New Metabolites and Bioactive Chlorinated Benzophenone Derivatives Produced by a Marine-Derived Fungus Pestalotiopsis heterocornis
Abstract
:1. Introduction
2. Results and Discussion
3. Experimental Section
3.1. General Experimental Procedures
3.2. Fungal Material
3.3. Fermentation, Extraction and Isolation
3.4. X-ray Crystallography of 1
3.5. Computational Work
3.6. Cytotoxicity Assay
3.7. Antimicrobial Assay
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Yang, X.Y.; Zhang, J.Z.; Luo, D.Q. The taxonomy, biology and chemistry of the fungal Pestalotiopsis genus. Nat. Prod. Rep. 2012, 29, 622–641. [Google Scholar] [CrossRef] [PubMed]
- Akone, S.H.; Amrani, M.E.; Lin, W.H.; Lai, D.W.; Proksch, P. Cytosporins F-K, new epoxyquinols from the endophytic fungus Pestalotiopsis theae. Tetrahedron Lett. 2013, 54, 6751–6754. [Google Scholar] [CrossRef]
- Wang, J.F.; Wei, X.Y.; Lu, X.; Xu, F.Q.; Wan, J.T.; Lin, X.P.; Zhou, X.F.; Liao, S.R.; Yang, B.; Tu, Z.C.; et al. Eight new polyketide metabolites from the fungus Pestalotiopsis vaccinii endogenous with the mangrove plant Kandelia candel (L.) Druce. Tetrahedron 2014, 70, 9695–9701. [Google Scholar] [CrossRef]
- Li, J.; Xie, J.; Yang, Y.H.; Li, X.L.; Zeng, Y.; Zhao, P.J. Pestalpolyols A-D, cytotoxic polyketides from Pestalotiopsis sp. cr013. Planta Med. 2015, 81, 1285–1289. [Google Scholar] [CrossRef] [PubMed]
