Res. 2007, 30, 816–819. [Google Scholar] [CrossRef] [PubMed]
- Motohashi, K.; Hashimoto, J.; Inaba, S.; Khan, S.T.; Komaki, H.; Nagai, A.; Takagi, M.; Shin-Ya, K. New sesquiterpenes, JBIR-27 and -28, isolated from a tunicate-derived fungus, Penicillium sp. SS080624SCf1. J. Antibiot. 2009, 62, 247–250. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.Y.S.; Borgeson, B.M.; Crews, P. Pitholides A–D, polyketides from a marine tunicate-derived culture of Pithomyces sp. Tetrahedron Lett. 1997, 38, 8449–8452. [Google Scholar] [CrossRef]
- Sumilat, D.A.; Yamazaki, H.; Endo, K.; Rotinsulu, H.; Wewengkang, D.S.; Ukai, K.; Namikoshi, M. A new biphenyl ether derivative produced by Indonesian ascidian-derived Penicillium albobiverticillium. J. Nat. Med. 2017, 71, 776–779. [Google Scholar] [CrossRef] [PubMed]
- Samson, R.A.; Yilmaz, N.; Houbraken, J.; Spierenburg, H.; Seifert, K.A.; Peterson, S.W.; Varga, J.; Frisvad, J.C. Phylogeny and nomenclature of the genus Talaromyces and taxa accommodated in Penicillium subgenus Biverticillium. Stud. Mycol. 2011, 70, 159–183. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, H.; Nakayama, W.; Takahashi, O.; Kirikoshi, R.; Izumikawa, Y.; Iwasaki, K.; Toraiwa, K.; Ukai, K.; Rotinsulu, H.; Wewengkang, D.S.; et al. Verruculides A and B, two new protein tyrosine phosphatase 1B inhibitors from an Indonesian ascidian-derived Penicillium verruculosum. Bioorg. Med. Chem. Lett. 2015, 25, 3087–3090. [Google Scholar] [CrossRef] [PubMed]
- Dewapriya, P.; Prasad, P.; Damodar, R.; Salim, A.A.; Capon, R.J. Talarolide A, a cyclic heptapeptide hydroxamate from an Australian marine tunicate-associated fungus, Talaromyces sp. (CMB-TU011). Org. Lett. 2017, 19, 2046–2049. [Google Scholar] [CrossRef] [PubMed]
- Garo, E.; Starks, C.M.; Jensen, P.R.; Fenical, W.; Lobkovsky, E.; Clardy, J. Trichodermamides A and B, cytotoxic modified dipeptides from the marine-derived fungus Trichoderma virens. J. Nat. Prod. 2003, 66, 423–426. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.J.; Koch, M.; Abdel, A.M.H.; Galindo-Murillo, R.; Tianero, M.D.; Cheatham, T.E.; Barrows, L.R.; Reilly, C.A.; Schmidt, E.W. Oxazinin A, a pseudodimeric natural product of mixed biosynthetic origin from a filamentous fungus. Org. Lett. 2014, 16, 4774–4777. [Google Scholar] [CrossRef] [PubMed]
- Shen, B. Polyketide biosynthesis beyond the type I, II and III polyketide synthase paradigms. Curr. Opin. Chem. Biol. 2003, 7, 285–295. [Google Scholar] [CrossRef]
- Riesenfeld, C.S.; Murray, A.E.; Baker, B.J. Characterization of the microbial community and polyketide biosynthetic potential in the palmerolide-producing tunicate Synoicum adareanum. J. Nat. Prod. 2008, 71, 1812–1818. [Google Scholar] [CrossRef] [PubMed]
