Role of Mitochondria in Regulating Lutein and Chlorophyll Biosynthesis in Chlorella pyrenoidosa under Heterotrophic Conditions
Abstract
1. Introduction
2. Results
2.1. Variation in Lutein and Chlorophyll Biosynthesis
2.2. Analysis of Redox and Energy States
2.3. SDS-PAGE of Total Proteins
2.4. 2-DE Analysis and Identification of Differentially Expressed Proteins
2.5. Functional Categorization
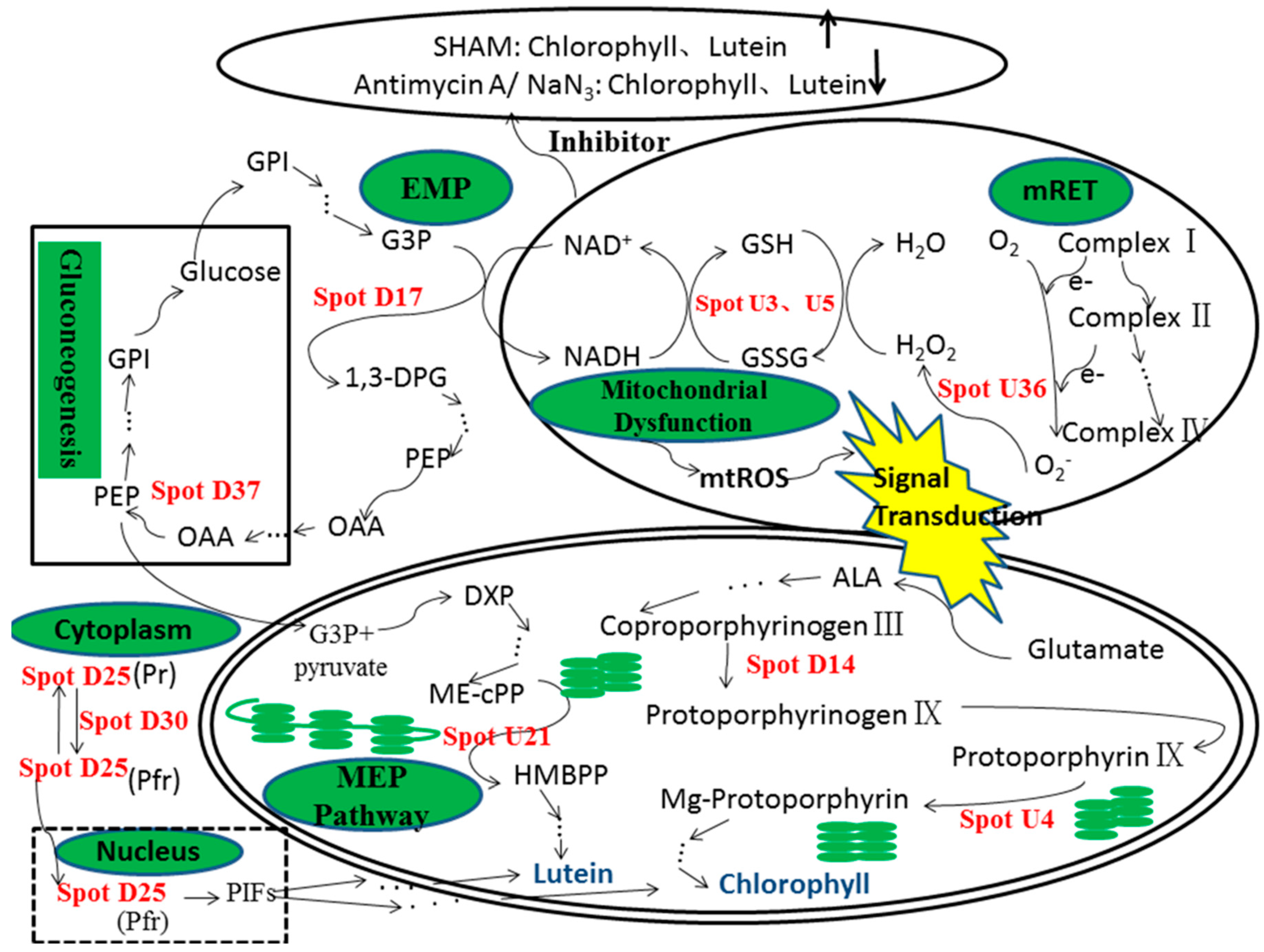
3. Discussion
3.1. Up-Regulation of Antioxidant Proteins
3.2. Variation in Chloroplast Protein Expression Following Mitochondrial Dysfunction
3.3. Variation in Transcription and Protein Fate-Related Protein Expression
3.4. Variation in Metabolism and Energy-Related Protein Expression
4. Materials and Methods
4.1. Microalga and Culture Conditions
4.2. Mitochondrial Dysfunction
4.3. Pigment Analysis
4.4. Redox and Energy State Evaluation
4.5. Protein Extraction
4.6. One-Dimensional SDS Gel Electrophoresis and 2-DE
4.7. Gel Silver-Staining and Image Analysis
4.8. In-Gel Trypsin Digestion, Mass Spectrometry, and Protein Identification
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Chen, F. High cell density culture of microalgae in heterotrophic growth. Trends Biotechnol. 1996, 14, 421–426. [Google Scholar] [CrossRef]
- Lee, Y.K. Commercial production of microalgae in the Asia-pacific rim. J. Appl. Phycol. 1997, 9, 403–411. [Google Scholar] [CrossRef]
- Qian, F.H. Study on the Optimization and Primary Scale-up of Chlorella Sequential Heterotrophic/Autotrophic Cultivation Technology for High-Density and High-Quality. Master’s Thesis, East China University of Science and Technology, Shanghai, China, 2007. [Google Scholar]
- Shi, X.M.; Chen, F.; Yuan, J.P.; Chen, H. Heterotrophic production of lutein by selected Chlorella strains. J. Appl. Phycol. 1997, 5, 445–450. [Google Scholar] [CrossRef]
- Wu, Z.Y.; Qu, C.B.; Shi, X.M. Biochemical system analysis of lutein production by heterotrophic Chlorella pyrenoidosa in a fermentor. Food Technol. Biotechnol. 2009, 47, 450–455. [Google Scholar]
- Johnson, E.J. Role of lutein and zeaxanthin in visual and cognitive function throughout the lifespan. Nutr. Rev. 2015, 72, 605–612. [Google Scholar] [CrossRef] [PubMed]
- Le, M.; Lin, X.M. Effects of lutein and zeaxanthin on aspects of eye health. J. Sci. Food Agric. 2010, 90, 2–12. [Google Scholar]
- Sangiovanni, J.P.; Neuringer, M. The putative role of lutein and zeaxanthin as protective agents against age-related macular degeneration: Promise of molecular genetics for guiding mechanistic and translational research in the field. Am. J. Clin. Nutr. 2012, 96, 1223S–1233S. [Google Scholar] [CrossRef] [PubMed]
- Mewborn, C.M.; Lindbergh, C.A.; Robinson, T.L.; Gogniat, M.A.; Terry, D.P.; Jean, K.R.; Hammond, B.R.; Renzi-Hammond, L.M.; Miller, L.S. Lutein and Zeaxanthin Are Positively Associated with Visual-Spatial Functioning in Older Adults: An fMRI Study. Nutrients 2018, 10, 458. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, M.; Yu, J. Mitochondrial retrograde regulation tuning fork in nuclear genes expressions of higher plants. J. Genet. Genom. 2008, 35, 65–71. [Google Scholar] [CrossRef]
- Matsuo, M.; Obokata, J. Remote control of photosynthetic genes by the mitochondrial respiratory chain. Plant J. 2006, 47, 873–882. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Huang, J.; Sandmann, G.; Chen, F. Glucose sensing and the mitochondrial alternative pathway are involved in the regulation of astaxanthin biosynthesis in the dark-grown Chlorella zofingiensis (chlorophyceae). Planta 2008, 228, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Padmasree, K.; Raghavendra, A. Response of photosynthetic carbon assimilation in mesophyll protoplasts to restriction on mitochondrial oxidative metabolism: Metabolites related to the redox status and sucrose biosynthesis. Photosynth. Res. 1999, 62, 231–239. [Google Scholar] [CrossRef]
- Saisho, D.; Nakazono, M.; Tsutsumi, N.; Hirai, A. ATP synthesis inhibitors as well as respiratory inhibitors increase steady-state level of alternative oxidase mRNA in Arabidopsis thaliana. J. Plant Physiol. 2001, 158, 241–245. [Google Scholar] [CrossRef]
- Li, N. Mitochondrial Complex I Inhibitor Rotenone Induces Apoptosis through Enhancing Mitochondrial Reactive Oxygen Species Production. J. Biol. Chem. 2002, 278, 8516–8525. [Google Scholar] [CrossRef] [PubMed]
- Rhoads, D.M.; Vanlerberghe, G.C. Plant Mitochondria: From Genome to Function; Springer Dordrecht: Dordrecht, The Netherlands, 2004; pp. 83–106. ISBN 978-1-4020-2400-9. [Google Scholar]
- Tang, X.Q.; Luo, Y.X.; Chen, H.Z.; Liu, D.P. Mitochondria, endothelial cell function, and vascular diseases. Front. Physiol. 2014, 5, 175. [Google Scholar] [CrossRef] [PubMed]
- Maxwell, D.P.; Nickels, R.; McIntosh, L. Evidence of mitochondrial involvement in the transduction of signals required for the induction of genes associated with pathogen attack and senescence. Plant J. 2002, 29, 269–279. [Google Scholar] [CrossRef] [PubMed]
- Popov, V.; Simonian, R.; Skulachev, V.; Starkov, A. Inhibition of the alternative oxidase stimulates H2O2 production in plant mitochondria. FEBS Lett. 1997, 415, 87–90. [Google Scholar] [CrossRef]
- Umbach, A.L.; Fiorani, F.; Siedow, J.N. Characterization of transformed Arabidopsis with altered alternative oxidase levels and analysis of effects on reactive oxygen species in tissue. Plant Physiol. 2005, 139, 1806–1820. [Google Scholar] [CrossRef] [PubMed]
- Claiborne, A.; Yeh, J.I.; Mallett, T.C.; Luba, J.; Crane, E.J.; Charrier, V.; Parsonage, D. Protein-sulfenic acids: Diverse roles for an unlikely player in enzyme catalysis and redox regulation. Biochemistry 1999, 38, 15407–15416. [Google Scholar] [CrossRef] [PubMed]
- Grene, R. The Arabidopsis Book; American Society of Plant Biologists: Rockville, MD, USA, 2002. [Google Scholar]
- Valko, M.; Leibfritz, D.; Moncol, J.; Cronin, M.T.; Mazur, M.; Telser, J. Free radicals and antioxidants in normal physiological functions and human disease. Int. J. Biochem. Cell Biol. 2007, 39, 44–84. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Noctor, G. Redox homeostasis and antioxidant signaling: A metabolic interface between stress perception and physiological responses. Plant Cell Online 2005, 17, 1866–1875. [Google Scholar] [CrossRef] [PubMed]
- Butow, R.A.; Avadhani, N.G. Mitochondrial signaling: The retrograde response. Mol. Cell 2004, 14, 1–15. [Google Scholar] [CrossRef]
- Yao, N.; Tada, Y.; Sakamoto, M.; Nakayashiki, H.; Park, P.; Tosa, Y.; Mayama, S. Mitochondrial oxidative burst involved in apoptotic response in oats. Plant J. 2002, 30, 567–579. [Google Scholar] [CrossRef] [PubMed]
- Ho, H.Y.; Lin, Y.T.; Lin, G.; Wu, P.R.; Cheng, M.L. Nicotinamide nucleotide transhydrogenase (NNT) deficiency dysregulates mitochondrial retrograde signaling and impedes proliferation. Redox Biol. 2017, 12, 916–928. [Google Scholar] [CrossRef] [PubMed]
- Gray, G.; Maxwell, D.; Villarimo, A.; McIntosh, L. Mitochondria/nuclear signaling of alternative oxidase gene expression occurs through distinct pathways involving organic acids and reactive oxygen species. Plant Cell Rep. 2004, 23, 497–503. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, K.; Beligni, M.V.; Prieto, S.; Haynes, P.A.; McDonald, W.H.; Yates, J.R.; Mayfield, S.P. Proteomic Characterization of the Chlamydomonas reinhardtii Chloroplast Ribosome identification of proteins unique to the 70s ribosome. J. Biol. Chem. 2003, 278, 33774–33785. [Google Scholar] [CrossRef] [PubMed]
- Rand, K.; Noll, C.; Schiebel, H.M.; Kemken, D.; Dülcks, T.; Kalesse, M.; Heinz, D.W.; Layer, G. The oxygen-independent coproporphyrinogen III oxidase HemN utilizes harderoporphyrinogen as a reaction intermediate during conversion of coproporphyrinogen III to protoporphyrinogen IX. Biol. Chem. 2010, 391, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Surpin, M.; Larkin, R.M.; Chory, J. Signal transduction between the chloroplast and the nucleus. Plant Cell Online 2002, 14, S327–S338. [Google Scholar] [CrossRef]
- Johanningmeier, U.; Howell, S.H. Regulation of light-harvesting chlorophyll-binding protein mRNA accumulation in Chlamydomonas reinhardi. Possible involvement of chlorophyll synthesis precursors. J. Biol. Chem. 1984, 259, 13541–13549. [Google Scholar] [PubMed]
- Oster, U.; Brunner, H.; Rüdiger, W. The greening process in cress seedlings. V. Possible interference of chlorophyll precursors, accumulated after thujaplicin treatment, with light-regulated expression of Lhc genes. J. Photochem. Photobiol. Biol. 1996, 36, 255–261. [Google Scholar] [CrossRef]
- Vinti, G.; Hills, A.; Campbell, S.; Bowyer, J.R.; Mochizuki, N.; Chory, J.; López-Juez, E. Interactions between hy1 and gun mutants of Arabidopsis, and their implications for plastid/nuclear signalling. Plant J. 2000, 24, 883–894. [Google Scholar] [CrossRef] [PubMed]
- Zavgorodnyaya, A.; Papenbrock, J.; Grimm, B. Yeast 5-aminolevulinate synthase provides additional chlorophyll precursor in transgenic tobacco. Plant J. 1997, 12, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Mochizuki, N.; Brusslan, J.A.; Larkin, R.; Nagatani, A.; Chory, J. Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc. Natl. Acad. Sci. USA 2001, 98, 2053–2058. [Google Scholar] [CrossRef] [PubMed]
- Walker, C.J.; Willows, R.D. Mechanism and regulation of Mg-chelatase. Biochem. J. 1997, 327, 321–333. [Google Scholar] [CrossRef] [PubMed]
- Kropat, J.; Oster, U.; Rüdiger, W.; Beck, C.F. Chloroplast signalling in the light induction of nuclear HSP70 genes requires the accumulation of chlorophyll precursors and their accessibility to cytoplasm/nucleus. Plant J. 2000, 24, 523–531. [Google Scholar] [CrossRef] [PubMed]
- La, R.N.; Rascio, N.; Oster, U.; Rüdiger, W. Amitrole treatment of etiolated barley seedlings leads to deregulation of tetrapyrrole synthesis and to reduced expression of Lhc and RbcS genes. Planta 2001, 213, 101–108. [Google Scholar]
- Moller, S.G.; Kunkel, T.; Chua, N.H. A plastidic ABC protein involved in intercompartmental communication of light signaling. Sci. Signal. 2001, 15, 90–103. [Google Scholar] [CrossRef]
- Estevez, J.M.; Cantero, A.; Reindl, A.; Reichler, S.; Leon, P. 1-deoxy-d-xylulose-5-phosphate synthase, a limiting enzyme for plastidic isoprenoid biosynthesis in plants. J. Biol. Chem. 2001, 276, 22901–22909. [Google Scholar] [CrossRef] [PubMed]
- Morris, W.L.; Ducreux, L.J.; Hedden, P.; Millam, S.; Taylor, M.A. Overexpression of a bacterial 1-deoxy-d-xylulose 5-phosphate synthase gene in potato tubers perturbs the isoprenoid metabolic network: Implications for the control of the tuber life cycle. J. Exp. Bot. 2006, 57, 3007–3018. [Google Scholar] [CrossRef] [PubMed]
- Reed, J.W. Phytochromes are Pr-ipatetic kinases. Curr. Opin. Plant Biol. 1999, 2, 393–397. [Google Scholar] [CrossRef]
- Kuno, N.; Furuya, M. Phytochrome regulation of nuclear gene expression in plants. Semin. Cell Dev. Biol. 2000, 11, 485–493. [Google Scholar] [CrossRef] [PubMed]
- Lintig, J.; Welsch, R.; Bonk, M.; Giuliano, G.; Batschauer, A.; Kleinig, H. Light-dependent regulation of carotenoid biosynthesis occurs at the level of phytoene synthase expression and is mediated by phytochrome in Sinapis alba and Arabidopsis thaliana seedlings. Plant J. 1997, 12, 625–634. [Google Scholar] [CrossRef]
- Ma, L.; Li, J.; Qu, L.; Hager, J.; Chen, Z.; Zhao, H.; Deng, X.W. Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell Online 2001, 13, 2589–2607. [Google Scholar] [CrossRef]
- Liu, R.; Xu, Y.H.; Jiang, S.C.; Lu, K.; Lu, Y.F.; Feng, X.J.; Wu, Z.; Liang, S.; Yu, Y.T.; Wang, X.F.; et al. Light-harvesting chlorophyll a/b-binding proteins, positively involved in abscisic acid signalling, require a transcription repressor, wrky40, to balance their function. J. Exp. Bot. 2013, 64, 5443–5456. [Google Scholar] [CrossRef] [PubMed]
- Toledoortiz, G.; Huq, E.; Rodríguez, C.M. Direct regulation of phytoene synthase gene expression and carotenoid biosynthesis by phytochrome-interacting factors. Proc. Natl. Acad. Sci. USA 2010, 107, 11626–11631. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Zhu, L.; Castillon, A.; Majee, M.; Downie, B.; Huq, E. Light-induced phosphorylation and degradation of the negative regulator phytochrome-interacting factor 1 from Arabidopsis depend upon its direct physical interactions with photoactivated phytochromes. Plant Cell Online 2008, 20, 1586–1602. [Google Scholar] [CrossRef] [PubMed]
- Leivar, P.; Monte, E. Pifs: Systems integrators in plant development. Plant Cell 2014, 26, 56–78. [Google Scholar] [CrossRef] [PubMed]
- Park, E.; Park, J.; Kim, J.; Nagatani, A.; Lagarias, J.C.; Choi, G. Phytochrome B inhibits binding of phytochrome-interacting factors to their target promoters. Plant J. Cell Mol. Biol. 2012, 72, 537–546. [Google Scholar] [CrossRef] [PubMed]
- Pires, N.; Dolan, L. Origin and Diversification of Basic-Helix-Loop-Helix Proteins in Plants. Mol. Biol. Evol. 2010, 27, 862–874. [Google Scholar] [CrossRef] [PubMed]
- Mummert, E.; Grimm, R.; Speth, V.; Eckerskorn, C.; Schiltz, E.; Gatenby, A.A.; Schäfer, E. ATCPl-related molecular chaperone from plants refolds phytochrome to its photoreversible form. Nature 1993, 363, 644–648. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.S.; Lagarias, J.C. Light-independent phytochrome signaling mediated by dominant GAF domain tyrosine mutants of Arabidopsis phytochromes in transgenic plants. Plant Cell Online 2007, 19, 2124–2139. [Google Scholar] [CrossRef] [PubMed]
- Mayfield, J.D.; Folta, K.M.; Paul, A.L.; Ferl, R.J. The 14-3-3 proteins µ and v influence transition to flowering and early phytochrome response. Plant Physiol. 2007, 145, 1692–1702. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, T.; Emi, T.; Tominaga, M.; Sakamoto, K.; Shigenaga, A.; Doi, M.; Shimazaki, K.I. Blue-light-and phosphorylation-dependent binding of a 14-3-3 protein to phototropins in stomatal guard cells of broad bean. Plant Physiol. 2003, 133, 1453–1463. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.P.; Adams, E.; Yanagawa, Y.; Matsui, M.; Shin, R. AtSKIP18 and AtSKIP31, F-box subunits of the SCF E3 ubiquitin ligase complex, mediate the degradation of 14-3-3 proteins in Arabidopsis. Biochem. Biophys. Res. Commun. 2017, 485, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, Y.; Zhao, H.; Huang, F.; Zhang, Z.; Lin, W. The important functionality of 14-3-3 isoforms in rice roots revealed by affinity chromatography. J. Proteom. 2017, 158, 20–30. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Zhao, X.; Guo, C.; Li, K. Spinach 14-3-3 protein interacts with the plasma membrane H(+)-ATPase and nitrate reductase in response to excess nitrate stress. Plant Physiol. Biochem. 2016, 106, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Delbaere, L.T.; Sudom, A.M.; Prasad, L.; Leduc, Y.; Goldie, H. Structure/function studies of phosphoryl transfer by phosphoenolpyruvate carboxykinase. Biochim. Biophys. Acta-Proteins Proteom. 2004, 1697, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Streatfield, S.J.; Weber, A.; Kinsman, E.A.; Häusler, R.E.; Li, J.; Post, B.D.; Kaiser, W.M.; Pyke, K.A.; Flügge, U.I.; Chory, J. The phosphoenolpyruvate/phosphate translocator is required for phenolic metabolism, palisade cell development, and plastid-dependent nuclear gene expression. Plant Cell Online 1999, 11, 1609–1621. [Google Scholar] [CrossRef]
- Cunningham, F.X. Regulation of carotenoid synthesis and accumulation in plants. Pure Appl. Chem. 2002, 74, 1409–1417. [Google Scholar] [CrossRef]
- Huang, W.; Ye, J.; Zhang, J.; Lin, Y.; He, M.; Huang, J. Transcriptome analysis of chlorella zofingiensis to identify genes and their expressions involved in astaxanthin and triacylglycerol biosynthesis. Algal Res. 2016, 17, 236–243. [Google Scholar] [CrossRef]
- Wu, X.; Jiang, L.; Yu, M.; An, X.; Ma, R.; Yu, Z. Proteomic analysis of changes in mitochondrial protein expression during peach fruit ripening and senescence. J. Proteom. 2016, 147, 197–211. [Google Scholar] [CrossRef] [PubMed]
- Baroli, I. Zeaxanthin Accumulation in the Absence of a Functional Xanthophyll Cycle Protects Chlamydomonas reinhardtii from Photooxidative Stress. Plant Cell Online 2003, 15, 992–1008. [Google Scholar] [CrossRef]
- Lazzarino, G.; Amorini, A.M.; Fazzina, G.; Vagnozzi, R.; Signoretti, S.; Donzelli, S.; Stasio, E.D.; Giardina, B.; Tavazzi, B. Single-sample preparation for simultaneous cellular redox and energy state determination. Anal. Biochem. 2003, 322, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Ganzera, M.; Vrabl, P.; Wörle, E.; Burgstaller, W.; Stuppner, H. Determination of adenine and pyridine nucleotides in glucose-limited chemostat cultures of Penicillium simplicissimum by one-step ethanol extraction and ion-pairing liquid chromatography. Anal. Biochem. 2006, 359, 132–140. [Google Scholar] [CrossRef] [PubMed]
- Hissin, P.J.; Hilf, R. A fluorometric method for determination of oxidized and reduced glutathione in tissues. Anal. Biochem. 1976, 74, 214–226. [Google Scholar] [CrossRef]
- Wang, S.B.; Hu, Q.; Sommerfeld, M.; Chen, F. An optimized protocol for isolation of soluble proteins from microalgae for two-dimensional gel electrophoresis analysis. J. Appl. Phycol. 2003, 15, 485–496. [Google Scholar] [CrossRef]
- Wessel, D.; Flügge, U.I. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 1984, 138, 141–143. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Wang, Y.; Cheung, Y.H.; Yang, Z.; Chiu, J.F.; Che, C.M.; He, Q.Y. Proteomic approach to study the cytotoxicity of dioscin (saponin). Proteomics 2006, 6, 2422–2432. [Google Scholar] [CrossRef] [PubMed]
- Ge, F.; Lu, X.P.; Zeng, H.L.; He, Q.Y.; Xiong, S.; Jin, L.; He, Q.Y. Proteomic and functional analyses reveal a dual molecular mechanism underlying arsenic-induced apoptosis in human multiple myeloma cells. J. Proteome Res. 2009, 6, 3006–3019. [Google Scholar] [CrossRef] [PubMed]
- Blanc, G.; Etten, J.L.V. The Chlorella variabilis NC64A genome reveals adaptation to photosymbiosis, coevolution with viruses, and cryptic sex. Plant Cell 2010, 22, 2943–2955. [Google Scholar] [CrossRef] [PubMed]
- Waridel, P.; Frank, A.; Thomas, H.; Surendranath, V.; Sunyaev, S.; Pevzner, P.; Shevchenko, A. Sequence similarity-driven proteomics in organisms with unknown genomes by LC-MS/MS and automated de novo sequencing. Proteomics 2007, 14, 2318–2329. [Google Scholar] [CrossRef] [PubMed]
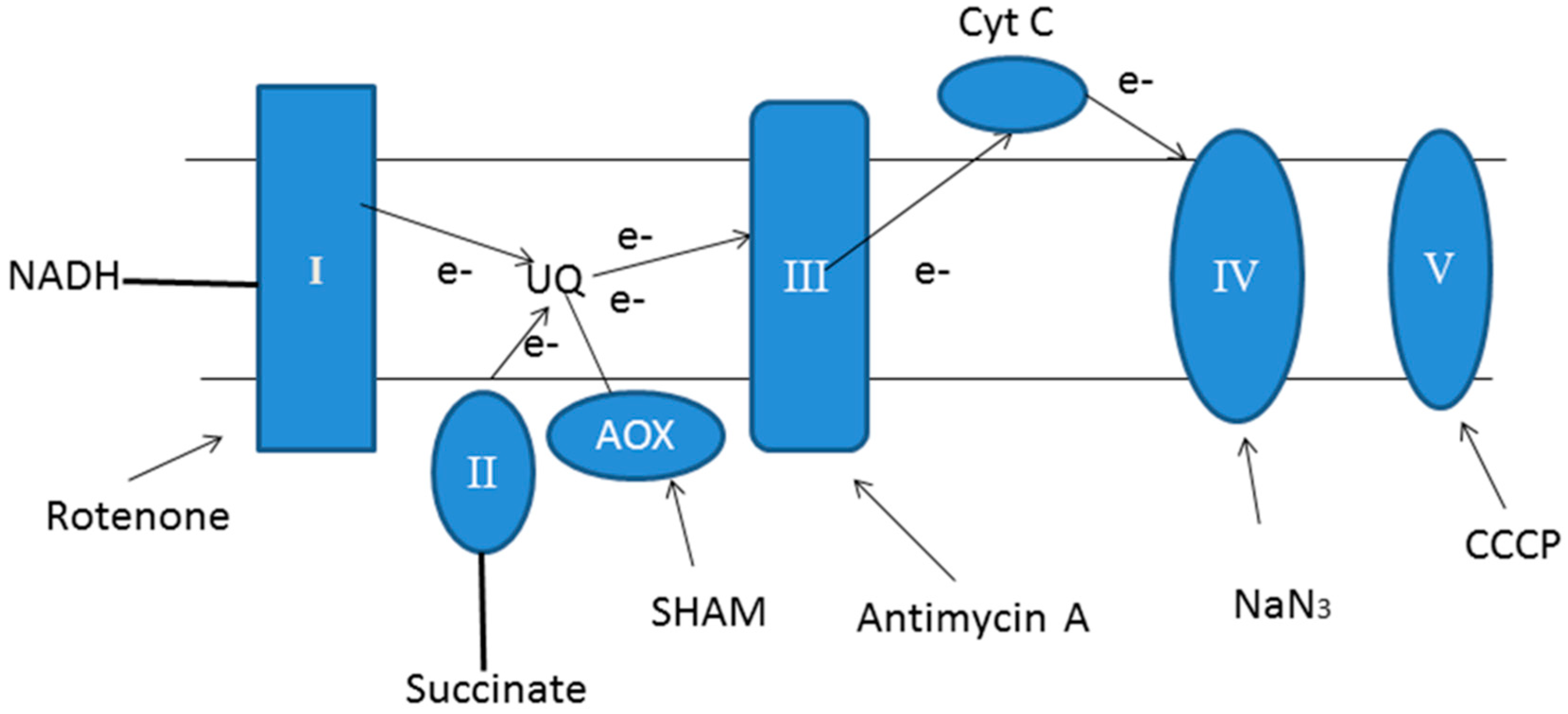
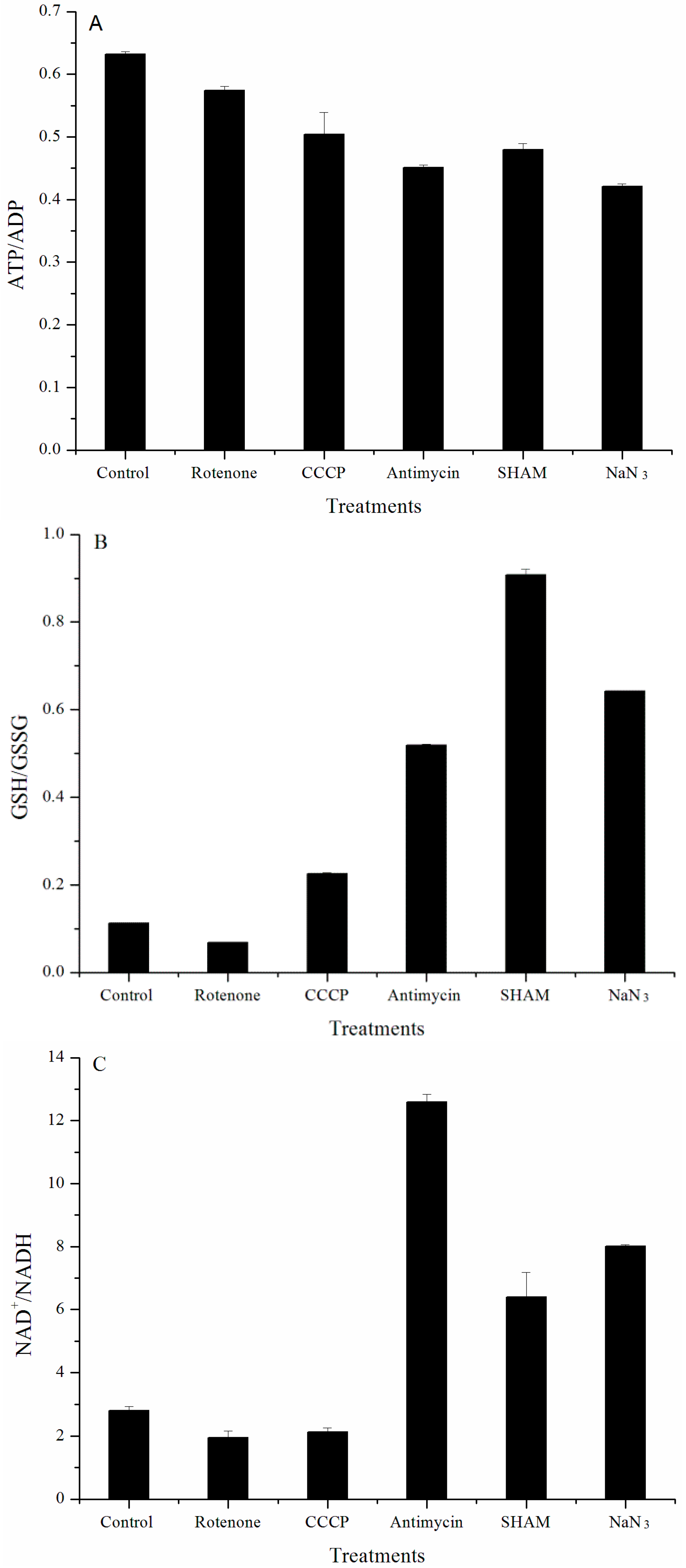
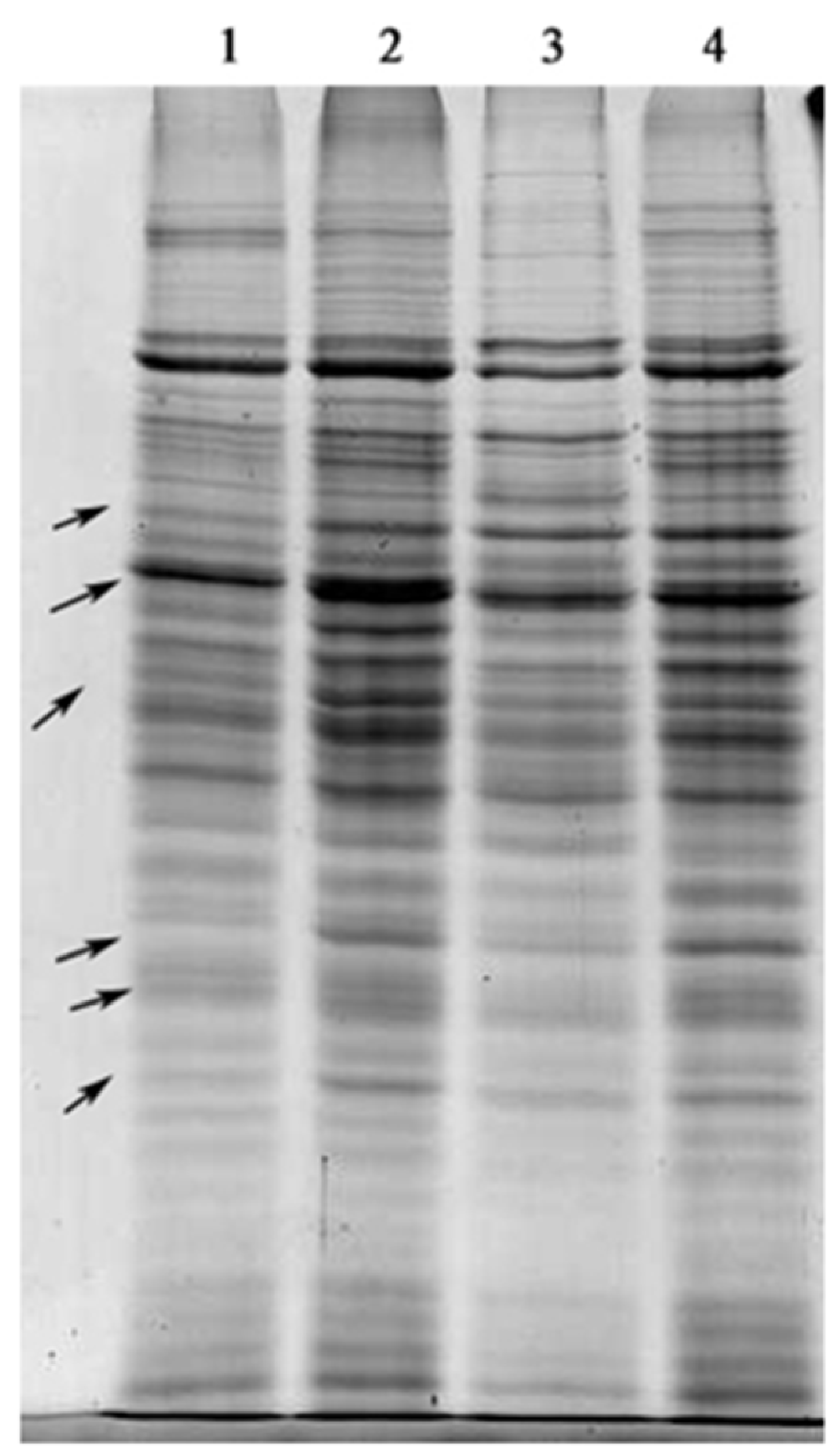

| Inhibitors | Biomass (%) | Lutein (%) | Chl. a (%) | Chl. b (%) |
|---|---|---|---|---|
| Antimycin A (0.1 mM) | 59.5 ± 3.2 | 87.7 ± 3.5 | 73.2 ± 2.3 | 73.4 ± 2.5 |
| NaN3 (0.05 mM) | 68.9 ± 9.6 | 49.1 ± 1.4 | 38.9 ± 0.9 | 45.2 ± 1.7 |
| SHAM (0.5 mM) | 46.1 ± 8.6 | 156.9 ± 7.2 | 121.6 ± 3.6 | 136.1 ± 5.4 |
| CCCP (0.032 mM) | 42.7 ± 2.6 | 106.4 ± 3.2 | 104.5 ± 1.8 | 128.6 ± 2.8 |
| Rotenone (0.032 mM) | 41.4 ± 3.1 | 107.1 ± 1.6 | 103.1 ± 1.0 | 104.9 ± 0.4 |
| Spot a | Protein Name b | Protein Function and Categorization c | Protein ID d | Species e | MW/pI | Peptides f | Score g |
|---|---|---|---|---|---|---|---|
| Down-regulated proteins | |||||||
| D2 | Autophagy-related protein 3 | Protein transport, Protein fate (folding, modification, destination) | gi|307105862 | Chlorella vulgaris | 35,252.9/4.42 | 1 | 35 |
| D3 | Adenylosuccinate synthetase | Purine nucleotide synthesis, Metabolism | gi|307108106 | Chlorella vulgaris | 53,409.7/6.62 | 3 | 64 |
| D4 | Hypothetical protein CHLNCDRAFT_144859 | GMP synthase, Metabolism | gi|307108123 | Chlorella vulgaris | 58,172.5/5.6 | 8 | 96 |
| D5 | Hypothetical protein CHLNCDRAFT_56182 | Adenylosuccinate synthetase, Metabolism | gi|307106668 | Chlorella vulgaris | 64,272.6/5.81 | 4 | 59 |
| D6 | Phosphoserine aminotransferase | L-glutamate synthesis, Metabolism | gi|307109471 | Chlorella vulgaris | 44,960.5/5.60 | 1 | 37 |
| D9 | Hypothetical protein CHLNCDRAFT_139931 | Alternative splicing factor SRp20/9G8, Transcription | gi|307103428 | Chlorella vulgaris | 19,849.2/11.52 | 6 | 53 |
| D10 | Hypothetical protein CHLNCDRAFT_134964 | SWI-SNF chromatin remodeling complex, Snf 5 subunit, Transcription | gi|307106629 | Chlorella vulgaris | 22,841.5/5.85 | 1 | 27 |
| D12 | Nitrite reductase | NO biosynthesis, Cell rescue, defense and virulence | gi|116265919 | Chlorella vulgaris | 22,954.7/9.11 | 1 | 60 |
| D14 | Coproporphyrinogen III oxidase | Key enzyme in heme synthesis, Metabolism | gi|71082810 | Candidatus Pelagibacter ubique | 32,383/9.83 | 1 | 68 |
| D15 | Hypothetical protein CHLNCDRAFT_30336 | ABC transporter superfamily, lipid transport, Protein with binding function or cofactor requirement | gi|307109169 | Chlorella vulgaris | 65,272.95/8.50 | 1 | 26 |
| D17 | Hypothetical protein CHLNCDRAFT_56437 | d-3-phosphoglycerate dehydrogenase, Metabolism | gi|307111670 | Chlorella vulgaris | 63,615.5/6.45 | 1 | 41 |
| D19 | Nitrite reductase | NO biosynthesis, Cell rescue, defense and virulence | gi|116265919 | Chlorella vulgaris | 22,954.7/9.11 | 1 | 69 |
| D21 | Malate dehydrogenase | Synthesis of oxaloacetate, Energy | gi|307103202 | Chlorella vulgaris | 35,063.6/8.2 | 6 | 104 |
| D22 | Hypothetical protein CHLNCDRAFT_57327 | Galactokinase activity, Protein with binding function or cofactor requirement | gi|307109337 | Chlorella vulgaris | 55,429.4/5.97 | 10 | 66 |
| D25 | Phytochrome A | G-protein coupled photoreceptor activity, Transcription | P42500 | Glycine max | 125,653.3/6.16 | 14 | 54 |
| D30 | Hypothetical protein CHLNCDRAFT_58635 | Chaperonin complex component, TCP-1 eta subunit (CCT7), Protein fate (folding, modification, destination) | gi|307105118 | Chlorella variabilis | 61,530.4/6.25 | 1 | 28 |
| D36 | Hypothetical protein CHLNCDRAFT_56384 | UDP-glucose 4-epimerase/UDP-sulfoquinovose synthase, Protein with binding function or cofactor requirement | gi|307103315 | Chlorella variabilis | 39,120.5/6.53 | 1 | 77 |
| D37 | Hypothetical protein CHLNCDRAFT_49080 | Phosphoenolpyruvate carboxykinase activity | gi|307104333 | Chlorella variabilis | 49,051.4/5.69 | 6 | 70 |
| D39 | Hypothetical protein CHLNCDRAFT_31785 | 14-3-3 family, multifunctional chaperone, Protein with binding function or cofactor requirement | gi|307106152 | Chlorella variabilis | 29,385.8/4.96 | 2 | 77 |
| D40 | Hypothetical protein CHLNCDRAFT_34933 | Prolyl-tRNA aminoacylation, Protein with binding function or cofactor requirement | gi|307109063 | Chlorella variabilis | 33,163.8/6.56 | 2 | 83 |
| D45 | Hypothetical protein CHLNCDRAFT_14282 | Aconitase/homoaconitase | gi|307110485 | Chlorella variabilis | 11,454.9/5.88 | 3 | 53 |
| D46 | Expressed protein | Unknown, Unknown | gi|307104059 | Chlorella variabilis | 19,035.7/7.02 | 1 | 28 |
| D50 | Aspartate carbamoyltransferase | Enzyme of the first committed step in pyrimidine synthesis, Protein activity regulation | P49077 | Arabidopsis thaliana | 43,139.3/6.21 | 8 | 55 |
| D51 | Hypothetical protein CHLNCDRAFT_35404 | Protein serine/threonine kinase, Protein fate (folding, modification, destination) | gi|307107220 | Chlorella variabilis | 40,979.4/9.01 | 6 | 56 |
| D53 | Hypothetical protein CHLNCDRAFT_52952 | Calcium ion binding, Protein with binding function or cofactor requirement | gi|307106250 | Chlorella variabilis | 245,122.6/7.28 | 1 | 27 |
| D54 | 14-3-3-like protein | Multifunctional chaperone, posttranslational modification, Protein with binding function or cofactor requirement | P52908 | Chlamydomonas reinhardtii | 29,495.8/4.9 | 1 | 32 |
| D60 | Elongation factor 2 (EF-2) | Catalyze GTP dependent ribosomal translocation step during translation elongation, Protein with binding function or cofactor requirement | gi|119167 | Parachlorella kessleri | 94,054.8/5.84 | 17 | 308 |
| D65 | Expressed protein | Splicing coactivator, RNA processing, Transcription | gi|307109910 | Chlorella variabilis | 84,395.1/10.28 | 1 | 28 |
| D69 | Citrate synthase | Citrate synthesis, located in mitochondria, Energy | gi|307105555 | Chlorella variabilis | 53,533.7/7.24 | 5 | 98 |
| D72 | Hypothetical protein CHLNCDRAFT_56456 | Dystonin, growth -arrest-specific protein, cytoskeletone, Subcellular localization | gi|307111694 | Chlorella variabilis | 311,562.1/4.77 | 8 | 66 |
| D73 | Ribosomal protein | Large ribosomal subunit, Protein synthesis | gi|307103203 | Chlorella variabilis | 25,401/8.65 | 10 | 138 |
| D80 | Hypothetical protein CHLNCDRAFT_138729 | Electron transport, Protein with binding function or cofactor requirement | gi|307104457 | Chlorella variabilis | 62,588.1/9.52 | 11 | 53 |
| D82 | OSJNBb0032E06.9 | Ribonuclease H activity, Cell cycle and DNA processing | gi|38344375 | Oryza stiva | 138,142.2/8.96 | 15 | 83 |
| D83 | Protein W01F3.3a (mlt-11) | Serine-type endopeptidase inhibitor activity, Development | gi|115534910 | Caenorhabditis elegans | 236,516.9/5.07 | 10 | 84 |
| D86 | Fumarate hydratase | Catalyze S-malate synthesis, mitochondria | gi|17549498 | Ralstonia solanacearum GMI1000 | 49,372.3/6.07 | 4 | 127 |
| D89 | Ribulose bisphosphate carboxylase small chain 1 | Carbon dioxide fixation, Energy | P00873 | Chlamydomonas reinhardtii | 20,606.4/9.36 | 2 | 66 |
| Up-regulated proteins | |||||||
| U1 | Chloroplast thioredoxin peroxidase | Peroxidase activity, Cell rescue, defense and virulence | gi|294845922 | Volvox carteri f. nagariensis | 17,421/5.15 | 2 | 193 |
| U2 | SMC domain protein | Chromosome structure maintenance, Unknown | gi|296427824 | Heliothis subflexa | 65,238/5.63 | 87 | |
| U3 | Peroxiredoxin TSA1 | Oxidoreductase, cell redox homeostasis, Cell rescue, defense and virulence | gi|126132194 | Scheffersomyces stipitis CBS 6054 | 21,761/4.92 | 2 | 82 |
| U4 | Magnesium chelatase subunit of protochlorophyllide reductase | Chlorophyll biosynthesis, Protein with binding function or cofactor requirement | gi|254798626 | Parachlorella kessleri | 39,567/5.08 | 6 | 310 |
| U5 | 2-Cys peroxiredoxin | Oxidoreductase, cell redox homeostasis, Cell rescue, defense and virulence | gi|327506370 | Dunaliella viridis | 22,235/5.74 | 1 | 51 |
| U6 | Hypothetical protein CHLNCDRAFT_48133 | Ornithine carbamoyltransferase, Protein with binding function or cofactor requirement | gi|307109894 | Chlorella variabilis | 38,764.5/6.68 | 9 | 51 |
| U7 | Hypothetical protein CHLNCDRAFT_53139 | Antioxidant activity, Cell rescue, defense and virulence | gi|307106076 | Chlorella variabilis | 21,778.9/8.35 | 1 | 40 |
| U8 | Hypothetical protein BAL199_15803 | Unknown, Unknown | gi|163792326 | alpha proteobacterium | 42,102.3/5.49 | 12 | 88 |
| U9 | Hypothetical protein CHLNCDRAFT_48477 | Membrane transport, Cellular transport, transport facilitation and transport routes | gi|307110872 | Chlorella vulgaris | 31,637.3/9.49 | 1 | 29 |
| U10 | tRNA(Ile)-lysidine synthase, chloroplastic | Ligase activity, translation, Transcription | Q32RX0 | Staurastrum punctulatum | 58,797/10.16 | 1 | 29 |
| U11 | Penecillin-binding protein 2 | Penicillin binding, Protein with binding function or cofactor requirement | gi|163752395 | Shewanella benthica | 68,820.33/9.54 | 67 | |
| U12 | Aldehyde dehydrogenase | Oxidation of aldehyde, Metabolism | gi|285018869 | Xanthomonas albilineans | 54,004.7/6.05 | 64 | |
| U13 | Hypothetical protein CHLNCDRAFT_30965 | Structural constitute of ribosome, Protein synthesis | gi|307107744 | Chlorella vulgaris | 21,281.2/10.33 | 8 | 51 |
| U14 | Hypothetical protein CHLNCDRAFT_143237 | Dystonin, growth arrest specific protein, Subcelluar location | gi|307109339 | Chlorella vulgaris | 56,981.7/5.56 | 13 | 53 |
| U15 | FG-GAP repeat protein | Ligand binding, Unknown | gi|40062562 | Uncultureed marine bacterium 159 | 136,477.9/4.18 | 66 | |
| U16 | Expressed protein | Unknown, Unknown | gi|307111048 | Chlorella variabilis | 20,888.8/10.28 | 6 | 57 |
| U17 | Hypothetical protein CHLNCDRAFT_59525 | nuclear receptor binding factor-1, Cell rescue, defense and virulence | gi|307111650 | Chlorella variabilis | 34,897.1/5.06 | 68 | |
| U18 | Hypothetical protein PEPMIC_01485 | Unknown, Unknown | gi|160947550 | Peptostreptococcus micros | 18,170.3/4.67 | 7 | 83 |
| U19 | Hypothetical protein CHLNCDRAFT_58231 | Ribosomal protein L22, Protein synthesis | gi|307105888 | Chlorella vulgaris | 67,650.2/10.05 | 1 | 29 |
| U20 | C protein alpha-antigen | Receptor, Protein with binding function or cofactor requirement | gi|307708369 | Streptococcus mitis NCTC 1226 | 34,6246.9/4.98 | 65 | |
| U21 | 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase | Oxidoreductase, terpenoids biosynthesis, Protein with binding function or cofactor requirement | Q5QYA9 | Idiomarina loihiensis | 40,462/5.68 | 1 | 58 |
| U22 | N-(5’-phosphoribosyl)anthranilate isomerase | Tryptophan biosynthesis, Metabolism | gi|307107003 | Chlorella vulgaris | 20,906.8/5.55 | 1 | 31 |
| U23 | Hypothetical protein CHLNCDRAFT_37743 | Aldehyde dehydrogenase, Metabolism | gi|307102335 | Chlorella variabilis | 45,285.3/6.11 | 3 | 111 |
| U24 | Hypothetical protein OsI_036678 | Calcium ion binding, Protein with binding function or cofactor requirement | gi|125536231 | Oryza sativa | 31,743.5/9.98 | 10 | 93 |
| U25 | Chloroplast 30S ribosomal protein S4 | Structural constituent of ribosome, Protein synthesis | P59137 | Catharomnion ciliatum | 23,589.9/10.3 | 1 | 30 |
| U26 | Hypothetical protein CHLNCDRAFT_140182 | Transcription initiation factor, Transcription | gi|307103188 | Chlorella variabilis | 63,894.8/5.19 | 13 | 54 |
| U27 | Nitrate reductase [NADH] 1 | Catalyze nitrite synthesis, Metabolism | P16081 | Oryza sativa | 101,447.9/6.19 | 1 | 29 |
| U28 | Ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | Carbon fixation, Energy | gi|164455027 | Chlorella variabilis | 52,496.3/5.99 | 1 | 32 |
| U29 | Hypothetical protein MGG_08723 | Unknown, Unknown | gi|145601241 | Magnaporthe grisea | 36,967.8/5.69 | 8 | 83 |
| U30 | Expressed protein | Large subunit of ribosome, Protein synthesis | gi|307108236 | Chlorella variabilis | 9805.4/11.71 | 4 | 69 |
| U31 | Hypothetical protein CHLNCDRAFT_59740 | 3-oxoacyl-(acyl-carrier-protein) synthase, lipid transport, Cellular transport, transport facilitation and transport routes | gi|307104988 | Chlorella variabilis | 32,230.3/6.19 | 1 | 28 |
| U32 | Hypothetical protein CHLNCDRAFT_18194 | Unknown, Unknown | gi|307111928 | Chlorella variabilis | 9084.4/4.37 | 1 | 28 |
| U33 | Ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | Carbon fixation, Energy | gi|164455037 | Chlorella variabilis | 52,496.3/6.0 | 18 | 193 |
| U34 | Glucose-6-phosphate isomerase | Isomerase, involved in glycolysis, Energy | gi|284434863 | Parachlorella kessleri | 27,035/5.35 | 1 | 48 |
| U35 | Hypothetical protein CHLNCDRAFT_143799 | Unknown, Unknown | gi|307108818 | Chlorella variabilis | 43,154.4/9.57 | 1 | 27 |
| U36 | Superoxide dismutase | Antioxidant enzyme, Cellular communication/signal transduction mechanism | gi|34558145 | Wolinella succinogenes DSM 1740 | 25,795.1/8.89 | 78 | |
| U37 | Hypothetical protein CHLNCDRAFT_138879 | FAP-dependent helicase activity, Transcription | gi|307104244 | Chlorella variabilis | 59,382.6/9.77 | 1 | 27 |
| U38 | Glucose -6-phosphate isomerase | Glycolysis enzyme, Energy | gi|307105594 | Chlorella variabilis | 72,048.5/6.41 | 7 | 53 |
| U39 | GTP-binding protein | Intracellular protein transport, Cellular communication/signal transduction mechanism | gi|307106020 | Chlorella variabilis | 25,468.9/6.66 | 2 | 66 |
| Spot a | F.D. h | ||
|---|---|---|---|
| SHAM | Antimycin A | NaN3 | |
| Down-regulated proteins | |||
| D2 | −4.95 ± 0.58 | −5.27 ± 2.16 | |
| D3 | −6.57 ± 0.59 | ||
| D4 | −3.01 ± 0.11 | −2.37 ± 0.22 | −2.29 ± 0.86 |
| D5 | −2.22 ± 0.30 | −1.93 ± 0.20 | |
| D6 | −2.18 ± 0.11 | ||
| D9 | −1.81 ± 0.35 | ||
| D10 | −1.77 ± 0.35 | ||
| D12 | −2.37 ± 0.74 | −3.12 ± 0.05 | |
| D14 | −1.95 ± 0.20 | −1.98 ± 0.43 | −2.29 ± 0.59 |
| D15 | −2.27 ± 0.66 | −6.95 ± 2.48 | |
| D17 | −2.45 ± 0.98 | −4.46 ± 1.59 | −3.87 ± 0.79 |
| D19 | −2.42 ± 0.98 | −2.08 ± 0.25 | |
| D21 | −2.26 ± 0.82 | ||
| D22 | −1.87 ± 0.26 | −6.32 ± 1.36 | −1.87 ± 0.26 |
| D25 | >−100 | >−100 | |
| D30 | >−100 | >−100 | |
| D36 | −3.03 ± 1.22 | −3.08 ± 0.91 | |
| D37 | −3.32 ± 0.65 | −5.66 ± 0.04 | |
| D39 | −2.27 ± 0.57 | ||
| D40 | >−100 | >−100 | |
| D45 | −2.03 ± 0.02 | ||
| D46 | −1.77 ± 0.37 | ||
| D50 | −1.98 ± 0.14 | ||
| D51 | −1.65 ± 0.24 | ||
| D53 | −1.89 ± 0.46 | −1.89 ± 0.46 | |
| D54 | −1.85 ± 0.33 | ||
| D60 | >−100 | ||
| D65 | >−100 | ||
| D69 | −3.55 ± 0.81 | ||
| D72 | −2.54 ± 0.97 | ||
| D73 | −4.20 ± 1.37 | ||
| D80 | −2.24 ± 0.52 | ||
| D82 | −1.89 ± 0.25 | ||
| D83 | −1.89 ± 0.30 | ||
| D86 | −1.59 ± 0.20 | ||
| D89 | −1.76 ± 0.38 | ||
| Up-regulated proteins | |||
| U1 | 1.65 ± 0.57 | ||
| U2 | 1.72 ± 0.48 | ||
| U3 | 1.53 ± 0.16 | 1.62 ± 0.08 | |
| U4 | 1.60 ± 0.09 | ||
| U5 | 1.84 ± 0.37 | ||
| U6 | 1.92 ± 0.46 | ||
| U7 | 1.85 ± 0.33 | ||
| U8 | 1.65 ± 0.57 | 1.59 ± 0.28 | 1.94 ± 0.68 |
| U9 | 1.54 ± 0.18 | ||
| U10 | 2.13 ± 0.37 | 2.75 ± 0.02 | |
| U11 | 1.68 ± 0.30 | ||
| U12 | 2.53 ± 0.78 | ||
| U13 | 2.56 ± 0.73 | 3.96 ± 2.05 | |
| U14 | 2.18 ± 0.10 | ||
| U15 | 3.08 ± 1.38 | ||
| U16 | 2.66 ± 0.45 | ||
| U17 | 2.21 ± 0.25 | ||
| U18 | 3.46 ± 0.93 | ||
| U19 | 4.38 ± 1.02 | ||
| U20 | 4.05 ± 0.09 | ||
| U21 | 5.36 ± 0.05 | 3.75 ± 0.01 | 1.53 ± 0.21 |
| U22 | 6.89 ± 1.10 | 3.61 ± 0.86 | |
| U23 | >100.00 | 5.39 ± 0.69 | |
| U24 | >100.00 | ||
| U25 | 1.79 ± 0.53 | 1.89 ± 0.39 | |
| U26 | 1.77 ± 0.39 | ||
| U27 | 1.60 ± 0.05 | ||
| U28 | 1.92 ± 0.48 | ||
| U29 | 1.92 ± 0.44 | ||
| U30 | 2.36 ± 0.71 | ||
| U31 | 2.56 ± 0.75 | ||
| U32 | 2.16 ± 1.08 | ||
| U33 | 3.02 ± 0.08 | ||
| U34 | >100 | ||
| U35 | 1.75 ± 0.21 | ||
| U36 | 1.60 ± 0.03 | ||
| U37 | 2.33 ± 0.20 | ||
| U38 | >100 | ||
| U39 | 3.75 ± 0.01 | ||
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Z.-h.; Li, T.; He, Q.-y.; Sun, Z.; Jiang, Y. Role of Mitochondria in Regulating Lutein and Chlorophyll Biosynthesis in Chlorella pyrenoidosa under Heterotrophic Conditions. Mar. Drugs 2018, 16, 354. https://doi.org/10.3390/md16100354
Liu Z-h, Li T, He Q-y, Sun Z, Jiang Y. Role of Mitochondria in Regulating Lutein and Chlorophyll Biosynthesis in Chlorella pyrenoidosa under Heterotrophic Conditions. Marine Drugs. 2018; 16(10):354. https://doi.org/10.3390/md16100354
Chicago/Turabian StyleLiu, Zhi-hui, Tao Li, Qing-yu He, Zheng Sun, and Yue Jiang. 2018. "Role of Mitochondria in Regulating Lutein and Chlorophyll Biosynthesis in Chlorella pyrenoidosa under Heterotrophic Conditions" Marine Drugs 16, no. 10: 354. https://doi.org/10.3390/md16100354
APA StyleLiu, Z.-h., Li, T., He, Q.-y., Sun, Z., & Jiang, Y. (2018). Role of Mitochondria in Regulating Lutein and Chlorophyll Biosynthesis in Chlorella pyrenoidosa under Heterotrophic Conditions. Marine Drugs, 16(10), 354. https://doi.org/10.3390/md16100354