- Hwang, I.H.; Swenson, D.C.; Gloer, J.B.; Wicklow, D.T. Disseminins and spiciferone analogues: Polyketide-derived metabolites from a fungicolous isolate of Pestalotiopsis disseminate. J. Nat. Prod. 2016, 79, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Li, Y.; Li, L.; Cao, Y.; Guo, L.D.; Liu, G.; Che, Y.S. Spiroketals of Pestalotiopsis fici provide evidence for a biosynthetic hypothesis involving diversified diels-alder reaction cascades. J. Org. Chem. 2013, 78, 2992–3000. [Google Scholar] [CrossRef] [PubMed]
- Hemberger, Y.; Xu, J.; Wray, V.; Proksch, P.; Wu, J.; Bringmann, G. Pestalotiopens A and B: Stereochemically challenging flexible sesquiterpene-cyclopaldic acid hybrids from Pestalotiopsis sp. Chem. Eur. J. 2013, 19, 15556–15564. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Wu, X.D.; Sun, M.; Li, M.H. Two novel tyrosinase inhibitory sesquiterpenes induced by CuCl2 from a marine-derived fungus Pestalotiopsis sp. Z233. Mar. Drugs 2013, 11, 2713–2721. [Google Scholar] [CrossRef] [PubMed]
- Luo, D.Q.; Chen, Y.P.; Zhang, J.; Shi, B.Z.; Yang, Z.Q.; Chen, C. A new glycine derivative and a new indole alkaloid from the fermentation broth of the plant endophytic fungus Pestalotiopsis podocarpi isolated from the Chinese podocarpaceae plant Podocarpus macrophyllus. Helv. Chim. Acta 2013, 96, 309–311. [Google Scholar] [CrossRef]
- Jia, Y.L.; Wei, M.Y.; Chen, H.Y.; Guan, F.F.; Wang, C.Y.; Shao, C.L. (+)- and (−)- Pestaloxazine A, a pair of antiviral enantiomeric alkaloid dimers with a symmetric spiro[oxazinane-piperazinedione] skeleton from Pestalotiopsis sp. Org. Lett. 2015, 17, 4216–4219. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.L.; Zhang, S.; Hu, Q.B.; Luo, D.Q.; Zhan, Y. Phthalide derivatives with antifungal activities against the plant pathogens isolated from the liquid culture of Pestalotiopsis photiniae. J. Antibiot. 2011, 64, 723–727. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wu, X.F.; Ding, G.; Feng, Y.; Jiang, X.J.; Guo, L.D.; Che, Y.S. α-Pyrones and pyranes from the plant pathogenic fungus Pestalotiopsis scirpina. Eur. J. Org. Chem. 2012, 12, 2445–2452. [Google Scholar] [CrossRef]
- Keith, L.M.; Velasquez, M.E.; Zee, F.T. Identification and characterization of Pestalotiopsis spp. causing scab disease of guava, Psidium guajava, in Hawaii. Plant Dis. 2006, 90, 16–23. [Google Scholar] [CrossRef]
- Wei, M.Y.; Li, D.; Shao, C.L.; Deng, D.S.; Wang, C.Y. (±)-Pestalachloride D, an antibacterial racemate of chlorinated benzophenone derivative from a soft coral-derived fungus Pestalotiopsis sp. Mar. Drugs 2013, 11, 1050–1060. [Google Scholar] [CrossRef] [PubMed]
- Koshino, H.; Yoshihara, T.; Okuno, M.; Sakamura, S.; Tajimi, A.; Shimanuki, T. Gamahonolides A, B, and gamahorin, novel antifungal compounds from stromata of epichloe typhina on phleum pratense. Biosci. Biotechnol. Biochem. 1992, 56, 1096–1099. [Google Scholar] [CrossRef] [PubMed]
- Li, E.; Jiang, L.H.; Guo, L.D.; Zhang, H.; Che, Y.S. Pestalachlorides A-C, antifungal metabolites from the plant endophytic fungus Pestalotiopsis adusta. Bioorg. Med. Chem. 2008, 16, 7894–7899. [Google Scholar] [CrossRef] [PubMed]
- Xing, Q.; Gan, L.S.; Mou, X.F.; Wang, W.; Wang, C.Y.; Wei, M.Y.; Shao, C.L. Isolation, resolution and biological evaluation of pestalachlorides E and F containing both point and axial chirality. RSC Adv. 2016, 6, 22653–22658. [Google Scholar] [CrossRef]
- Shao, C.L.; Wang, C.Y.; Xing, Q.; Wang, K.L. Manufacture of Chlorinated Benzophenone Compounds with Pestalotiopsis for Use as Marine Antifouling Agents. CN Patent 104,163,805, 26 November 2014. [Google Scholar]
- Slavov, N.; Cvengros, J.; Neudorfl, J.M.; Schmalz, H.G. Total synthesis of the marine antibiotic pestalone and its surprisingly facile conversion into pestalalactone and pestalachloride A. Angew. Chem. Int. Ed. 2010, 49, 7588–7591. [Google Scholar] [CrossRef] [PubMed]
- Rukachaisirikul, V.; Rodglin, A.; Sukpondma, Y.; Phongpaichit, S.; Buatong, J.; Sakayaroj, J. Phthalide and isocoumarin derivatives produced by an Acremonium sp. isolated from a mangrove Rhizophora apiculata. J. Nat. Prod. 2012, 75, 853–858. [Google Scholar] [PubMed]
- Wang, S.; Li, Z.H.; Dong, Z.J.; Liu, J.K.; Feng, T. Norbisabolane and eremophilane sesquiterpenoids from cultures of the basidiomycete Polyporus ellisii. Fitoterapia 2013, 91, 194–198. [Google Scholar] [CrossRef] [PubMed]
- Mazzeo, G.; Santoro, E.; Andolfi, A.; Cimmino, A.; Troselj, P.; Petrovic, A.G.; Superchi, S.; Evidente, A.; Berova, N. Absolute configurations of fungal and plant metabolites by chiroptical methods. ORD, ECD, and VCD studies on phyllostin, scytolide, and oxysporone. J. Nat. Prod. 2013, 76, 588–599. [Google Scholar] [CrossRef] [PubMed]
- Bruhn, T.; Schaumloeffel, A.; Hemberger, Y.; Bringmann, G. SpecDis: Quantifying the comparison of calculated and experimental electronic circular dichroism spectra. Chirality 2013, 25, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.; Han, L.; Li, Y.Q.; Li, J.; Rong, H.; Leng, Q.; Jiang, Y.; Zhao, L.X.; Huang, X.S. Isostreptazolin and sannaphenol, two new metabolites from Streptomyces sannanensis. Molecules 2012, 17, 836–842. [Google Scholar] [CrossRef] [PubMed]
- Ding, N.; Jiang, Y.; Han, L.; Chen, X.; Ma, J.; Qu, X.; Mu, Y.; Liu, J.; Li, L.; Jiang, C.; et al. Bafilomycins and odoriferous sesquiterpenoids from Streptomyces albolongus isolated from Elephas maximus Feces. J. Nat. Prod. 2016, 79, 799–805. [Google Scholar] [CrossRef] [PubMed]
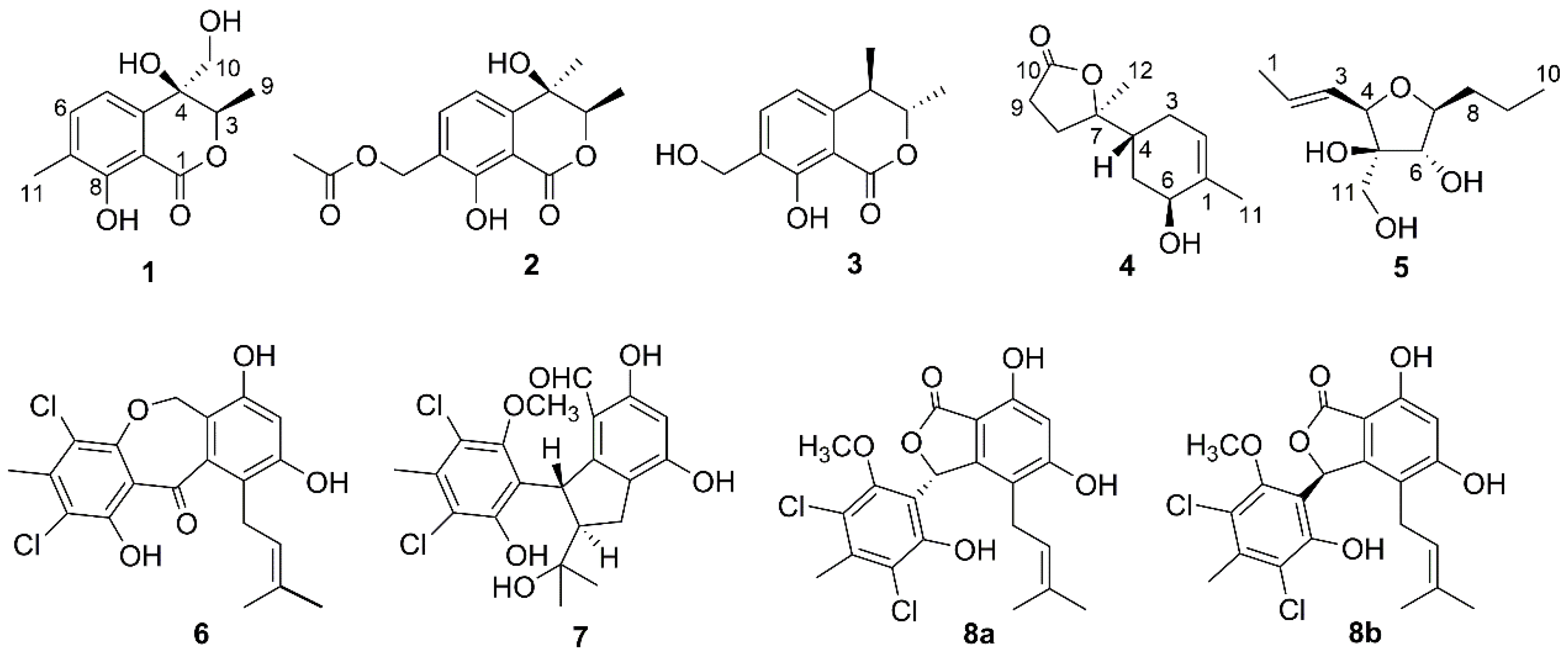
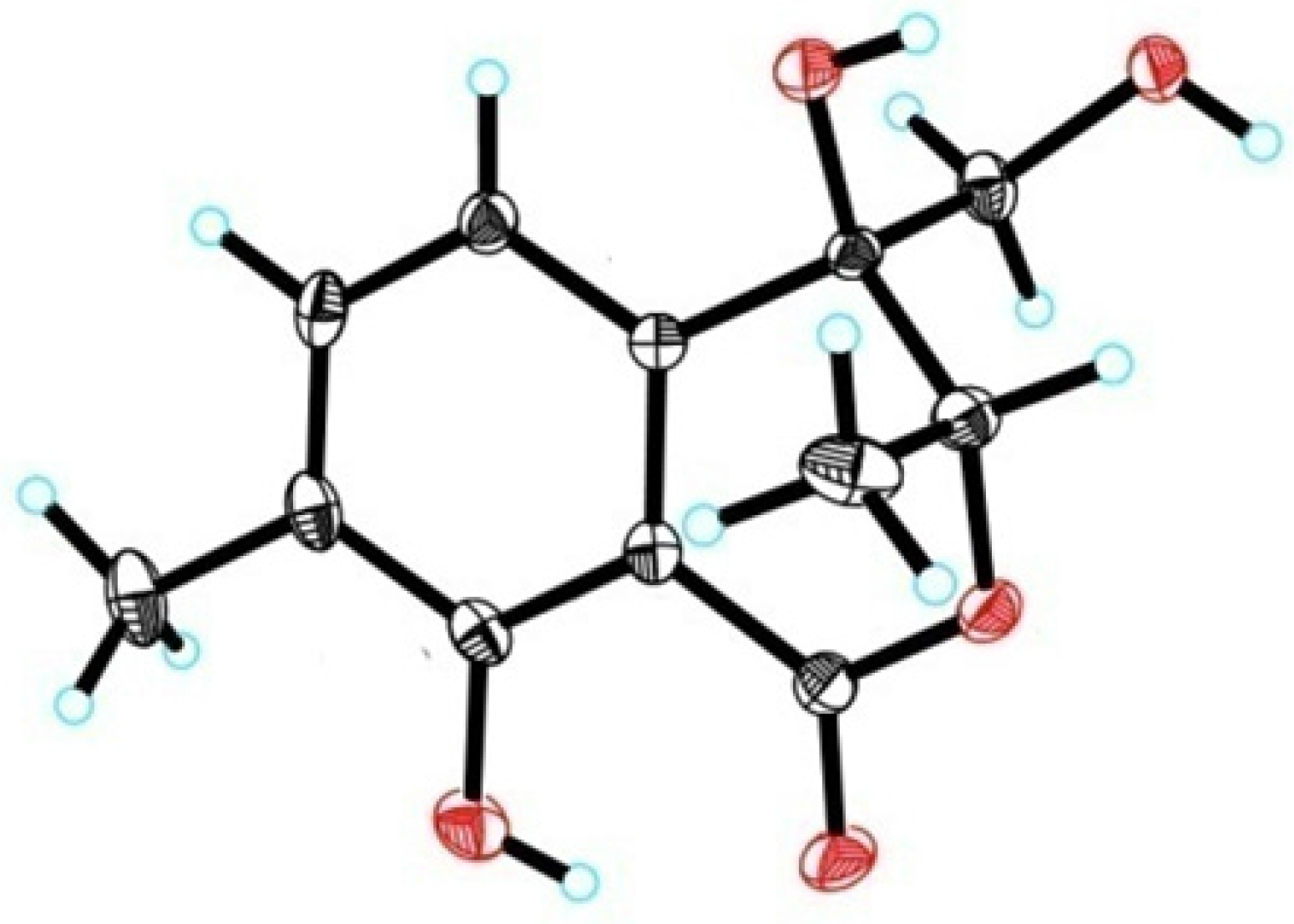
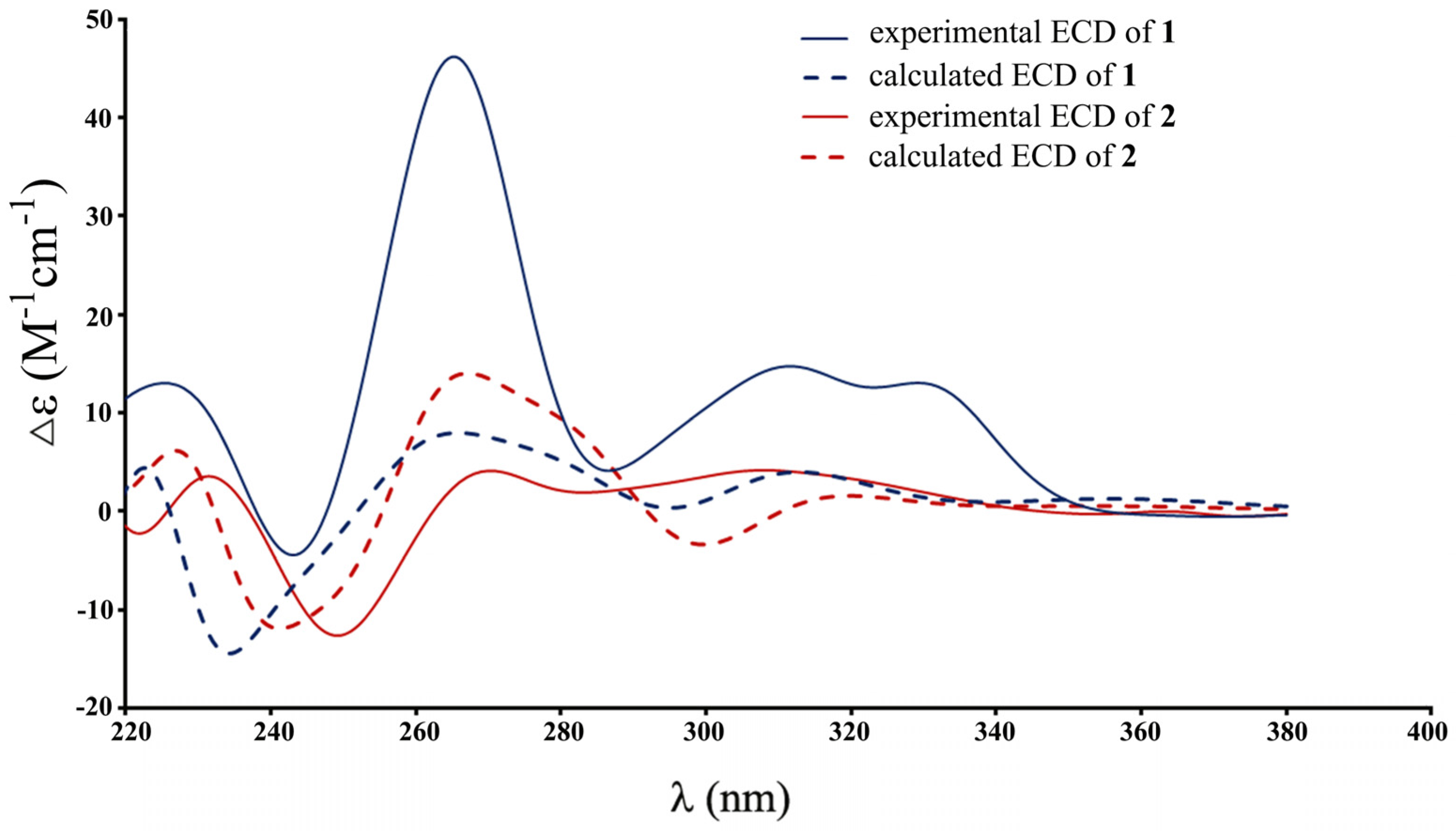
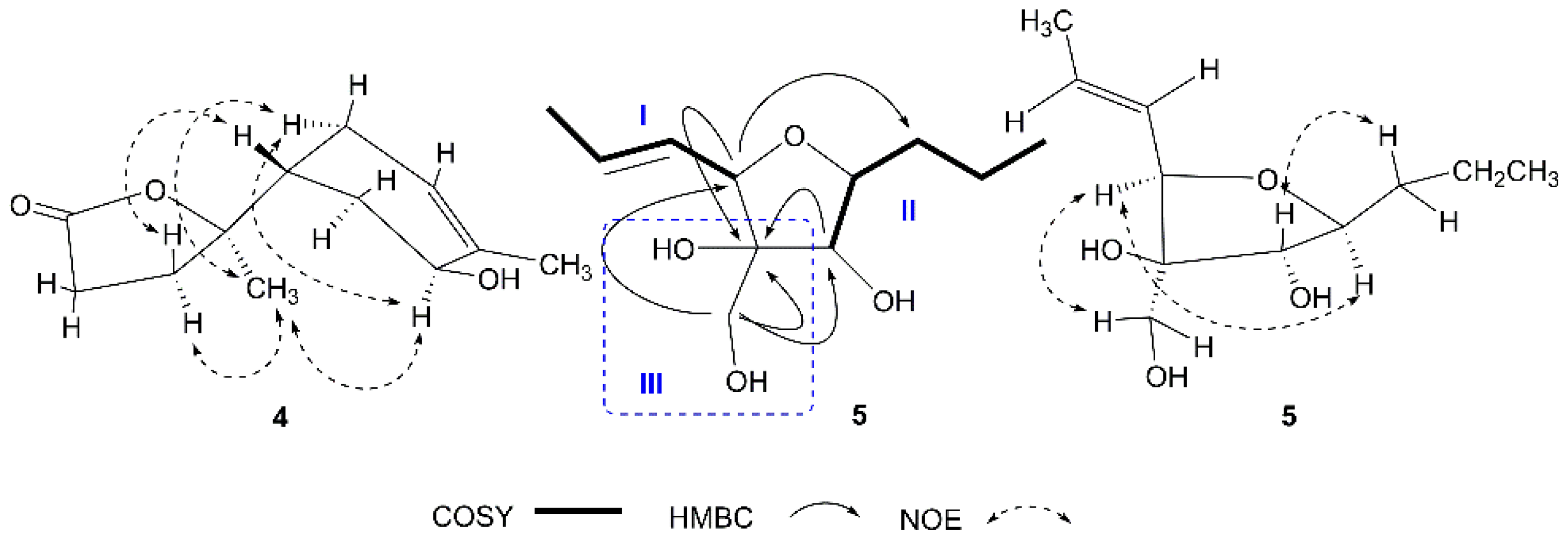
| 1 | 2 | |||
|---|---|---|---|---|
| Position | δC, type | δH (J in Hz) | δC, type | δH (J in Hz) |
| 1 | 168.9, C | 169.0, C | ||
| 3 | 78.0, CH | 4.90, q (6.6) | 82.5, CH | 4.60, q (6.6) |
| 4 | 71.3, C | 67.9, C | ||
| 4a | 139.2, C | 145.3, C | ||
| 5 | 115.3, CH | 7.05, d (7.5) | 114.2, CH | 7.16, d (7.7) |
| 6 | 136.9, CH | 7.47, d (7.5) | 136.2, CH | 7.64, d (7.7) |
| 7 | 125.7, C | 123.6, C | ||
| 8 | 159.5, C | 159.4, C | ||
| 8a | 106.1, C | 106.4, C | ||
| 9 | 13.6, CH3 | 1.36, d (6.6) | 13.5, CH3 | 1.43, d (6.6) |
| 10 | 65.7, CH2 | 3.81, d (11.7) 3.55, d (11.7) | 23.6, CH3 | 1.57, s |
| 11 | 14.0, CH3 | 2.24, s | 60.3, CH2 | 5.17, s |
| AcO | 171.1, C | |||
| 19.2, CH3 | 2.08, s | |||
| 4 (in CD3OD) | 4 (in CDCl3) | 5 (in CD3OD) | ||||
|---|---|---|---|---|---|---|
| Position | δC, type | δH (J in Hz) | δC, type | δH (J in Hz) | δC, type | δH (J in Hz) |
| 1 | 134.4, C | 134.7, C | 16.7, CH3 | 1.75, dd (6.4, 1.5) | ||
| 2 | 123.5, CH | 5.57, brd (5.0) | 124.2, CH | 5.57, brd (4.4) | 130.4, CH | 5.78, dd (14.7, 6.6) |
| 3 | 26.1, CH2 | 2.10, m 1.81, m | 26.5, CH2 | 2.12, m 1.82, m | 126.3, CH | 5.60, brdd (14.7, 7.2) |
| 4 | 37.2, CH | 2.06, m | 37.4, CH | 2.03, m | 82.9, CH | 4.13, d (7.9) |
| 5 | 32.3, CH2 | 1.95, brd (13.0) 1.48, td (13.0, 4.0) | 32.5, CH2 | 1.97, brd (13.2) 1.49, td (13.2, 3.7) | 81.6, C | |
| 6 | 67.1, CH | 4.00, brs | 68.0, CH | 4.07, brs | 82.7, CH | 3.75, d (4.5) |
| 7 | 89.0, C | 88.3, C | 84.7, CH | 3.58, td (6.6, 4.5) | ||
| 8 | 30.5, CH2 | 2.24, ddd (13.0, 10.0, 8.8) 1.98, ddd (13.0, 10.2, 4.7) | 31.2, CH2 | 2.19, ddd (13.2, 10.1, 9.0) 1.94, ddd (13.2, 10.0, 4.6) | 35.9, CH2 | 1.65, m |
| 9 | 28.4, CH2 | 2.72, ddd (18.3, 10.2, 8.8) 2.58, ddd (18.3, 10.0, 4.7) | 29.0, CH2 | 2.65, ddd (18.2, 10.0, 9.0) 2.57, ddd (18.2, 10.1, 4.6) | 18.9, CH2 | 1.50, m 1.42, m |
| 10 | 178.1, C | 176.7, C | 13.0, CH3 | 0.96, t (7.4) | ||
| 11 | 19.6, CH3 | 1.78, s | 20.8, CH3 | 1.80, s | 62.6, CH2 | 3.69, d (11.3) 3.51, d (11.3) |
| 12 | 21.5, CH3 | 1.37, s | 23.1, CH3 | 1.37, s | ||
| OH | 3.65, brs | |||||
| Compound | BGC-823 | H460 | PC-3 | SMMC-7721 |
|---|---|---|---|---|
| 6 | 6.8 | 23.6 | 28.1 | 7.9 |
| 7 | 48.0 | 87.8 | 55.1 | 40.2 |
| 8a/8b | 53.8 | 48.2 | 66.1 | 41.5 |
| Adriamycin | 1.5 | 1.0 | 1.8 | 2.2 |
| MIC | 1 | 2 | 3 | 6 | 7 | 8a/8b | Control |
|---|---|---|---|---|---|---|---|
| Bacillus subtilis | 50 | 25 | 100 | 3 | 50 | 50 | 0.25 a |
| Staphylococcus aureus | 25 | 25 | 100 | 3 | 25 | 50 | 0.13 a |
| Escherichia coli | - | - | - | - | - | - | 0.13 a |
| Candida albicans | 100 | - | - | - | - | - | 1.0 b |
| Candidad parapsilosis | 100 | - | 100 | - | - | - | 2.0 b |
| Cryptococcus neoformans | 100 | 100 | 100 | - | - | - | 2.0 b |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lei, H.; Lin, X.; Han, L.; Ma, J.; Ma, Q.; Zhong, J.; Liu, Y.; Sun, T.; Wang, J.; Huang, X. New Metabolites and Bioactive Chlorinated Benzophenone Derivatives Produced by a Marine-Derived Fungus Pestalotiopsis heterocornis. Mar. Drugs 2017, 15, 69. https://doi.org/10.3390/md15030069
Lei H, Lin X, Han L, Ma J, Ma Q, Zhong J, Liu Y, Sun T, Wang J, Huang X. New Metabolites and Bioactive Chlorinated Benzophenone Derivatives Produced by a Marine-Derived Fungus Pestalotiopsis heterocornis. Marine Drugs. 2017; 15(3):69. https://doi.org/10.3390/md15030069
Chicago/Turabian StyleLei, Hui, Xiuping Lin, Li Han, Jian Ma, Qingjuan Ma, Jialiang Zhong, Yonghong Liu, Tiemin Sun, Jinhui Wang, and Xueshi Huang. 2017. "New Metabolites and Bioactive Chlorinated Benzophenone Derivatives Produced by a Marine-Derived Fungus Pestalotiopsis heterocornis" Marine Drugs 15, no. 3: 69. https://doi.org/10.3390/md15030069
APA StyleLei, H., Lin, X., Han, L., Ma, J., Ma, Q., Zhong, J., Liu, Y., Sun, T., Wang, J., & Huang, X. (2017). New Metabolites and Bioactive Chlorinated Benzophenone Derivatives Produced by a Marine-Derived Fungus Pestalotiopsis heterocornis. Marine Drugs, 15(3), 69. https://doi.org/10.3390/md15030069