- Diyabalanage, T.; Amsler, C.D.; McClintock, J.B.; Baker, B.J. Palmerolide A, a cytotoxic macrolide from the Antarctic tunicate Synoicum adareanum. J. Am. Chem. Soc. 2006, 128, 5630–5631. [Google Scholar] [CrossRef] [PubMed]
- Diyabalanage, T. Chemical Investigation of Two Antarctic Invertebrates, Synoicum adareanum (Chordata: Ascidiaceae; Enterogona; Polyclinidae) and Austrodoris kergulenensis (Molusca; Gastropoda; Nudibranchia; Dorididae). Ph.D. Thesis, University of South Florida, Tallahassee, FL, USA, 2006. [Google Scholar]
- Richardson, A.D.; Aalbersberg, W.; Ireland, C.M. The patellazoles inhibit protein synthesis at nanomolar concentrations in human colon tumor cells. Anticancer Drugs 2005, 16, 533–541. [Google Scholar] [CrossRef] [PubMed]
- Kwan, J.C.; Schmidt, E.W. Bacterial endosymbiosis in a chordate host: Long-term co-evolution and conservation of secondary metabolism. PLoS ONE 2013, 8, e80822. [Google Scholar] [CrossRef] [PubMed]
- Ueno, T.; Takahashi, H.; Oda, M.; Mizunuma, M.; Yokoyama, A.; Goto, Y.; Mizushina, Y.; Sakaguchi, K.; Hayashi, H. Inhibition of human telomerase by rubromycins: Implication of spiroketal system of the compounds as an active moiety. Biochemistry 2000, 39, 5995–6002. [Google Scholar] [CrossRef] [PubMed]
- Mehta, G.; Pan, S.C. First total synthesis of yanuthones: Novel farnesylated epoxycyclohexenoid marine natural products. Tetrahedron Lett. 2005, 46, 5219–5223. [Google Scholar] [CrossRef]
- Krick, A.; Kehraus, S.; Gerhäuser, C.; Klimo, K.; Nieger, M.; Maier, A.; Fiebig, H.H.; Atodiresei, I.; Raabe, G.; Fleischhauer, J.; König, G.M. Potential cancer chemopreventive in vitro activities of monomeric xanthone derivatives from the marine algicolous fungus Monodictys putredinis. J. Nat. Prod. 2007, 70, 353–360. [Google Scholar] [CrossRef] [PubMed]
- Geris, R.; Simpson, T.J. Meroterpenoids produced by fungi. Nat. Prod. Rep. 2009, 26, 1063–1094. [Google Scholar] [CrossRef] [PubMed]
- Shirai, M.; Okuda, M.; Motohashi, K.; Imoto, M.; Furihata, K.; Matsuo, Y.; Katsuta, A.; Shizuri, Y.; Seto, H. Terpenoids produced by actinomycetes: Isolation, structural elucidation and biosynthesis of new diterpenes, gifhornenolones A and B from Verrucosispora gifhornensis YM28-088. J. Antibiot. 2010, 63, 245–250. [Google Scholar] [CrossRef] [PubMed]
- Caboche, S.; Pupin, M.; Leclère, V.; Fontaine, A.; Jacques, P. NORINE: A database of nonribosomal peptides. Nucleic Acids Res. 2008, 36, D326–D331. [Google Scholar] [CrossRef] [PubMed]
- Chun, H.G.; Davies, B.; Hoth, D.; Suffness, M.; Plowman, J.; Flora, K.; Grieshaber, C.; Leyland-Jones, B. Didemnin B: The first marine compound entering clinical trials as an antineoplastic agent. Investig. New Drugs 1986, 4, 279–284. [Google Scholar] [CrossRef]
