Abstract
Sterol 14α-demethylases from Cytochrome P450 family (CYP51s) are essential enzymes in sterol biosynthesis and well-known as the target of antifungal drugs. The 3D structure of CYP51A from Penicillium italicum (PiCYP51A) was constructed through homology modeling based on the crystal structure of human CYP51A (PDB: 3LD6). Molecular dynamics (MD) simulation was operated to relax the initial model and followed by quality assessment using PROCHECK program. On the basis of the docking information on the currently available CYP51s with the patent demethylase inhibitors (DMIs), pharmacophore-based virtual screening combined with docking analysis was performed to pick out twelve new compounds from ZINC database. Six hits revealed in the ligand database suggested potential ability to inhibit PiCYP51A. Compared to patent fungicide triazolone, the top three lead compounds had similar or higher affinity with the target enzyme, and accordingly, exhibited comparable or lower EC50 values to P. italicum isolates. The results could provide references for de novo antifungal drug design.
1. Introduction
Citrus is the world’s most common crop grown in over 100 countries worldwide [1]. The fungal diseases of blue mold on post-harvested citrus fruits, caused by Penicillium italicum, have led to 30%–50% loss of annual citrus economy in China [2]. Application of synthetic fungicides is the preferred method to control post-harvest diseases caused by fungal phytopathogens in fruits and vegetables. Sterol 14α-demethylases from Cytochrome P450 family (CYP51s) are essential enzymes in sterol biosynthesis and well-known as the target of antifungal drugs, which was membrane-bound and difficult to solve its crystal structure [3,4]. Demethylase inhibitors (DMIs) have been exploited as azole-antifungal drugs to inhibit lanosterol 14α-demethylase (CYP51), impairing ergosterol/lanosterol metabolisms and finally resulting in fungal growth inhibition [5]. DMI fungicides have been widely used in medicine and agriculture, but the resistance to DMIs has been reported in many fungal species [6,7]. It is imperative to design new and efficient fungicides for the treatment of pathogens of plants based on the CYP51 structure.
Natural compounds extracted from marine fungi can be used as fungicides for phytopathogenic fungal pathogens [8,9,10]. Based on their structure characteristics, these natural antibiotics from marine fungi can be divided into several typical categories including aminoglycosides, macrolides antibiotics, microlides antibiotics, quinones, peptides, enzymes and heterocyclic [11,12,13]. At present, Penicillium italicum has raised increasing concerns due to its serious threat to citrus fruit storage and transportation, and identifying novel drugs against the pathogen is of great significance for the control of blue mold. Currently, the effective fungicides reported in the control of blue mold include imazali-, triazolone- and prochloraz-related chemical drugs, all classified as benzene and nitrogen heterocyclic ring compounds, sharing structural similarities with the marine anti-fungi drugs [14,15]. This makes it possible to find valuable lead compounds by high throughput virtual screening based on the pharmacophores generated from particular marine natural fungicides (Figure 1).
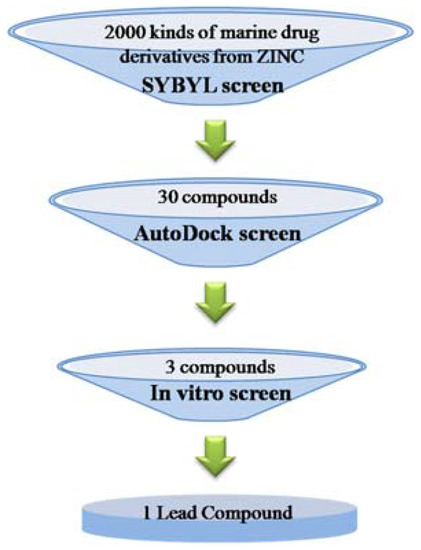
Figure 1.
Schematic for the high throughput virtual screening protocol in the present study.
