From Discovery to Production: Biotechnology of Marine Fungi for the Production of New Antibiotics
Abstract
:1. Need for New Antibiotics
2. Marine Fungi as a Promising Source to Meet the Need for New Antibiotics
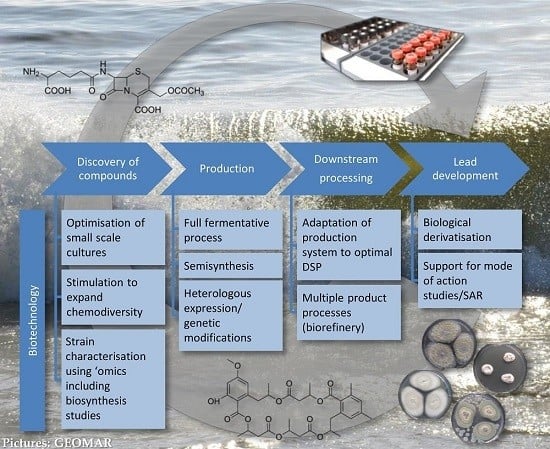
3. Biotechnology for Sustainable Production of New Antibiotics
3.1. Role of Biotechnology in Discovery
- Controlled miniaturisation for increased screening throughput. Cultivation in small-scale fermentation (mL ranges) systems, e.g., microtiter plates, has been proven to be feasible for marine fungi [61,62,63] and used in screening for antibiotics of filamentous fungi (in deep well microtiter plates [64]). Controlled process development as the biotechnological approach is possible in specialised miniaturised fermentation systems such as “System Duetz” [65] or “BioLector” [66,67]. They allow enlarging the number of tested cultivation conditions, as well as to screen strains or mutant libraries in a targeted and efficient way. A mutant library established from a marine fungus has recently been screened for anticancer natural products [63]. This bioactivity-independent approach can be easily transferred to the antibiotic field. For detailed information on the application of microtiter plates as mini-bioreactors the reader is referred to the review by Duetz [68]. Such systems are necessary for the efficient application of statistical approaches for process optimisation (Design of Experiment, DoE, [69,70]).
- Targeted stimulation of strains to expand chemodiversity. A proper understanding of fungal producers and their ecological role can help to find the appropriate production conditions and extend the chemical diversity of their constituents [71]. Standard approaches in recent years focussed on triggering a strain by as many parameters (randomly chosen) as possible (OSMAC) [21]. More strategic approaches would include targeted mixed fermentations based on genetic and ecological knowledge as used in the food biotechnology to enhance enzyme production. The full potential of such approaches of ecologically or genetically-based biotechnology needs to be proven in the future.
- Strain characterisation using ‘omics’ techniques. Comprehensive knowledge from genome to metabolome level contributes to a general understanding of the fungal potential in drug discovery but also to concrete optimisation strategies for biotechnological processes. Especially the analyses on proteome level (proteomics) may deliver valuable insights into the producing cell, underlying regulatory processes and angles for metabolic engineering [72]. Transcriptomics, proteomics, and secretomics can be applied to elucidate the metabolic state of a cell on all levels of gene regulation and to indicate regulation sites on DNA, RNA, and protein level. Based on such knowledge, conditions required to induce expression of the full biosynthetic potential of an organism can be established and further be controlled [60]. Until now, no such example is available for production of antibiotics by marine fungi, but this approach is considered as one of the major directions for future research. Current examples from a related field, anticancer compounds, already show how powerful these tools may be: A comparative proteome study on a marine Microascus brevicaulis revealed how the biotechnological fermentation process should be controlled in order to increase the production of the anti-cancer compounds scopularides A and B [73]. Furthermore, fluxomics, which determine the metabolic flux of primary molecules during primary and secondary metabolism in a quantitative manner, is a powerful tool to display the conversation of nutrient source into products or by-products [74]. This knowledge can be used to design fermentation conditions or to engineer the underlying pathways by means of genetic modification. Metabolomics, finally, identifies the global metabolite profile in both qualitative and quantitative manner.