- Ireland, C.M.; Durso, A.R.; Newman, R.A.; Hacker, M.P. Antineoplastic cyclic peptides from the marine tunicate Lissoclinum patella. J. Org. Chem. 1982, 47, 1807–1811. [Google Scholar] [CrossRef]
- Williams, A.B.; Jacobs, R.S. A marine natural product, patellamide D, reverses multidrug resistance in a human leukemic cell line. Cancer Lett. 1993, 71, 97–102. [Google Scholar] [CrossRef]
- Mcdonald, L.A.; Ireland, C.M. Patellamide E: A new cyclic peptide from the ascidian Lissoclinum patella. J. Nat. Prod. 1992, 55, 376–379. [Google Scholar] [CrossRef] [PubMed]
- Rashid, M.A.; Gustafson, K.R.; Cardellina, J.H.I.; Boyd, M.R. Patellamide F, a new cytotoxic cyclic peptide from the colonial ascidian Lissoclinum patella. J. Nat. Prod. 1995, 58, 594–597. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, T.; Takagi, M.; Shin-Ya, K. Three new depsipeptides, JBIR-113, JBIR-114 and JBIR-115, isolated from a marine sponge-derived Penicillium sp. fS36. J. Antibiot. 2011, 65, 147–150. [Google Scholar] [CrossRef] [PubMed]
- Zotchev, S.B. Alkaloids from marine bacteria. Adv. Bot. Res. 2013, 68, 301–333. [Google Scholar] [CrossRef]
- Powell, R.G.; Smith, C.R., Jr.; Weisleder, D.; Matsumoto, G.; Clardy, J.; Kozlowski, J. Sesbanimide, a potent antitumor substance from Sesbania drummondii seed. J. Am. Chem. Soc. 1983, 105, 3739–3741. [Google Scholar] [CrossRef]
- Powell, R.G.; Smith, C.R., Jr.; Weisleder, D. Sesbanimide A and related tumor inhibitors from Sesbania drummondii: Structure and chemistry. Phytochemistry 1984, 23, 2789–2796. [Google Scholar] [CrossRef]
- Cirillo, P.F.; Panek, J.S. Studies directed toward the synthesis of (+)-Sesbanimide A: Construction of the AB-ring system (a formal total synthesis). J. Org. Chem. 1994, 59, 3055–3063. [Google Scholar] [CrossRef]
- Atmaca, H.; Bozkurt, E. Trabectedin (ET-743) from Marine Tunicate for Cancer Treatment. In Handbook of Anticancer Drugs from Marine Origin; Springer: Berlin, Germany, 2015; pp. 397–412. [Google Scholar]
- Brodowicz, T. Trabectedin in soft tissue sarcomas. Future Oncol. 2014, 10, s1–s5. [Google Scholar] [CrossRef] [PubMed]
- Tamaoki, T.; Nomoto, H.; Takahashi, I.; Kato, Y.; Morimoto, M.; Tomita, F. Staurosporine, a potent inhibitor of phospholipid/Ca++ dependent protein kinase. Biochem. Biophys. Res. Commun. 1986, 135, 397–402. [Google Scholar] [CrossRef]
- Schupp, P.; Eder, C.; Proksch, P.; Wray, V.; Schneider, B.; Herderich, M.; Paul, V. Staurosporine derivatives from the ascidian Eudistoma toealensis and its predatory flatworm Pseudoceros sp. J. Nat. Prod. 1999, 62, 959–962. [Google Scholar] [CrossRef] [PubMed]
- Hayakawa, Y.; Shirasaki, S.; Shiba, S.; Kawasaki, T.; Mtsuo, Y.; Adachi, K.; Shizuri, Y. Piericidins C7 and C8, new cytotoxic antibiotics produced by a marine Streptomyces sp. J. Antibiot. 2007, 60, 196–200. [Google Scholar] [CrossRef] [PubMed]
- Mahyudin, N.A.; Blunt, J.W.; Cole, A.L.J.; Munro, M.H.G. The isolation of a new S-methyl benzothioate compound from a marine-derived Streptomyces sp. J. Biomed. Biotechnol. 2012, 2012, 894708. [Google Scholar] [CrossRef] [PubMed]
- Anahit, P.; Staffan, K.; Suhelen, E. Development of novel drugs from mrine surface associated microorganisms. Mar. Drugs 2010, 8, 438–459. [Google Scholar] [CrossRef]
- Schmidt, E.W.; Donia, M.S.; McIntosh, J.A.; Fricke, W.F.; Ravel, J. Origin and variation of tunicate secondary metabolites. J. Nat. Prod. 2012, 75, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, J.; Marchesi, J.R.; Dobson, A.D. Marine metagenomics: Strategies for the discovery of novel enzymes with biotechnological applications from marine environments. Microb. Cell Fact. 2008, 7, 27. [Google Scholar] [CrossRef] [PubMed]
- Zengler, K.; Walcher, M.; Clark, G.; Haller, I.; Toledo, G.; Holland, T.; Mathur, E.J.; Woodnutt, G.; Short, J.M.; Keller, M. High-throughput cultivation of microorganisms using microcapsules. Methods Enzymol. 2005, 397, 124–130. [Google Scholar] [CrossRef] [PubMed]
- Ben-Dov, E.; Kramarsky-Winter, E.; Kushmaro, A. An in situ method for cultivating microorganisms using a double encapsulation technique. FEMS Microbiol. Ecol. 2009, 68, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Steinert, G.; Whitfield, S.; Taylor, M.W.; Thoms, C.; Schupp, P.J. Application of diffusion growth chambers for the cultivation of marine sponge-associated bacteria. Mar. Biotechnol. 2014, 16, 594–603. [Google Scholar] [CrossRef] [PubMed]
- Palanisamy, S.K.; Rajendran, N.M.; Marino, A. Natural products diversity of marine ascidians (Tunicates; Ascidiacea) and successful drugs in clinical development. Nat. Prod. Bioprospect. 2017, 7, 1–111. [Google Scholar] [CrossRef] [PubMed]
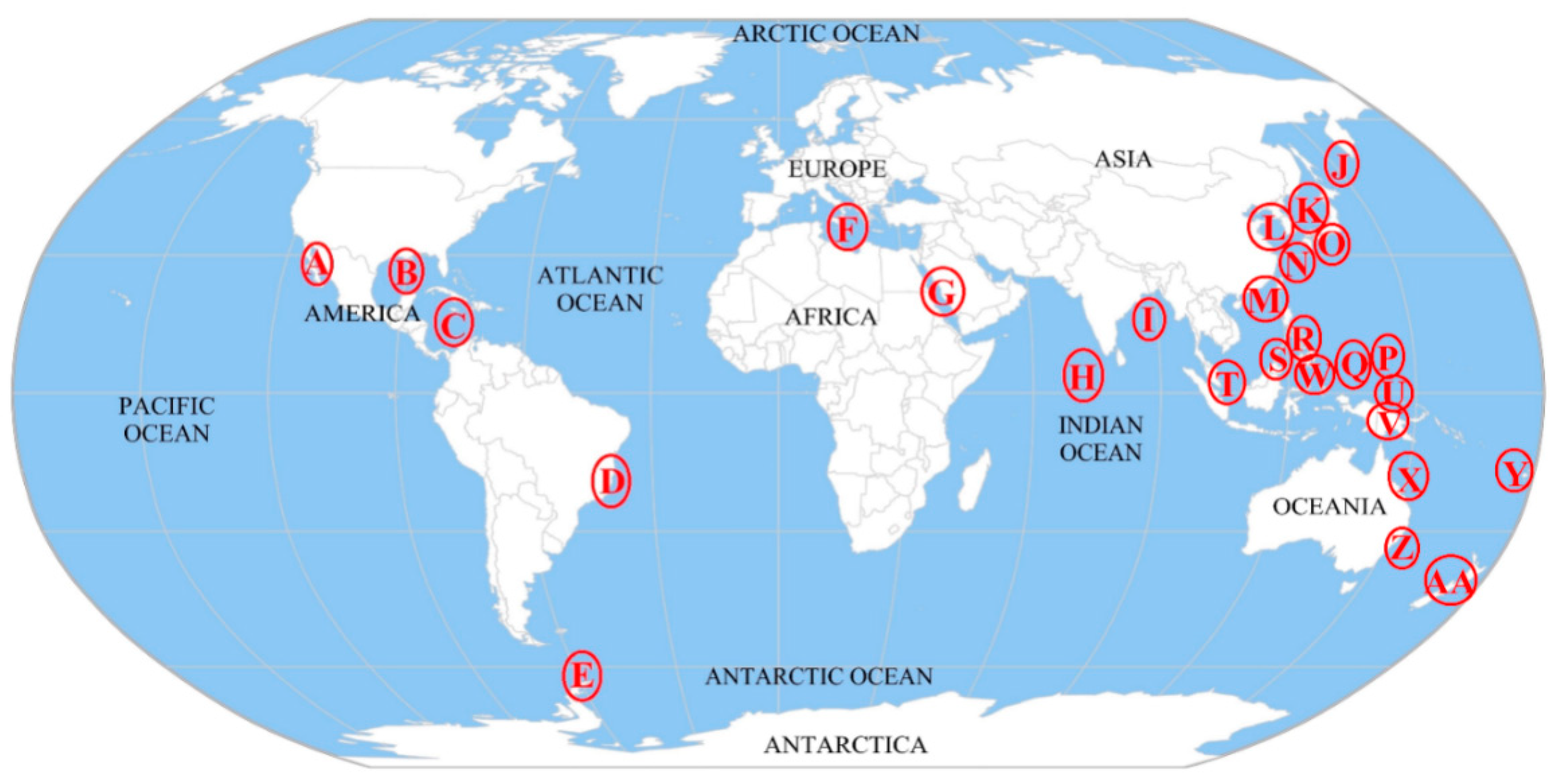
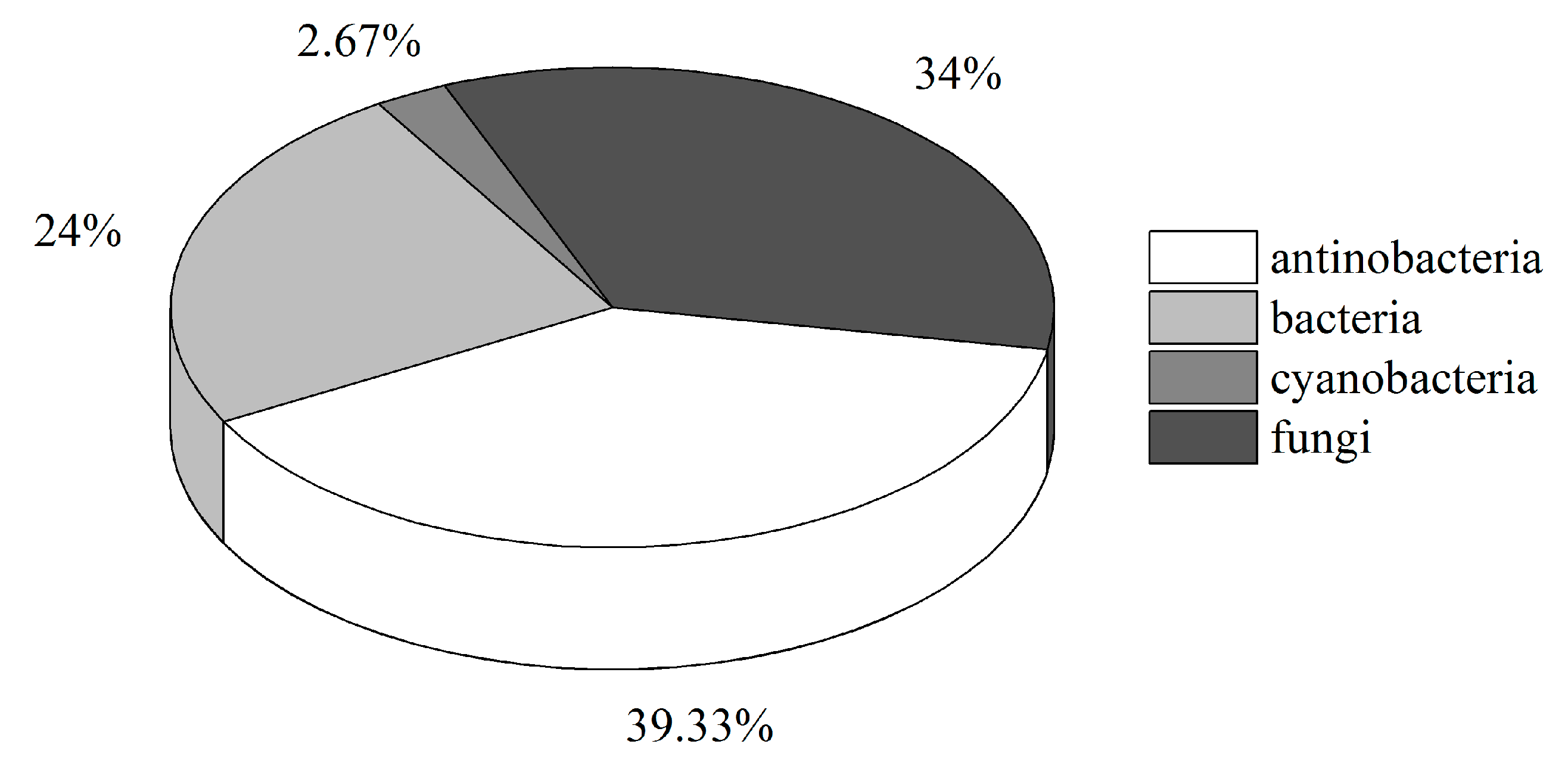
| Microorganism | Host Ascidian | Geographical Location | Reference |
|---|---|---|---|
| Bacteria | |||
| Acinetobacter sp. | Stomozoa murrayi | AO: Yucatan Peninsula, Mexico | [12] |
| Didemnum ligulum | AO: São Paulo, Brazil | [30] | |
| Agrobacterium sp. | Ecteinascidia turbinata | AO: mangroves of the Florida peninsula, US | [31] |
| Polycitonidae sp. | AO: Turkish coast | [31] | |
| Bacillus pumilus | Halocynthia aurantium | PO: Sea of Japan | [32,33] |
| Bacillus sp. | Didemnum sp. | AO: São Paulo, Brazil | [30] |
| Didemnum ligulum | AO: São Paulo, Brazil | [30] | |
| Candidatus Endoecteinascidia frumentensis | Ecteinascidia turbinate | AO: Florida Keys | [16] |