Virtual screening supported by large-scale chemical libraries has been developed to be an important tool in novel lead molecules discovery [16,17]. Ligand-based screening techniques have the advantage of finding lead molecules based on a set of pharmacophore elements derived from a specific functional ligand family [15,18,19,20]. Owing to the membrane-associated characteristics for all the eukaryotic CYP51s, it remains a challenge to solve their crystal structures. Homology modeling has been widely applied in three-dimensional model building of CYP51s to understand molecular interactions between inhibitors and target enzymes [17,21]. The crystal structure of M. tuberculosis CYP51 (MtCYP51) has been used in many CYP51 modeling studies [4,22,23]. However, MtCYP51 has only 27.2% sequence identity with the PiCYP51A. The structure of human CYP51A, with 37.7% sequence identity to the PiCYP51A [19,24], is a more suitable template to construct the PiCYP51A structure model in the fungicide screening. In the present study, based on PiCYP51A target protein, we use the natural fungicidal components of marine fungi as reference compounds for the design of novel antifungal agents based on pharmacophores.
In this study, the three-dimensional model of CYP51A from P. italicum isolate (HS-1) was constructed based on the crystal structure of human CYP51A (PDB: 3LD6). The protein model further relaxed by molecular dynamics (MD) simulations and evaluated by PROCHECK program was used in pharmacphore-based virtual screening to identify new head antifungal compound(s) from ZINC database (http://zinc.docking.org/).
2. Results
2.1. Homology Modeling of PiCYP51A
The human CYP51 has an identity of 37.66% on amino acid sequences with PiCYP51A, and thus is suitable to be the template for the homology modeling in this study. The first 39 residues in the constructed model were truncated due to the lack of N-terminal residues of human CYP51 deposited in the PDB database. The initial model was relaxed by MD simulations to achieve the stable 3D structure of PiCYP51A (Figure 2). The quality of PiCYP51A model was evaluated to be rational with 86.4% in favored core regions, 12.4% in allowed regions, 0.7% in generously allowed regions, and only 0.5% in disallowed regions.
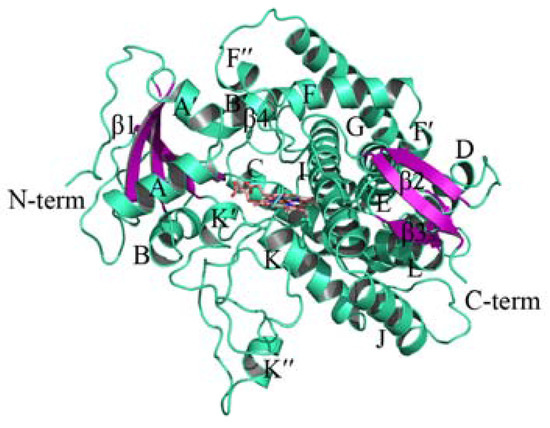
Figure 2.
3D model of Penicillium italicum (PiCYP51A) shown in ribbon representation.
2.2. Virtual Screening
Based on PiCYP51A active site characteristics, virtual screening in ZINC database (about 2000 compounds at beginning) exported the head 30 compounds with desirable query-fit (Qfit) values. Among the 30 compounds, twelve molecules exhibited similar structure-characteristics to some typical marine antibiotics, as shown in Figure 3. Using Qfit 10.0 as cutoff value, six molecules were chosen as candidates in further docking analysis, including ZINC72242441 (Qfit 12.33), ZINC73667465 (Qfit 14.19), ZINC81316574 (Qfit 15.56), ZINC61974481 (Qfit 18.86), ZINC74431162 (Qfit 23.71), and ZINC65393574 (Qfit 27.04).
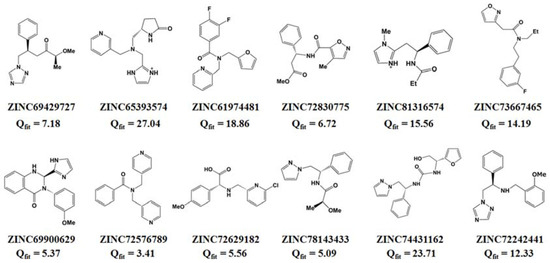
Figure 3.