3.2. Role of Biotechnology in Production
3.2.1. Production Using Full Fermentative Processes
3.2.2. Production Using Semi-Synthesis
3.2.3. Production Using Heterologous Systems and Genetic and Metabolic Engineering
3.3. Role of Biotechnology in Downstream Processing
3.4. Role of Biotechnology in Lead Development
4. Conclusions: “Success Factors” for Bridging the Gap from Discovery to Production
- Use of marine fungi. The biodiversity of filamentous fungi from marine sources is mostly untapped. Projects should extend knowledge on both biodiversity and chemodiversity of these unique organisms.
- Innovation in technology. There is a clear requirement for the “Next Generation Biotechnology”, which includes the new methodological approaches and the understanding of the underlying biological and technological processes.
- Transdisciplinarity. The transdisciplinary and integrative approach of developmental projects should encompass research and development partners comprising all stages of the early drug discovery pipeline, integrating academia, SMEs, and industry. Thus, important gaps in the knowledge of marine fungi relevant for the production of bioactive compounds should be actively addressed and made relevant to pharmaceutical drug discovery pipelines.
- Bridging the innovation gap. Researchers can promote innovation directly by employing novel techniques that promote the biosynthesis/production of new metabolites in early stage drug discovery (lead finding) programs and by enriching the pool of metabolites available for screening programs.
Author Contributions
Conflicts of Interest
Abbreviations
| ADAPT | Antibiotic Development to Advance Patient Treatment |
| DoE | Design of Experiment |
| DSP | Downstream Processing |
| EMF | Erlenmeyer Flask |
| MIC | Minimal Inhibitory Concentration |
| MIC | Minimal Inhibitory Concentration |
| MR | Methicillin-Resistant |
| OSMAC | One-Strain-Many-Compounds |
| Ref. | References |
| SAR | Structure-Activity Relationship |
| SME | Small and Medium Size Enterprises |
| STR | Stirred Tank Reactor |
| USP | Upstream Processing |
References
- Zhu, H.; Swierstra, J.; Wu, C.; Girard, G.; Choi, Y.H.; van Wamel, W.; Sandiford, S.K.; van Wezel, G.P. Eliciting antibiotics active against the ESKAPE pathogens in a collection of actinomycetes isolated from mountain soils. Microbiology 2014, 160, 1714–1725. [Google Scholar] [CrossRef] [PubMed]
- Antibacterial drugs market (By Class—Aminoglycosides, B-Lactams, Tetracyclines, Sulfonamides, Quinolones/ Fluoroquinolones, Macrolides, Phenicols and Miscellaneous Antibacterials, and Pipeline Drugs)—Global Industry Analysis, Size, Share, Growth, Trends and Forecast 2015–2023; Transparency Market Research: Albany, NY, USA, 2014.
- Hamad, B. The antibiotics market. Nat. Rev. Drug Discov. 2010, 9, 675–676. [Google Scholar] [CrossRef] [PubMed]
- Davies, J.; Davies, D. Origins and evolution of antibiotic resistance. MMBR 2010, 74, 417–433. [Google Scholar] [CrossRef] [PubMed]
- World Health Organisation. Antimicrobial Resistance: Global Report on Surveillance; WHO: Geneva, Switzerland, 2014; p. 256. [Google Scholar]
- WHO. World Health Statistics 2015; World Health Organization, Department of Health Statistics and Information Systems of the Health Systems and Innovation Cluster: Geneva, Switzerland, 2015; p. 161. [Google Scholar]
- Forsyth, C. Repairing the antibiotic pipeline: Can the gain act do it? Wash. J. Law Technol. Arts 2013. Available online: http://hdl.handle.net/1773.1/1267 (accessed on 13 July 2016). [Google Scholar]
- Demain, A.L. Importance of microbial natural products and the need to revitalise their discovery. J. Ind. Microbiol. Biotechnol. 2014, 41, 185–201. [Google Scholar] [CrossRef] [PubMed]
- Kong, D.X.; Jiang, Y.-Y.; Zhang, H.-Y. Marine natural products as sources of novel scaffolds: Achievement and concern. Drug Discov. Today 2010, 15, 884–886. [Google Scholar] [CrossRef] [PubMed]
- Song, F.; Ren, B.; Yu, K.; Chen, C.; Guo, H.; Yang, N.; Gao, H.; Liu, X.; Liu, M.; Tong, Y.; et al. Quinazolin-4-one coupled with pyrrolidin-2-iminium alkaloids from marine-derived fungus Penicillium aurantiogriseum. Mar. Drugs 2012, 10, 1297–1306. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Wang, Y.; Liu, D.; Proksch, P.; Yu, S.; Lin, W. Antioxidative phenolic compounds from a marine-derived fungus Aspergillus versicolor. Tetrahedron 2016, 72, 50–57. [Google Scholar] [CrossRef]
- Bhakuni, D.S.; Rawat, D.S. Bioactive Marine Natural Products; Springer: Rotterdam, The Netherlands, 2005; p. 382. [Google Scholar]
- Himaya, S.W.A.; Kim, S.-K. Marine symbiotic microorganisms: A new dimension in natural products research. In Marine Microbiology—Bioactive Compounds and Biotechnological Applications; Kim, S.-K., Ed.; Wiley-VCH: Singapore, 2013; pp. 295–306. [Google Scholar]
- Rateb, M.E.; Ebel, R. Secondary metabolites of fungi from marine habitats. Nat. Prod. Rep. 2011, 28, 290–344. [Google Scholar] [CrossRef] [PubMed]
- Thomas, T.R.; Kavlekar, D.P.; LokaBharathi, P.A. Marine drugs from sponge-microbe association—A review. Mar. Drugs 2010, 8, 1417–1468. [Google Scholar] [CrossRef] [PubMed]
- Richter, L.; Wanka, F.; Boecker, S.; Storm, D.; Kurt, T.; Vural, Ö.; Süßmuth, R.; Meyer, V. Engineering of Aspergillus niger for the production of secondary metabolites. Fungal Biol. Biotechnol. 2014, 1, 1–13. [Google Scholar] [CrossRef]
- Verma, V.C.; Gange, A.C. Advances in Endophytic Research; Springer: New Delhi, India, 2014; p. 454. [Google Scholar]
- Manohar, C.S.; Raghukumar, C. Fungal diversity from various marine habitats deduced through culture-independent studies. FEMS Microbiol. Lett. 2013, 341, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Brakhage, A.A.; Schroeckh, V. Fungal secondary metabolites—Strategies to activate silent gene clusters. Fungal Genet. Biol. 2011, 48, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Bugni, T.S.; Ireland, C.M. Marine-derived fungi: A chemically and biologically diverse group of microorganisms. Nat. Prod. Rep. 2004, 21, 143–163. [Google Scholar] [CrossRef] [PubMed]
- Bode, H.B.; Bethe, B.; Höfs, R.; Zeeck, A. Big effects from small changes: Possible ways to explore nature’s chemical diversity. ChemBioChem 2002, 3, 619–627. [Google Scholar] [CrossRef]
- Kusari, S.; Hertweck, C.; Spiteller, M. Chemical ecology of endophytic fungi: Origins of secondary metabolites. Chem. Biol. 2012, 19, 792–798. [Google Scholar] [CrossRef] [PubMed]
- Scherlach, K.; Hertweck, C. Triggering cryptic natural product biosynthesis in microorganisms. Org. Biomol. Chem. 2009, 7, 1753–1760. [Google Scholar] [CrossRef] [PubMed]
- Bode, H.B. No need to be pure: Mix the cultures! Chem. Biol. 2006, 13, 1245–1246. [Google Scholar] [CrossRef] [PubMed]