| Candidatus Endolissoclinum faulkneri | Lissoclinum patella | PO: Papua New Guinea, Solomon Islands and Fiji | [34] |
| Endozoicomonas sp. | Didemnum sp. | AO: São Paulo, Brazil | [30] |
| Exiguobacterium sp. | Didemnum ligulum | AO: São Paulo, Brazil | [30] |
| Halomonas halocynthiae | Halocynthia aurantium | PO: Sea of Japan | [35] |
| Hasllibacter halocynthiae | Halocynthia roretzi | PO: the coast of Gangneung, Korea | [36,37] |
| Labilibacter aurantiacus | Styela clava | PO: the Yellow Sea, China | [38] |
| Paenibacillus sp. | Didemnum ligulum | AO: São Paulo, Brazil | [30] |
| Paucisalibacillus sp. | Didemnum ligulum | AO: São Paulo, Brazil | [30] |
| Pseudomonas stutzeri | Didemnum sp. | IO: Maldives | [39] |
| Pseudomonas xanthomarina | Halocynthia aurantium | PO: Troitsa Bay, Peter the Great Bay, the Sea of Japan, Russia | [39] |
| Pseudovibrio sp. | Lissoclinum patella | AO: São Paulo, Brazil | [30] |
| Rubritalea halochordaticola | Unidentified | PO: Himezu Port, Sado Island, Niigata Prefecture, Japan | [40] |
| Ruegeria halocynthiae | Halocynthia roretzi | PO: the South Sea, Korea | [41] |
| Ruegeria sp. | Didemnum sp. | AO: São Paulo, Brazil | [30] |
| Didemnum ligulum | AO: São Paulo, Brazil | [30] | |
| Staphylococus sp. | Didemnum ligulum | AO: São Paulo, Brazil | [30] |
| Stappia sp. | Didemnum sp. | AO: São Paulo, Brazil | [30] |
| Didemnum ligulum | AO: São Paulo, Brazil | [30] | |
| Tenacibaculum halocynthiae | Halocynthia roretzi | PO: the South Sea, Korea | [42] |
| Tistrella mobilis | Trididemnum solidum | PO: Tateyama cove, Chiba, Japan | [18] |
| IO: the Red Sea | [19] | ||
| Vibrio sp. | Polyclinum glabrum | IO: Tuticorin coast | [43] |
| Didemnum sp. | AO: São Paulo, Brazil | [30] | |
| Unidentified | Unidentified | PO: Fiji | [44] |
| Didemnum sp. | AO: São Paulo, Brazil | [30] | |
| Didemnum ligulum | AO: São Paulo, Brazil | [30] | |
| Ciona intestinalis | Not mentioned | [45] | |
| Actinobacteria | |||
| Actinomadura sp. | Ecteinascidia turbinata | AO: Florida Keys | [46] |
| Ecteinascidia turbinata | Not mentioned | [47] | |
| Aeromicrobium halocynthiae | Halocynthia roretzi | PO: the coast of Gangneung, Korea | [48] |
| Arthrobacter sp. | Didemnum ligulum | AO: São Paulo, Brazil | [30] |
| Brevibacterium sp. | Didemnum ligulum | AO: São Paulo, Brazil | [30] |
| Curtobacterium sp. | Didemnum sp. | AO: São Paulo, Brazil | [30] |
| Didemnum ligulum | AO: São Paulo, Brazil | [30] | |
| Gordonia didemni | Didemnum sp. | AO: São Paulo, Brazil | [49] |
| Gordonia sp. | Didemnum sp. | AO: São Paulo, Brazil | [30] |
| Kocuria sp. | Didemnum ligulum | AO: São Paulo, Brazil | [30] |
| Micrococcus sp. | Didemnum sp. | AO: São Paulo, Brazil | [30] |
| Didemnum ligulum | AO: São Paulo, Brazil | [30] | |
| Micromonospora spp. | Eudistoma vannamei | AO: Taiba Beach northeastern coast of Brazi | [50] |
| Nocardia sp. | Trididemnum orbiculatum | AO: Florida Keys | [51] |
| Unidentified | PO: Simushir Island, Kuril Islands | [52] | |
| Didemnum ligulum | AO: São Paulo, Brazil | [30] | |
| Nocardiopsis dassonvillei | Botryllus schlosseri | PO: the Yellow Sea, China | [53] |
| Saccharopolyspora sp. | Unidentified | PO: Tateyama City, Chiba Prefecture, Japan | [54] |
| Salinispora arenicola | Ecteinascidia turbinata | AO: Sweetings Cay, Grand Bahama Island | [14] |
| Salinispora pacifica | Polysyncraton lithostrotum | Not mentioned | [55,56] |
| Salinispora sp. | Eudistoma toealensis | PO: Islands of Chuuk and Pohnpei, Micronesia | [57] |
| Solwaraspora sp. | Trididemnum orbiculatum | AO: Florida Keys | [58] |
| Streptomyces hyaluromycini | Molgula manhattensis | PO: Tokyo Bay, Japan | [59] |
| Streptomyces sp. | Aplidium lenticulum | PO: Heron Island, Queensland, Australia | [60] |
| Aplidium lenticulum | PO: Great Barrier Reef, Australia | [61] | |
| Didemnum sp. | IO: Obhur, Saudi Arabia | [62] | |
| Ecteinascidia turbinata | AO: La Parguera, Puerto Rico | [13] | |
| Styela clava | PO: the Yellow Sea, China | [53] | |
| Styela canopus | AO: the Bastimentos National Park in Bocas del Toro, Panama | [63] | |
| Verrucosispora sp. | Eudistoma toealensis | PO: Islands of Chuuk and Pohnpei, Micronesia | [56] |
| Cyanobacteria | |||
| Prochloron didemni | Lissoclinum patella | PO: Palau | [64] |
| Prochloron sp. | Didemnum etiolum | PO: nothren Great Barrier Reef and Philippine | [27,65] |
| Didemnum molle | PO: Fiji, Philippine, Palau Island, Lizard Island, northern Great Barrier Reef, Guam and Caroline Islands | [27,65] | |
| Diplosoma multipapillata | PO: Fiji | [27,65] | |
| Diplosoma similis | PO: Caroline Islands, Philippine, Palau, Guam, Norhern Great Barrier Reef and Singapore | [65] | |
| Diplosoma virens | PO: Caroline Islands, Philippine, Palau and Norhern Great Barrier Reef | [65] | |
| Echinoclinum triangulum | PO: Philippine | [27,65] | |
| Lissoclinum patella | PO: Davies Reef, Great Barrier Reef, Australia | [66] | |
| Lissoclinum patella | PO: Philippine, Palau and Guam | [27,65] | |
| Lissoclinum punctatum | PO: Palau and Singapore | ||
| Lissoclinum voeltzkowi | PO: Caroline Islands, Philippine, Palau and Guam | [65] | |
| Trididemnum clinides | PO: Philippine and Guam | [27,65] | |
| Trididemnum cyclops | PO: Palau and Caroline Islands | [65] | |
| Trididemnum miniatum | PO: Norhern Great Barrier Reef | [27,65] | |
| Trididemnum nubilum | PO: Philippine, Fiji and Great Barrier Reef | [27,65] | |
| Trididemnum paraclinides | PO: Palau | [27,65] | |
| Trididemnum paracyclops | PO: Palau, Philippine and Guam | [65] | |
| Trididemnum strigosum | PO: Philippine | [27,65] | |
| Prochloron spp. | Diplosoma simile | PO: Crawl Key; Isla Cristobal | [67] |
| Lissoclinum patella | PO: Palau; Palau New Guinea | [68] | |
| Lissoclinum verrilli | PO: Isla Cristobal | [67] | |
| Synechocystis didemin | Didemnum spp. | PO: Baja, California, Mexico | [69] |
| Synechocystis sp. | Didemnum viride | PO: Philippine and Palau | [27,70] |
| Trididemnum cyanophorum | PO: Panama and Guadaloupe | [27,70] | |
| Trididemnum solidum | AO: Galeta, Panama | [27] | |
| Unidentified | Trididemnum clinides | PO: Okinawajima Island, Ryukyu, Archipelago, Japan | [71] |
| Fungi | |||
| Acremonium sp. | Ecteinascidia turbinata | AO: Bahamas | [72] |
| Alternaria sp. | Cystodytes dellechiajei | AO: Mediterranean Sea | [29] |
| Didemnum sp. | AO: São Paulo, Brazil | [30] | |
| Aspergillus candidus | Unidentified | Not mentioned | [73] |
| Aspergillus fumigatus | Pycnoclavella communis | AO: Mediterranean Sea | [29] |
| Aspergillus niger | Aplidium sp. | PO: Caesar’s Rock in Benga, Fiji | [74] |
| Aspergillus sp. | Cystodytes dellechiajei | AO: Mediterranean Sea | [29] |
| Didemnum sp. | AO: São Paulo, Brazil | [30] | |
| Eudistoma vannamei | AO: Northeast Brazil | [75] | |
| Bionectria sp. | Didemnum sp. | AO: São Paulo, Brazil | [30] |
| Pycnoclavella communis | AO: Mediterranean Sea | [29] | |
| Botryosphaeria sp. | Didemnum sp. | AO: São Paulo, Brazil | [30] |
| Botrytis cinerea | Cystodytes dellechiajei | AO: Mediterranean Sea | [29] |
| Cladosporium sp. | Cystodytes dellechiajei | AO: Mediterranean Sea | [29] |
| Didemnum fulgens | AO: Mediterranean Sea | [29] | |
| Didemnum sp. | AO: São Paulo, Brazil | [30] | |
| Pycnoclavella communis | AO: Mediterranean Sea | [29] | |
| Clonostachys sp. | Didemnum fulgens | AO: Mediterranean Sea | [29] |
| Cochliobolus sp. | Didemnum sp. | AO: São Paulo, Brazil | [30] |
| Cunninghamella sp. | Didemnum sp. | AO: São Paulo, Brazil | [30] |
| Epicoccum nigrum | Cystodytes dellechiajei | AO: Mediterranean Sea | [29] |
| Fusarium sp. | Cystodytes dellechiajei | AO: Mediterranean Sea | [29] |
| Didemnum sp. | AO: São Paulo, Brazil | [30] | |
| Humicola fuscoatra | Unidentified | PO: Shikotan island, the Kuril isles | [76] |
| Meyerozyma sp. | Ciona intestinalis | PO: the Yellow Sea, China | [77] |
| Microdiplodia sp. | Didemnum fulgens | AO: L’Escala, Spain ‘La Depuradora’, Mediterranean Sea | [29] |
| Mucor sp. | Didemnum sp. | AO: São Paulo, Brazil | [30] |
| Penicillium brevicompactum | Cystodytes dellechiajei | AO: Mediterranean Sea | [29] |
| Didemnum fulgens | AO: Mediterranean Sea | [29] | |
| Penicillium rubens | Didemnum fulgens | AO: Mediterranean Sea | [29] |
| Penicillium steckii | Unidentified | AO: Mochima Bay, Mochima National Park and Paria Bay, Irapa, Venezuela | [78] |
| Penicillium stoloniferum | Unidentified | PO: Jiaozhou Bay, Qingdao, China | [79] |
| Penicillium sp. | Cystodytes dellechiajei | AO: Mediterranean Sea | [29] |
| Didemnum fulgens | AO: Mediterranean Sea | [29] | |
| Didemnum molle | PO: Ishigaki Island, Okinawa Prefecture, Japan | [80] | |
| Didemnum sp. | AO: São Paulo, Brazil | [30] | |
| Pycnoclavella communis | AO: Mediterranean Sea | [29] | |
| Pestalotiopsis sp. | Didemnum sp. | AO: São Paulo, Brazil | [30] |
| Phoma sp. | Cystodytes dellechiajei | AO: Mediterranean Sea | [29] |
| Pycnoclavella communis | AO: Mediterranean Sea | [29] | |
| Didemnum sp. | AO: São Paulo, Brazil | [30] | |
| Pithomyces sp. | Oxycorynia fascicularis | IO and PO | [81] |
| Plectosphaerella sp. | Cystodytes dellechiajei | AO: Mediterranean Sea | [29] |
| Rhizopus sp. | Didemnum sp. | AO: São Paulo, Brazil | [30] |
| Scopulariopsis sp. | Didemnum fulgens | AO: Mediterranean Sea | [29] |
| Talaromyces albobiverticillius (basionym: Penicillium albobiverticillium) | Unidentified | PO: Manado, Indonesia | [82,83] |
| Talaromyces verruculosus (basionym: Penicillium verruculosum) | Polycarpa aurata | PO: Manado, Indonesia | [83,84] |
| Talaromyces sp. | Pycnoclavella communis | AO: Mediterranean Sea | [29] |
| Unidentified | PO: Tweed Heads, NSW, Australia | [85] | |
| Trichoderma harzianum | Pycnoclavella communis | AO: Mediterranean Sea | [29] |
| Trichoderma virens | Didemnum molle | PO: Madang, Papua New Guinea | [86] |
| Trichoderma sp. | Didemnum fulgens | AO: Mediterranean Sea | [29] |
| Didemnum sp. | AO: São Paulo, Brazil | [30] | |
| Unidentified | Didemnum sp. | AO: São Paulo, Brazil | [30] |
| Unidentified (A fungus in the class Eurotiomycetes) | Lissoclinum patella | PO: Papua New Guinea | [87] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, L.; Hu, J.-S.; Xu, J.-L.; Shao, C.-L.; Wang, G.-Y. Biological and Chemical Diversity of Ascidian-Associated Microorganisms. Mar. Drugs 2018, 16, 362. https://doi.org/10.3390/md16100362
Chen L, Hu J-S, Xu J-L, Shao C-L, Wang G-Y. Biological and Chemical Diversity of Ascidian-Associated Microorganisms. Marine Drugs. 2018; 16(10):362. https://doi.org/10.3390/md16100362
Chicago/Turabian StyleChen, Lei, Jin-Shuang Hu, Jia-Lei Xu, Chang-Lun Shao, and Guang-Yu Wang. 2018. "Biological and Chemical Diversity of Ascidian-Associated Microorganisms" Marine Drugs 16, no. 10: 362. https://doi.org/10.3390/md16100362
APA StyleChen, L., Hu, J.-S., Xu, J.-L., Shao, C.-L., & Wang, G.-Y. (2018). Biological and Chemical Diversity of Ascidian-Associated Microorganisms. Marine Drugs, 16(10), 362. https://doi.org/10.3390/md16100362