Chemical structures of the twelve compounds retrieved from the ZINC database.
2.3. Molecular Docking with Ligands
To explore the binding patterns of the active site of PiCYP51A with its ligands, molecular docking analysis was carried out by using FlexX software (University of Hamburg, Center for Bioinformatics, Hamburg, Germany). The docking analysis revealed six ligands of hits with desirable rank scores obtained from FlexX evaluation. The putative conformations of the six candidate compounds in the binding sites of PiCYP51A were represented in Figure 4. The binding energy of the ligands with the receptor was shown in Table 1. According to the binding energy, the top three lead compounds were selected to evaluate fungicidal activities in vitro using binding spectrum and EC50 assay.
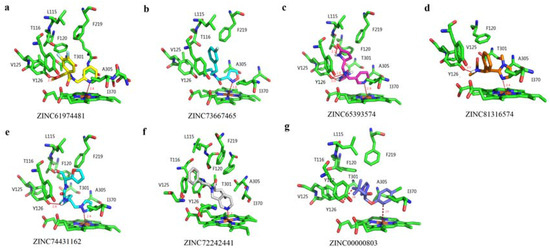
Figure 4.
3D-conformation of PiCYP51A docking with six candidate compounds. The six candidate compounds’ IDs are listed as (a) ZINC61974481, (b) ZINC73667465, (c) ZINC65393574, (d) ZINC81316574, (e) ZINC74431162, (f) ZINC72242441, and (g) ZINC00000803.

Table 1.
The molecular binding energy of PiCYP51A with candidate compounds.
2.4. Assay of Binding Constant (Kd) and EC50
The binding constants (Kd) of the selected top three lead compounds with PiCYP51A were determined by type II binding spectrum analysis. As shown in Figure 5A, among the four compounds including the patent fungicide triazolone, the lead compound c (ZINC65393574) had the lowest Kd value, i.e., the highest affinity with PiCYP51A target. In contrast, the other two lead compounds a (ZINC61974481) and e (ZINC74431162) had lower or similar affinity with the target comparing with triazolone (Figure 5A). The inhibitory effects of lead compounds on P. italicum isolates were experimentally investigated by EC50 assays (Figure 5B). Using the triazolone (EC50 17.74 g/mL) as control, the compound c exhibited significantly higher ability to inhibit HS-1 growth with EC50 11.37 μg/mL, and the compound e had a similar inhibiting effect on the HS-1 growth with EC50 22.96 μg/mL.
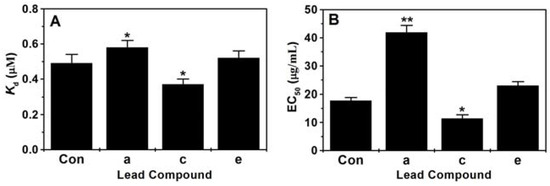
Figure 5.
In vitro binding constants (Kd) and EC50 of the selected lead compounds. (A) In vitro analysis of binding constants (Kd); (B) EC50 assay; Con: Triazolone, a patent fungicide used as control. (* p < 0.05; ** p < 0.01).
3. Discussion
For decades, synthetic fungicides based on cytochrome P450 target enzymes have been widely used to control post-harvest diseases caused by fungal phytopathogens [3,4]. Marine drug research has revealed a great number of natural fungicides from marine fungi such as Ascochyta salicorniae, Preussia aurantiaca and Penicillium chrysogenum [8,9,10]. Recently, a set of marine drugs have been reported to have potent activities against Pyricularia oryzae in agriculture production [10], which highlights a chance to design novel fungicides based on functional structure similarity. Currently, virtual screening supported by large-scale chemical libraries has been evaluated as a powerful tool to discover novel lead molecules that initiate synthesis of effective antifungal drugs [14,16]. The structure core(s) in the previously reported marine drugs to interfere with certain protein target(s) might provide a good choice of pharmacophore design that could lead a virtual screening to achieve desirable antifungal compound(s). In this study, we report the homolog modeling of CYP51A from P. italicum isolate (HS-1) and the further pharmacophore-based virtual screening to identify novel lead antifungal compound(s).
In the current study, we have constructed a PiCYP51A model for the first time with the core region of PiCYP51 proteins (Figure 2), containing the most conserved fold structure reported in the other P450 proteins [16,17,18,21,22,23], including helices E, I, J and L. Each of these helices has at least two aromatic-ring-containing residues (Phe and/or Tyr) involved in the hydrophobic interacting with some specific receptor(s). On the other hand, the present PiCYP51A model also suggested the critical role of nitrogen (N) atoms in the azole and aromatic rings in the coordination bond formation, as experimentally reported for the ligand-heme Fe(II) interaction in PiCYP51A active site [4].
The typical noncovalent bonds interactions have been well documented to contribute to enzyme catalysis, including hydrogen bonds, hydrophobic interaction and van der Waals force [20,25]. All these structure-relating data have been exploited in an efficient pharmacophore-based virtual screening to relax target ligand structure, producing desirable lead compounds with optimal binding energy. The previous reports have described in vitro antibacterial activities of asperflavin ribofuranoside, anthracene glycoside, flavoglaucin and citrinin by certain ligand-target interfering [12]. The similar anti-fungi effects were also reported in the antibiotic activity of diketopiperazine against Pyricularia oryzae [13]. In the present pharmacophore-based virtual screening system, the hydrophobic amino acid residues Y112, Y126, F120, F219 and V125 composed a large hydrophobic cleft in the PiCYP51A active site (Figure 4). The aromatic group of the ligands formed hydrophobic interactions with the hydrophobic cleft which fixed the orientation of the inhibitors in the active site [21,26]. The N atoms of the azole ring or hexatomic ring of ligands were bound to heme iron with distances from 2.2 Å to 2.9 Å, respectively (Figure 4). The unique carbonyl (C=O) oxygen atom in compound a, the hydrogen atom attached to amide nitrogen in compound d, and the hydrogen atom attached to N16 (amide nitrogen) in compound e formed H-bond with Y126 about 2.9 Å, 2.0 Å and 2.6 Å, respectively (Figure 4a,d,e). For ligand c, H39 and N1 involved in H-bond interactions with Y126, while another H-bond with a length of 2.1 Å was found between H41 and T116 (Figure 4c). The binding energies of the six ligands were shown in Table 1, indicating the strength of binding interactions of these compounds in the active site. For the selected top three lead compounds a, c and e, the EC50 results were well consistent with the Kd values and also with the binding energy obtained from virtual docking analysis, indicating the effectiveness of the reported pharmacophore-based virtual screening.
4. Materials and Methods
4.1. Preparation of PiCYP51A Model
The homology model of PiCYP51 was built based on the crystal structure of the human CYP51 (PDB: 3LD6) by SWISS-MODEL, as previously described [22,27]. The model was further relaxed by molecular dynamic (MD) simulations using GROMACS 4.5.5 program package [23,28]. The 5 ns MD simulation was processed under GROMOS96 43a1 force field, within SPC3 water model, and at 300 K/1 atm conditions. The van der Waals and electrostatic interactions were constrained to 0.9 and 1.4 nm reign for the defined long-range interactions. The relaxed 3D model of PiCYP51A was checked by PROCHECK program [24,29]. In addition, the RMSD value between the superimposed model and crystal structure was calculated by Dali sever [25,30].
4.2. Virtual Screening
Pharmacophore-based virtual screening was used to find novel potential leads from a ligand database and the general protocol was summarized in Figure 1. DISCOtech module of the SYBYL 7.3 program package [26,31] was used to identify structural features. Further, 3D alignments of the pharmacophore features in desirable molecules with benzene ligand(s) and/or structure core of nitrogen heterocyclic ring, i.e. with the expected structure properties similar to the previously reported marine natural fungicides [10,11,12,13], are found by automatically iterating through experimentally adjustable parameters. The distance tolerance between the pharmacophore features in the overlaid structures was set to 0.25 Å, and gradually increased to 2.5 Å if no model was available with the lower tolerance setting (step size 0.25). The head 30 compounds that matched the main features of the pharmacophore model were exported as candidate hits for following molecular docking. Molecules, structurally matching all the features of ligand model, were selected as hits, and the percentage of the similarity was defined as query fit (Qfit) value.
4.3. Molecular Docking
In order to explore binding patterns of the active sites of PiCYP51A with its ligands, molecular docking analysis was carried out by using FlexX module of SYBYL, a fast and automated docking method. The 3D structures of each compound were built through the SKETCH option in SYBYL with the default setting. Energy minimizations were performed using the Gasteiger–Hückel charge and Tripos force field with a distance-dependent dielectric and the Powell conjugate gradient algorithm with a convergence criterion of 0.05 kcal·mol−1. The amino acid residues with atoms located within 9.0 Å distance around the Fe atom in heme group were considered as the essential elements in PiCYP51A active sites.
4.4. Analysis of Binding Constants (Kd) and EC50 for Fungicidal Compounds
Potential effective compounds were purchased from the capable vendors (Enamine Chemical Market), and these commercial lead compounds were exploited to investigate fungicidal activities towards PiCYP51A target. Recombinant PiCYP51A-39, a truncated version lacking N-terminal 39 residues, was heterogeneously expressed in isopropyl-β-d-1-thiogalactopyranoside (IPTG)-induced Escherichia coli BL21 (DE3) cells, and the cell lysates containing soluble PiCYP51A were subjected to carbon monoxide (CO) differential spectrometry analysis to ensure enzyme activity and further to make Kd determination for the commercial lead compounds by binding spectra assay, as described by Li et al. [32]. EC50 (the effective concentration to reduce growth by 50%) for each compound was evaluated according to previously reported method [33]. Three replicates were used for each EC50 calculation by software SPSS 10.0. Data are expressed as mean ± SE, and differences between means were evaluated by one-way analysis of variance (ANOVA) test.
Acknowledgments
This work was supported by grants from the Natural Science Foundation of China (31071653, 31371893). We sincerely thank Huazhong Shi (Department of Chemistry and Biochemistry, Texas Tech University, USA) for English language editing.
Author Contributions
D.L., Y.Y. and R.H. conceived and designed the experiments; J.Y., Y.Y., R.H., T.Z. and Y.N. performed the experiments; Y.Y. and Q.C. analyzed the data; Q.C., R.H., S.W., J.M. contributed reagents/materials/analysis tools; Y.Y and D.L. wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Smilanick, J.; Mansour, M.; Margosan, D.; Gabler, F.M.; Goodwine, W. Influence of pH and NaHCO3 on effectiveness of imazalil to inhibit germination of Penicillium digitatum and to control postharvest green mold on citrus fruit. Plant Dis. 2005, 89, 640–648. [Google Scholar] [CrossRef]
- Sharma, R.R.; Singh, D.; Singh, R. Biological control of postharvest diseases of fruits and vegetables by microbial antagonists: A review. Biol. Control 2009, 50, 205–221. [Google Scholar] [CrossRef]
- Lepesheva, G.I.; Waterman, M.R. Sterol 14α-demethylase cytochrome P450 (CYP51), a P450 in all biological kingdoms. Biochim. Biophys. Acta 2007, 1770, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Li, D.; Wei, P.; Zhang, J.; Wan, J.; Ren, Y.; Chen, Z.; Liu, D.; Yu, Z.; Feng, L. Structure-based rational screening of novel hit compounds with structural diversity for cytochrome P450 sterol 14α-demethylase from Penicillium digitatum. J. Chem. Inf. Model. 2010, 50, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Lepesheva, G.I.; Hargrove, T.Y.; Kleshchenko, Y.; Nes, W.D.; Villalta, F.; Waterman, M.R. CYP51: A major drug target in the cytochrome P450 superfamily. Lipids 2008, 43, 1117–1125. [Google Scholar] [CrossRef] [PubMed]
- Sombardier, A.; Dufour, M.C.; Blancard, D.; Corio-Costet, M.F. Sensitivity of Podosphaera aphanis isolates to DMI fungicides: Distribution and reduced cross-sensitivity. Pest Manag. Sci. 2010, 66, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yu, J.; Liu, J.; Yuan, Y.; Li, N.; He, M.; Qi, T.; Hui, G.; Xiong, L.; Liu, D. Novel mutations in CYP51B from Penicillium digitatum involved in prochloraz resistance. J. Microbiol. 2014, 52, 762–770. [Google Scholar] [CrossRef] [PubMed]
- Osterhage, C.; Kaminsky, R.; König, G.M.; Wright, A.D. Ascosalipyrrolidinone A, an antimicrobial alkaloid, from the obligate marine fungus Ascochyta salicorniae. J. Org. Chem. 2000, 65, 6412–6417. [Google Scholar] [CrossRef] [PubMed]
- Poch, G.K.; Gloer, J.B. Auranticins A and B: Two new depsidones from a mangrove isolate of the fungus Preussia aurantiaca. J. Nat. Prod. 1991, 54, 213–217. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Chen, H.Y.; Li, W.S.; Zhu, X.W.; Ding, W.J.; Li, C.Y. Bioactive chaetoglobosins from the mangrove endophytic fungus Penicillium chrysogenum. Mar. Drugs 2016, 14, 172. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Brauers, G.; Ebel, R.; Wray, V.; Berg, A.; Proksch, P. Novel chromone derivatives from the fungus Aspergillus versicolor isolated from the marine sponge Xestospongia exigua. J. Nat. Prod. 2003, 66, 57–61. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, X.; Lee, U.; Kang, J.S.; Hong, D.C.; Son, B.W. A new radical scavenging anthracene glycoside, asperflavin ribofuranoside, and polyketides from a marine isolate for the fungus microsporum. Chem. Pharm. Bull. 2006, 54, 882–883. [Google Scholar] [CrossRef] [PubMed]
- Byun, H.G.; Zhang, H.; Mochizuki, M.; Adachi, K.; Shizuri, Y.; Lee, W.J.; Kim, S.K. Novel antifungal diketopiperazine from marine fungus. ChemInform 2003, 34, 102–106. [Google Scholar] [CrossRef]
- Neto, A.C.; Maraschin, M.; Piero, R.M. Antifungal activity of salicylic acid against Penicillium expansum and its possible mechanisms of action. Int. J. Food Microbiol. 2015, 215, 64–70. [Google Scholar] [CrossRef] [PubMed]
- El-Megharbel, S.M.; Adam, A.M.; Megahed, A.S.; Refat, M.S. Synthesis and molecular structure of moxifloxacin drug with metal ions as a model drug against some kinds of bacteria and fungi. Russ. J. Gen. Chem. 2015, 85, 2366–2373. [Google Scholar] [CrossRef]
- Kitchen, D.B.; Decornez, H.; Furr, J.R.; Bajorath, J. Docking and scoring in virtual screening for drug discovery: Methods and applications. Nat. Rev. Drug Discov. 2004, 3, 935–949. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Paliwal, S.K.; Sharma, M.; Mittal, A.; Sharma, S. In silico and in vitro screening to identify structurally diverse non-azole CYP51 inhibitors as potent antifungal agent. J. Mol. Graph. Model. 2016, 63, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Cavasotto, C.N.; Abagyan, R.A. Protein flexibility in ligand docking and virtual screening to protein kinases. J. Mol. Biol. 2004, 337, 209–225. [Google Scholar] [CrossRef] [PubMed]
- Elshaier, Y.A.; Barakat, A.; Al-Qahtany, B.M.; Al-Majid, A.M.; Al-Agamy, M.H. Synthesis of pyrazole-thiobarbituric acid derivatives: Antimicrobial activity and docking studies. Molecules 2016, 21, 1337. [Google Scholar] [CrossRef] [PubMed]
- Al-Dabbagh, M.M.; Salim, N.; Himmat, M.; Ahmed, A.; Saeed, F. Quantum probability ranking principle for ligand-based virtual screening. J. Comput. Aided Mol. Des. 2017. [Google Scholar] [CrossRef] [PubMed]
- Sheng, C.Q.; Zhang, W.N.; Zhang, M.; Song, Y.L.; Ji, H.T.; Zhu, J.; Yao, J.Z.; Yu, J.X.; Yang, S.; Zhou, Y.J. Homology modeling of lanosterol 14α-demethylase of Candida albicans and Aspergillus fumigatus and insights into the enzyme-substrate interactions. J. Biomol. Struct. Dyn. 2004, 22, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Sheng, C.; Miao, Z.; Ji, H.; Yao, J.; Wang, W.; Che, X.; Dong, G.; Lü, J.; Guo, W.; Zhang, W. Three-dimensional model of lanosterol 14α-demethylase from Cryptococcus neoformans: Active-site characterization and insights into azole binding. Antimicrob. Agents Chemother. 2009, 53, 3487–3495. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Liu, D.; Zhang, Q.; Zhang, S.; Wan, J.; Xiao, W. Expression and homology modeling of sterol 14α-demethylase from Penicillium digitatum. FEMS Microbiol. Lett. 2007, 277, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhang, J.; Cao, S.; Han, R.; Yuan, Y.; Yang, J.; Yan, Y.; Liu, D. Homology modeling, molecular docking and spectra assay studies of sterol 14α-demethylase from Penicillium digitatum. Biotechnol. Lett. 2011, 33, 2005–2011. [Google Scholar] [CrossRef] [PubMed]
- Vijayakumar, T.M.; Kumar, R.M.; Agrawal, A.; Dubey, G.P.; Ilango, K. Comparative inhibitory potential of selected dietary bioactive polyphenols, phytosterols on CYP3A4 and CYP2D6 with fluorometric high-throughput screening. J. Food Sci. Technol. 2015, 52, 4537–4543. [Google Scholar] [CrossRef] [PubMed]
- Su, B.H.; Tu, Y.S.; Lin, C.; Shao, C.Y.; Lin, O.A.; Tseng, Y.J. Rule-based prediction models of cytochrome P450 inhibition. J. Chem. Inf. Model. 2015, 55, 1426–1434. [Google Scholar] [CrossRef] [PubMed]
- Arnold, K.; Bordoli, L.; Kopp, J.; Schwede, T. The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics 2006, 22, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Hess, B.; Kutzner, C.; Van Der Spoel, D.; Lindahl, E. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008, 4, 435–447. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Holm, L.; Rosenstrom, P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010, 38, 545–549. [Google Scholar] [CrossRef] [PubMed]
- SYBYL7.3. Tripos Inc.: St. Louis, MO, USA, 2003. Available online: http://www.tripos.com (accessed on 21 January 2014).
- Li, W.J.; Li, Q.; Liu, D.L.; Ding, M.W. Synthesis, fungicidal activity, and sterol 14α-demethylase binding interaction of 2-azolyl-3,4-dihydroquinazolines on Penicillium digitatum. J. Agric. Food Chem. 2013, 61, 1419–1426. [Google Scholar] [CrossRef] [PubMed]
- Holmes, G.J.; Eckert, J.W. Sensitivity of Penicillium digitatum and P. italicum to postharvest citrus fungicides in California. Phytopathology 1999, 89, 716–721. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).