- Abraham, E.P.; Newton, G.G.; Crawford, K.; Burton, H.S.; Hale, C.W. Cephalosporin N: A new type of penicillin. Nature 1953. [Google Scholar] [CrossRef]
- Newton, G.G.F.; Abraham, E.P. Cephalosporin C, a new antibiotic containing sulphur and d-α-aminoadipic acid. Nature 1955, 175, 548–548. [Google Scholar] [CrossRef]
- Okutani, K. Gliotoxin produced by a strain of Aspergillus isolated from marine mud. Bull. Jap. Soc. Sci. Fish. 1977, 43, 995–1000. [Google Scholar] [CrossRef]
- Biabani, M.A.F.; Laatsch, H. Advances in chemical studies on low-molecular weight metabolites of marine fungi. J. Prakt Chem. 1998, 340, 589–607. [Google Scholar] [CrossRef]
- Hiort, J. Neue Naturstoffe aus schwamm-assoziierten Pilzen des Mittelmeeres. PhD thesis, Heinrich Heine University Düsseldorf, Düsseldorf, Germany, 2003. [Google Scholar]
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Munro, M.H.G.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2015, 32, 116–211. [Google Scholar] [CrossRef] [PubMed]
- Wei, M.Y.; Li, D.; Shao, C.L.; Deng, D.S.; Wang, C.Y. (±)-Pestalachloride D, an antibacterial racemate of chlorinated benzophenone derivative from a soft coral-derived fungus Pestalotiopsis sp. Mar. Drugs 2013, 11, 1050–1060. [Google Scholar] [CrossRef] [PubMed]
- Cuomo, V.; Palomba, I.; Perretti, A.; Guerriero, A.; D’Ambrosio, M.; Pietra, F. Antimicrobial activities from marine fungi. J. Mar. Biotech. 1995, 2, 199–204. [Google Scholar]
- Pruksakorn, P.; Arai, M.; Kotoku, N.; Vilcheze, C.; Baughn, A.D.; Moodley, P.; Jacobs, W.R.; Kobayashi, M. Trichoderins, novel aminolipopeptides from a marine sponge-derived Trichoderma sp., are active against dormant mycobacteria. Bioorg. Med. Chem. Lett. 2010, 20, 3658–3663. [Google Scholar] [CrossRef] [PubMed]
- Cueto, M.; Jensen, P.R.; Kauffman, C.; Fenical, W.; Lobkovsky, E.; Clardy, J. Pestalone, a new antibiotic produced by a marine fungus in response to bacterial challenge. J. Nat. Prod. 2001, 64, 1444–1446. [Google Scholar] [CrossRef] [PubMed]
- Waites, M.J.; Morgan, N.L.; Rockey, J.S.; Higton, G. Industrial Microbiology: An Introduction; Blackwell Science Ltd.: New York City, NY, US, 2001; p. 288. [Google Scholar]
- Bhadury, P.; Mohammad, B.T.; Wright, P.C. The current status of natural products from marine fungi and their potential as anti-infective agents. J. Ind. Microbiol. Biotechnol. 2006, 33, 325–37. [Google Scholar] [CrossRef] [PubMed]
- Quian, P.-Y.; Li, Y.; Kwong, F.N.; Yang, L.H.; Dobretsov, S.V. Use of Marine Fungus Originated Compounds as Antifouling Agents. U.S. Patent US2006/0147410 A1, 6 July 2006. [Google Scholar]
- Abbanat, D.; Leighton, M.; Maiese, W.; Jones, E.B.; Pearce, C.; Greenstein, M. Cell wall active antifungal compounds produced by the marine fungus Hypoxylon oceanicum LL-15G256. I. Taxonomy and fermentation. J. Antibiot. 1998, 51, 296–302. [Google Scholar] [CrossRef] [PubMed]
- Kanoh, K.; Okada, A.; Adachi, K.; Imagawa, H.; Nishizawa, M.; Matsuda, S.; Shizuri, Y.; Utsumi, R. Ascochytatin, a novel bioactive spirodioxynaphthalene metabolite produced by the marine-derived fungus, Ascochyta sp. NGB4. J. Antibiot. 2008, 61, 142–148. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Wiese, J.; Labes, A.; Kramer, A.; Schmaljohann, R.; Imhoff, J.F. Lindgomycin, an unusual antibiotic polyketide from a marine fungus of the Lindgomycetaceae. Mar. Drugs 2015, 13, 4617–4632. [Google Scholar] [CrossRef] [PubMed]
- Qi, S.-H.; Xu, Y.; Xiong, H.-R.; Qian, P.-Y.; Zhang, S. Antifouling and antibacterial compounds from a marine fungus Cladosporium sp. F14. World J. Microbiol. Biotechnol. 2008, 25, 399–406. [Google Scholar] [CrossRef]
- Silber, J.; Ohlendorf, B.; Labes, A.; Erhard, A.; Imhoff, J.F. Calcarides A–E, antibacterial macrocyclic and linear polyesters from a Calcarisporium strain. Mar. Drugs 2013, 11, 3309–3323. [Google Scholar] [CrossRef] [PubMed]
- Elander, R.P. Industrial production of β-lactam antibiotics. Appl. Microbiol. Biotechnol. 2003, 61, 385–392. [Google Scholar] [CrossRef] [PubMed]
- Naik, C.G.; Devi, P.; Rodrigues, E. Chrysogenazine obtained from Fungus Penicillium chrysogenum Having Antibacterial Activity. U.S. Patent US2005/0143392, 30 June 2005. [Google Scholar]
- Mikolasch, A.; Hessel, S.; Salazar, M.G.; Neumann, H.; Manda, K.; Gōrdes, D.; Schmidt, E.; Thurow, K.; Hammer, E.; Lindequist, U.; et al. Synthesis of new N-analogous corollosporine derivatives with antibacterial activity by laccase-catalyzed amination. Chem. Pharm. Bull. 2008, 56, 781–786. [Google Scholar] [CrossRef] [PubMed]
- Liberra, K.; Jansen, R.; Lindequist, U. Corollosporine, a new phtalide derivative from the marine fungus Corollospora maritima Werderm. 1069. Pharmazie 1998, 53, 578–581. [Google Scholar] [PubMed]
- Zobel, S.; Boecker, S.; Kulke, D.; Heimbach, D.; Meyer, V.; Süssmuth, R.D. Reprogramming the biosynthesis of cyclodepsipeptide synthetases to obtain new enniatins and beauvericins. ChemBioChem 2016, 17, 283–287. [Google Scholar] [CrossRef] [PubMed]
- Doshida, J.; Hasegawa, H.; Onuki, H.; Shimidzu, N. Exophilin A, a new antibiotic from a marine microorganism Exophilia pisciphila. J. Antibiot. 1996, 49, 1105–1109. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.D.; Savard, M.E. Antibiotic activity of the marine fungus Leptosphaeria oraemaris. Proc. N. S. Inst. Sci. 1989, 39, 51–58. [Google Scholar]
- Yin, Y.; Gao, Q.; Zhang, F.; Li, Z. Medium optimization for the high yield production of single (+)-terrein by Aspergillus terreus strain PF26 derived from marine sponge Phakellia fusca. Process Biochem. 2012, 47, 887–891. [Google Scholar] [CrossRef]
- Xiong, H.; Qi, S.; Xu, Y.; Miao, L.; Qian, P.-Y. Antibiotic and antifouling compound production by the marine-derived fungus Cladosporium sp. F14. J. Hydro Environ. Res. 2009, 2, 264–270. [Google Scholar] [CrossRef]
- Tamminen, A.; Wang, Y.; Wiebe, M.G. Production of calcaride A by Calcarisporium sp. in shaken flasks and stirred bioreactors. Mar. Drugs 2015, 13, 3992–4005. [Google Scholar] [CrossRef] [PubMed]
- Skatrud, P.L.; Tietz, A.J.; Ingolia, T.D.; Cantwell, C.A.; Fisher, D.L.; Chapman, J.L.; Queener, S.W. Use of recombinant DNA to improve production of cephalosporin C by Cephalosporium acremonium. Nature 1989, 7, 477–485. [Google Scholar] [CrossRef]
- Masuma, R.; Yamaguchi, Y.; Noumi, M.; Omura, S.; Namikoshi, M. Effect of sea water concentration on hyphal growth and antimicrobial metabolite production in marine fungi. Mycoscience 2001, 42, 455–459. [Google Scholar] [CrossRef]
- Xu, B.; Yin, Y.; Zhang, F.; Li, Z.; Wang, L. Operating conditions optimization for (+)-terrein production in a stirred bioreactor by Aspergillus terreus strain PF-26 from marine sponge Phakellia fusca. