Biological and Chemical Diversity of Bacteria Associated with a Marine Flatworm
Abstract
1. Introduction
2. Results and Discussion
2.1. Sample Identification
2.2. Isolation and Taxonomy of Bacteria from the Marine Flatworm Paraplanocera sp.
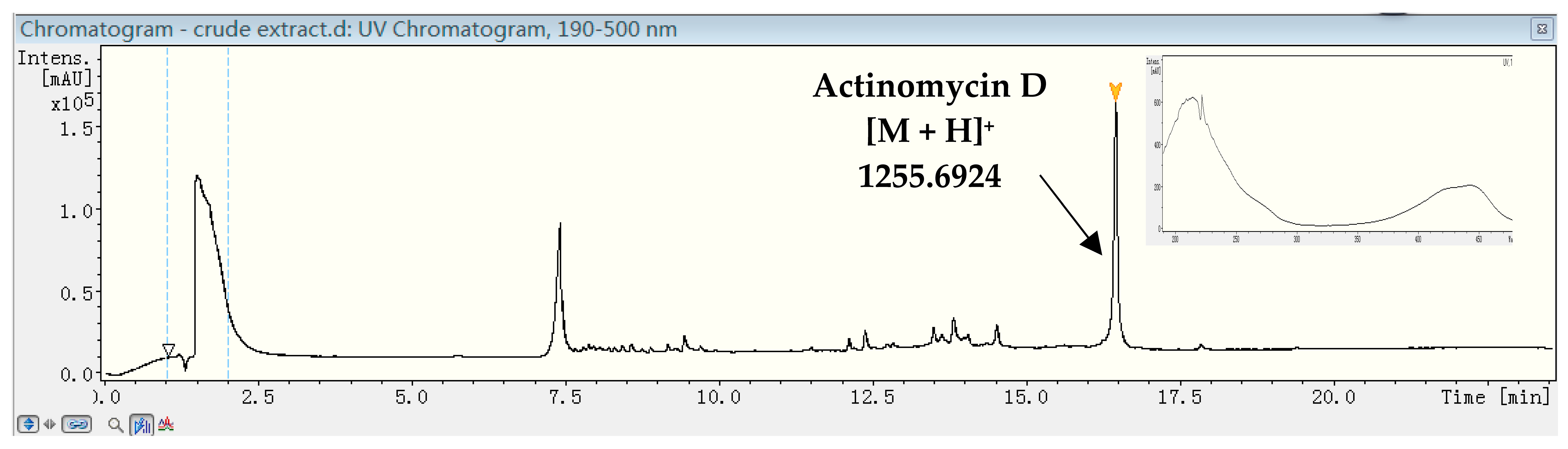
2.3. The Bioactive and UPLC-MS Chemical Screening of the Isolated Bacteria
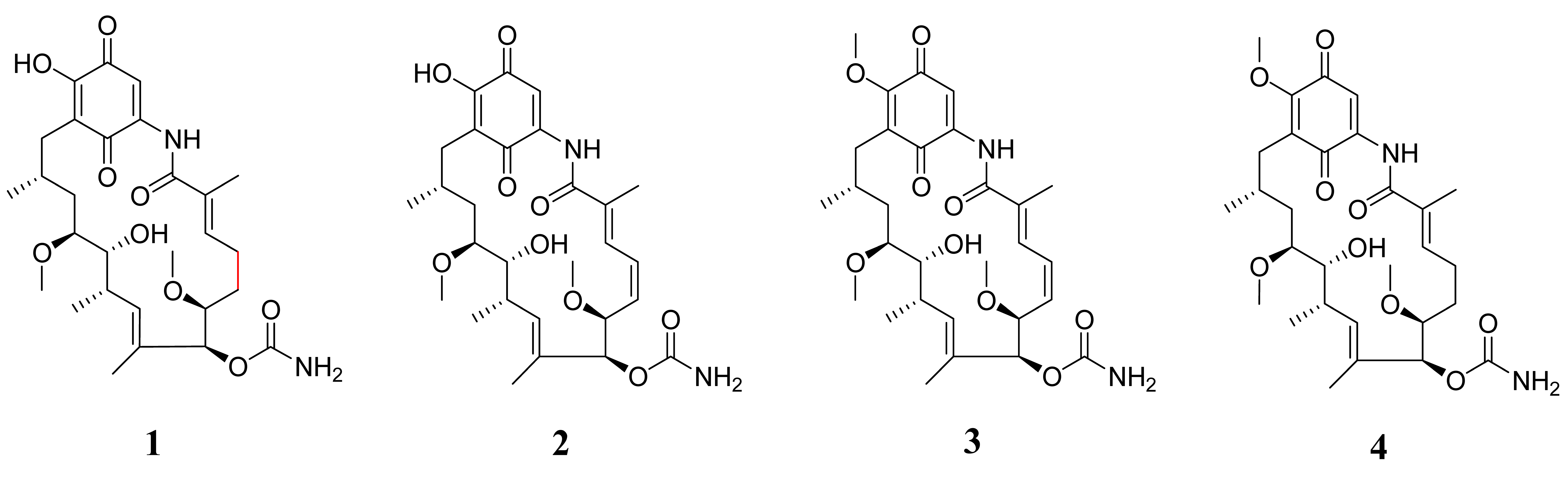
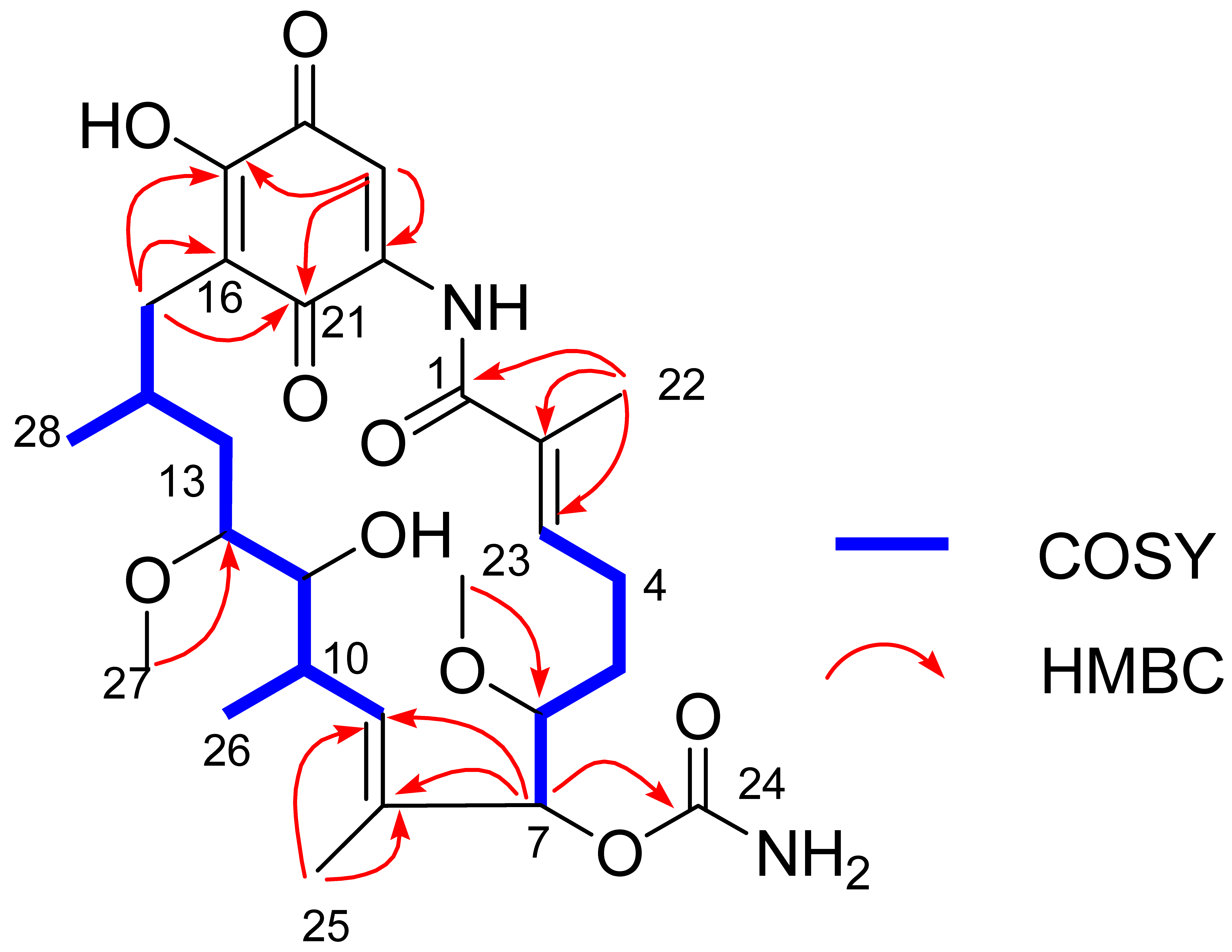
2.4. Structural Elucidation of the Isolated Compounds
2.5. Bioactive Evaluation and Structure–Activity Relationship
3. Materials and Methods
3.1. Bacteria Isolation and Identification
3.2. Bacteria Fermentation
3.3. Bioactive and Chemical Screening
3.4. Extraction and Compounds Isolation
3.5. Bioactive Assays
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Rawlinson, K.A. The diversity, development and evolution of polyclad flatworm larvae. Evodevo 2014, 5, 9. [Google Scholar] [CrossRef] [PubMed]
- Pfister, D.; Mulder, K.D.; Philipp, I.; Kuales, G.; Hrouda, M.; Eichberger, P.; Borgonie, G.; Hartenstein, V.; Ladurner, P. The exceptional stem cell system of Macrostomum lignano: Screening for gene expression and studying cell proliferation by hydroxyurea treatment and irradiation. Front. Zool. 2007, 4, 9. [Google Scholar] [CrossRef] [PubMed]
- Mouton, S.; Willemsa, M.; Braeckmanb, B.P.; Eggerc, B.; Ladurnerc, P.; Schärerd, L.; Borgoniea, G. The free-living flatworm Macrostomum lignano: A new model organism for ageing research. Exp. Gerontol. 2009, 44, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Lengerer, B.; Pjeta, R.; Wunderer, J.; Rodrigues, M.; Arbore, R.; Schärer, L.; Berezikov, E.; Hess, M.W.; Pfaller, K.; Egger, B.; et al. Biological adhesion of the flatworm Macrostomum lignano relies on a duo-gland system and is mediated by a cell type-specific intermediate filament protein. Front. Zool. 2014, 11, 12. [Google Scholar] [CrossRef] [PubMed]
- Rivera-Ingraham, G.A.; Nommick, A.; Blondeau-Bidet, E.; Ladurner, P.; Lignot, J.H. Salinity stress from the perspective of the energy-redox axis: Lessons from a marine intertidal flatworm. Redox Biol. 2016, 10, 3–64. [Google Scholar] [CrossRef] [PubMed]
- Rivera-Ingraham, G.A.; Bickmeyer, U.; Abele, D. The physiological response of the marine platyhelminth Macrostomum lignano to different environmental oxygen concentrations. J. Exp. Biol. 2013, 216, 2741–2751. [Google Scholar] [CrossRef] [PubMed]
- Pawlik, J.R. Marine invertebrate chemical defenses. Chem. Rev. 1993, 5, 1911–1922. [Google Scholar] [CrossRef]
- Holmström, C.; Kjelleberg, S. Marine Pseudoalteromonas species are associated with higher organisms and produce biologically active extracellular agents. FEMS Microbiol. Ecol. 1999, 30, 285–293. [Google Scholar] [CrossRef]
- Bernan, V.S.; Greensten, M.; Maiese, W.M. Marine microorganisms as a source of new natural products. Adv. Appl. Microbiol. 1997, 43, 57–90. [Google Scholar] [PubMed]
- Fenical, W. Chemical studies of marine bacteria: Developing a new resource. Chem. Rev. 1993, 93, 1673–1682. [Google Scholar] [CrossRef]
- Tanu, M.B.; Mahmud, Y.; Arakawa, O.; Takatani, T.; Kajihara, H.; Kawatsu, K.; Hamano, Y.; Asakawa, M.; Miyazawa, K.; Noguchi, T. Immunoenzymatic visualization of tetrodotoxin (TTX) in Cephalothrix species (Nemertea: Anopla: Palaeonemertea: Cephalotrichidae) and Planocera reticulata (Platyhelminthes: Turbellaria: Polycladida: Planoceridae). Toxicon 2004, 41, 515–520. [Google Scholar] [CrossRef] [PubMed]
- Yamada, R.; Tsunashima, T.; Takei, M.; Sato, T.; Wajima, Y.; Kawase, M.; Oshikiri, S.; Kajitani, Y.; Kosoba, K.; Ueda, H.; et al. Seasonal changes in the Tetrodotoxin content of the flatworm Planocera multitentaculata. Mar. Drugs 2017, 15, 56. [Google Scholar] [CrossRef] [PubMed]
- Ritson-Williams, R.; Yotsu-Yamashita, M.; Paul, V.J. Ecological functions of tetrodotoxin in a deadly polyclad flatworm. Proc. Natl. Acad. Sci. USA 2006, 103, 3176–3179. [Google Scholar] [CrossRef] [PubMed]
- Gruber-Vodicka, H.R.; Dirks, U.; Leisch, N. Paracatenula, an ancient symbiosis between thiotrophic Alphaproteobacteria and catenulid flatworms. Proc. Natl. Acad. Sci. USA 2011, 108, 12078–12083. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Li, Q.; Tian, R.M.; Lai, Q.L.; Zhang, Y. Pseudovibrio hongkongensis sp. nov., isolated from a marine flatworm. Antonie Van Leeuwenhoek 2015, 108, 127–132. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, Q.; Tian, R.M.; Lai, Q.L.; Xu, Y. Pseudovibrio stylochi sp. nov., isolated from a marine flatworm. Int. J. Syst. Evolut. Microbiol. 2016, 66, 2025–2029. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.N.; Wang, Y.; Huang, J.M.; Lai, Q.L.; Xu, Y. Deinococcus planocerae sp. nov., isolated from a marine flatworm. Antonie Van Leeuwenhoek 2017, 110, 811–817. [Google Scholar] [CrossRef] [PubMed]
- Martín-Durán, J.M.; Egger, B. Developmental diversity in free-living flatworms. Evodevo 2012, 3, 7. [Google Scholar] [CrossRef] [PubMed]
- Deboer, C.; Meulman, P.A.; Wnuk, R.J.; Peterson, D.H. Geldanamycin, a new antibiotic. J. Antibiot. 1970, 23, 442–447. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, J.Y.; Ni, S.Y.; Wu, L.Z.; Wang, H.Y.; Lin, L.; He, W.Q.; Wang, Y.G. A pair of sulfur-containing geldanamycin analogs, 19-S-methylgeldanamycin and 4,5-dihydro-19-S-methylgeldanamycin, from Streptomyces hygroscopicus 17997. J. Antibiot. 2011, 64, 519–522. [Google Scholar] [CrossRef] [PubMed]
- Fukuyo, Y.; Hunt, C.R.; Horikoshi, N. Geldanamycin and its anti-cancer activities. Cancer Lett. 2010, 290, 24–35. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Carbonero, R.; Carnero, A.; Paz-Ares, L. Inhibition of HSP90 molecular chaperones: Moving into the clinic. Lancet Oncol. 2013, 14, e358–e369. [Google Scholar] [CrossRef]
- Lian, F.; Lin, D.; Gu, J. Novel research progress of anti-tumor activities of geldanamycin and its derivations. World Notes Antibiot. 2010, 5, 202–206. [Google Scholar]
- Li, S.F.; Cui, J.; Lu, X.H.; Zheng, Z.H.; Liu, X.; Ni, S.Y.; Wang, Y.G.; Wu, L.Z. Methanethiol as a catabolite of methionine provides methylthio—Group for chemical formation of 19-S-methylgeldanamycin and 17,19-dimethylthioherbimycin A. J. Antibiot. 2013, 66, 499–503. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Sun, G.Z.; Li, X.; Pan, H.Y.; Zhang, Y.S. A New Geldanamycin Analogue from Streptomyces hygroscopicus. Molecules 2010, 15, 1161–1167. [Google Scholar] [CrossRef] [PubMed]
- Li, S.F.; Ni, S.Y.; Wu, L.Z.; Li, L.; Jiang, B.Y.; Wang, H.Y.; Sun, G.Z.; Gan, M.L.; Li, J.Y.; He, W.Q.; et al. 19-[(1′S,4′R)-4′-Hydroxy-1′-methoxy-2′-oxopentyl] geldanamycin, a Natural Geldanamycin Analogue from Streptomyces hygroscopicus 17997. J. Nat. Prod. 2013, 76, 969–973. [Google Scholar] [CrossRef] [PubMed]
- Bondarev, V.; Richter, M.; Romano, S.; Piel, J.; Schwedt, A.; Schulz-Vogt, H.N. The genus Pseudovibrio contains metabolically versatile bacteria adapted for symbiosis. Environ. Microbiol. 2013, 15, 2095–2113. [Google Scholar] [CrossRef] [PubMed]
- Brooks, B.W.; Murray, R.G.E. Nomenclature for “Micrococcus radiodurans” and other radiation-resistant cocci: Deinococcaceae fam. nov. and Deinococcus gen. nov., including five species. Int. J. Syst. Bacteriol. 1981, 31, 353–360. [Google Scholar] [CrossRef]
- Meienhofer, J.; Atherton, E. Structure-Activity Relationship in the Actinomycins. Adv. Appl. Microbiol. 1973, 16, 203–300. [Google Scholar] [PubMed]
- Sousa, M.F.V.Q.; Lopes, C.E.; Pereira, N., Jr. Development of a bioprocess for the production of actinomycin-D. Braz. J. Chem. Eng. 2002, 19, 277–285. [Google Scholar] [CrossRef]
- Sazak, A.; Şahin, N.; Güven, K.; Işık, K.; Goodfellow, M. Streptomyces samsunensis sp. nov., a member of the Streptomyces violaceusniger clade isolated from the rhizosphere of Robinia pseudoacacia. Int. J. Syst. Evolut. Microbiol. 2011, 61, 1309–1314. [Google Scholar] [CrossRef] [PubMed]
- Yin, M.; Lu, T.; Zhao, L.X.; Chen, Y.; Huang, S.X.; Lohman, J.R.; Xu, L.H.; Jiang, C.L.; Shen, B. The missing C-17-O-methyltransferase in geldanamycin biosynthesis. Org. Lett. 2011, 13, 3726–3729. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Wu, L.; Lin, L.; He, W.; Sun, G.; Wang, Y. Identification and detection of 4,5-dihydrogeldanamycin produced by Streptomyces hygroscopicus 17997. Chin. J. Antibiot. 2009, 34, 267–271. [Google Scholar]
- Buchanan, G.; Regentin, R.; Piagentini, M.; Andreas, R.; Robert, M.; Jorge, L.G.; Peter, J.L. Production of 8-Demethylgeldanamycin and 4,5-Epoxy-8-demethylgeldanamycin from a Recombinant Strain of Streptomyces hygroscopicus. J. Nat. Prod. 2005, 68, 607–610. [Google Scholar] [CrossRef] [PubMed]
- Khomsan, S.; Paranee, S.; Somboon, T.; Kannawat, D.; Pranee, R.; Pattama, P. Investigation on antimicrobial agents of the terrestrial Streptomyces sp. BCC71188. Appl. Microbiol. Biotechnol. 2017, 101, 533–543. [Google Scholar]
- Hong, Y.S.; Lee, D.; Kim, W.; Jeong, J.K.; Kim, C.G.; Sohng, J.K.; Lee, J.H.; Paik, S.G.; Lee, J.J. Inactivation of the carbamoyltransferase gene refines post-polyketide synthase modification steps in the biosynthesis of the antitumor agent geldanamycin. J. Am. Chem. Soc. 2004, 126, 11142–11143. [Google Scholar] [CrossRef] [PubMed]
- Ni, S.; Zhang, K.; Wang, Y.; He, W.; Wang, Y.; He, J.; Wu, L. Analysis of geldanamycin analogues in trace amounts by LC-MS/MS. Chin. J. Biotechnol. 2009, 25, 847–853. [Google Scholar]
- Schnur, R.C.; Corman, M.L.; Gallaschun, R.J.; Cooper, B.A.; Dee, M.F.; Doty, J.L.; Mumi, M.L.; DiOrio, C.I.; Barbacci, E.G.; Miller, P.E.; et al. erbB-2 oncogene inhibition by geldanamycin derivatives: Synthesis, mechanism of action, and structure-activity relationships. J. Med. Chem. 1995, 38, 3813–3820. [Google Scholar] [CrossRef] [PubMed]
- Tadtong, S.; Meksuriyen, D.; Tanasupawat, S.; Isobe, M.; Suwanboriruxa, K. Geldanamycin derivatives and neuroprotective effect on cultured P19-derived neurons. Bioorg. Med. Chem. Lett. 2007, 17, 2939–2943. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xu, Y.; Liu, L.; Han, Z.; Lai, P.Y.; Guo, X.; Zhang, X.; Lin, W.; Qian, P.Y. Five new amicoumacins isolated from a marine-derived bacterium Bacillus subtilis. Mar. Drugs 2012, 10, 319–328. [Google Scholar] [CrossRef] [PubMed]


| Species | Isolate ID | Accession Number of the Most Similar Strain | Species | Isolate ID | Accession Number of the Most Similar Strain | |
|---|---|---|---|---|---|---|
| Actinobacteria | Streptomyces libani | XY-FW38 | JN180219 | Mycobacterium pallens | XY-FW60 | KF378757 |
| S. rutgersensis | XY-FW46 | FJ99174 | M. parafortuitum | XY-FW63 | NZ_MVID01000062 | |
| S. samsunensis | XY-FW47 | MF664376 | M. peregrinum | XY-FW64 | AF537362 | |
| S. sudanensis | XY-FW121 | EF515876 | M. kumamotonense | XY-FW71 | AB239925 | |
| S. tendae | XY-FW57W | D63873 | M. poriferae | XY-FW80 | JN627174 | |
| S. luridus | XY-FW100W | AB184150 | M. chubuense | XY-FW83 | JYNX01000034 | |
| S. alboflavus | XY-FW107 | JNXT01000131 | M. bacteremicum | XY-FW90 | FJ172308 | |
| S. cirratus | XY-FW109 | AY999794 | M. iranicum | XY-FW62 | HQ009482 | |
| S. parvulus | XY-FW120 | AB184326 | XY-FW77 | HQ009482 | ||
| S. bacillaris | XY-FW137 | AB184439 | Microbacterium saperdae | XY-FW102 | AB004719 | |
| S. cavourensis | XY-FW153 | AB184264 | ||||
| S. daliensis | XY-FW154 | AY785161 | M. resistens | XY-FW145 | BCRA01000173 | |
| S. phytohabitans | XY-FW134 | JQ345722 | Tsukamurella tyrosinosolvens | XY-FW86 | FJ643549 | |
| XY-FW142 | JQ345722 | |||||
| Micromonospora eburnea | XY-FW94 | AB107231 | T. strandjordii | XY-FW101 | AF283283 | |
| M. schwarzwaldensis | XY-FW95 | KC517406 | Brevibacterium iodinum | XY-FW82 | X76567 | |
| M. carbonacea | XY-FW99 | KM370042 | Micrococcus yunnanensis | XY-FW48A | FJ214355 | |
| M. tulbaghiae | XY-FW123 | EU196562 | XY-FW75 | FJ214355 | ||
| M. echinospora | XY-FW130 | KY818670 | Arthrobacter soli | XY-FW120A | EF660748 | |
| M. marina | XY-FW132 | KM370074 | Pseudonocardia carboxydivorans | XY-FW148 | EF114314 | |
| M. wenchangensis | XY-FW65 | JQ768361 | ||||
| M. aurantiaca | XY-FW69 | CP002162 | XY-FW149 | EF114314 | ||
| XY-FW97 | CP002162 | |||||
| XY-FW122 | CP002162 | |||||
| Non-actinobacteria | Bacillus lehensis | XY-FW24 | KX082867 | M. variabilis | XY-FW48B | KP640585 |
| B. pumilus | XY-FW37 | AF492815 | XY-FW54 | KP640585 | ||
| B. qibsonii | XY-FW43 | KM036072 | Ruegeria arenilitoris | XY-FW31 | JQ807219 | |
| B. aerius | XY-FW50 | KY243945 | XY-FW32 | JQ807219 | ||
| B. berkeleyi | XY-FW53 | KR476448 | XY-FW40 | JQ807219 | ||
| B. algicola | XY-FW55 | KY580789 | XY-FW45 | JQ807219 | ||
| B. altitudinis | XY-FW66 | ASJC01000029 | XY-FW58 | JQ807219 | ||
| B. amyloliquefaciens subsp. plntruim | XY-FW105 | FN597644 | XY-FW72 | JQ807219 | ||
| XY-FW96 | JQ807219 | |||||
| B. aerophilus | XY-FW112 | KX951942 | XY-FW138 | JQ807219 | ||
| B. flecus | XY-FW125 | NZ_JANV01000041 | Pseudovibrio hongkongenesis | UST20140214-015B | KP207599 | |
| B. invictus | XY-FW131 | KF060662 | [15] | |||
| B. megaterium | XY-FW1A | MF597792 | P. stylochi [16] | UST20140214-052 | KP207600 | |
| B. aquimaris | XY-FW5 | MF429570 | P. ascidiaceicola | XY-FW21B | LN812993 | |
| XY-FW23 | KY777466 | XY-FW51 | LN812993 | |||
| XY-FW49 | KY753251 | XY-FW57 | LN812993 | |||
| B. marisflavi | XY-FW30 | MF062965 | Fictibacillus barbaricus | XY-FW78 | KY436215 | |
| XY-F W59 | MF062965 | F. phosphorivorans | XY-FW42 | KY471632 | ||
| B. aryabhattai | XY-FW110 | EF114313 | Photobacterium swingsii | XY-FW3 | KY229808 | |
| XY-FW117 | EF114313 | Arcobacter nitrofigilis | XY-FW17 | EU106662 | ||
| B. nealsonii | XY-FW116 | EU656111 | Staphylococcus epidermidis | XY-FW21A | MF429388 | |
| XY-FW126 | EU656111 | Aquimarina mueller | XY-FW21C | AY608408 | ||
| XY-FW133 | EU656111 | Tenacibaculum aiptasiae | XY-FW28 | KC178948 | ||
| XY-FW139 | EU656111 | Roseovarius aestuarii | XY-FW56 | EU156066 | ||
| B. idriensis | XY-FW113 | AY904033 | Cupriavidus campinensis | XY-FW67 | KY010351 | |
| XY-FW115 | AY904033 | Oceanobacillus picturae | XY-FW73 | KX068643 | ||
| XY-FW118 | AY904033 | Deinococcus planocerae [17] | XY-FW106 | KT886059 | ||
| XY-FW129 | AY904033 | Pseudomonas libanensis | XY-FW111 | KY933473 | ||
| XY-FW136 | AY904033 | Paenibacillus cineris | XY-FW114 | AJ575658 | ||
| Vibrio cyclitrophicus | XY-FW7B | KY382786 | Stenotrophomonas rhizophila | XY-FW119 | CP007597 | |
| V. chagasii | XY-FW19 | LN832958 | Paracoccus honmiensis | XY-FW124 | DQ342239 | |
| H. locisalis | XY-FW34A | JQ799098 | Alcaligenes aquatilis subsp. phenolicus | XY-FW141 | JX986974 | |
| H. halophilus | XY-FW34B | KX507262 | XY-FW146 | AUBT01000026 | ||
| H. trueperi | XY-FW68 | LT635432 | ||||
| H. alkaliphilus | XY-FW10 | NZ_FOOG01000089 | ||||
| XY-FW16 | NZ_FOOG01000089 | |||||
| Microbulbifer elongates | XY-FW27 | KY176867 | ||||
| M. epialgicus | XY-FW39 | KT758460 |
| Bacteria Associated with Paraplanocera sp. | Tested Pathogen | |
|---|---|---|
| Isolate ID | Closest Described Species | MRSA ATCC43300 |
| XY-FW47 | Streptomyces samsunensis | +++ |
| XY-FW142 | Streptomyces phytohabitans | +++ |
| XY-FW153 | Streptomyces cavourensis | +++ |
| XY-FW120 | Streptomyces parvulus | +++ |
| XY-FW120A | Arthrobacter soli | ++ |
| XY-FW105 | Bacillus siamensis | ++ |
| XY-FW56 | Roseovarius aestuarii | + |
| XY-FW48A | Micrococcus yunnanensis | + |
| Bacteria Associated with Paraplanocera sp. | Cell Line | |
|---|---|---|
| Isolate ID | Closest Described Species | HeLa Cell |
| XY-FW47 | Streptomyces samsunensis | +++ |
| XY-FW120 | Streptomyces parvulus | +++ |
| XY-FW142 | Streptomyces phytohabitans | ++ |
| XY-FW153 | Streptomyces cavourensis | ++ |
| XY-FW124 | Paracoccus honmiensis | ++ |
| XY-FW105 | Bacillus siamensis | + |
| XY-FW100W | Streptomyces luridus | + |
| Position | δC | δH (J in Hz) | Position | δC | δH (J in Hz) |
|---|---|---|---|---|---|
| 1 | 168.3, C | 16 | 117.2, C | ||
| 2 | 133.1, C | 17 | 152.8, C | ||
| 3 | 138.4, CH | 6.25, t (6.7) | 18 | 183.0, C | |
| 4 | 24.4, CH2 | 2.42, m | 19 | 107.4, CH | 7.28, s |
| 5 | 29.6, CH2 | 1.77, m | 20 | 140.8, C | |
| 6 | 80.9, CH | 3.37, m | 21 | 184.3, C | |
| 7 | 80.3, CH | 5.18, d (4.2) | NH | 8.99, s | |
| 8 | 131.0, C | 22-CH3 | 12.3, CH3 | 1.91, s | |
| 9 | 133.8, CH | 5.68, d (9.6) | 23-OCH3 | 59.0, CH3 | 3.41, s |
| 10 | 32.5, C | 2.77, m | 24-OCONH2 | 156.1, C | |
| 11 | 73.3, CH | 3.58, d (7.8) | 25-CH3 | 12.5, CH3 | 1.68, s |
| 12 | 82.5, CH | 3.38, m | 26-CH3 | 12.7, CH3 | 0.96, d (7.5) |
| 13 | 34.9, CH2 | 1.73, m | 27-OCH3 | 57, CH3 | 3.35, s |
| 14 | 28.3, CH | 1.75, m | 28-CH3 | 22.6, CH3 | 0.98, d (7.5) |
| 15 | 32.0, CH2 | 2.40, m 2.48, dd (13.2, 3.2) |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, H.-N.; Wang, K.-L.; Wu, Z.-H.; Tian, R.-M.; Liu, G.-Z.; Xu, Y. Biological and Chemical Diversity of Bacteria Associated with a Marine Flatworm. Mar. Drugs 2017, 15, 281. https://doi.org/10.3390/md15090281
Lin H-N, Wang K-L, Wu Z-H, Tian R-M, Liu G-Z, Xu Y. Biological and Chemical Diversity of Bacteria Associated with a Marine Flatworm. Marine Drugs. 2017; 15(9):281. https://doi.org/10.3390/md15090281
Chicago/Turabian StyleLin, Hui-Na, Kai-Ling Wang, Ze-Hong Wu, Ren-Mao Tian, Guo-Zhu Liu, and Ying Xu. 2017. "Biological and Chemical Diversity of Bacteria Associated with a Marine Flatworm" Marine Drugs 15, no. 9: 281. https://doi.org/10.3390/md15090281
APA StyleLin, H.-N., Wang, K.-L., Wu, Z.-H., Tian, R.-M., Liu, G.-Z., & Xu, Y. (2017). Biological and Chemical Diversity of Bacteria Associated with a Marine Flatworm. Marine Drugs, 15(9), 281. https://doi.org/10.3390/md15090281






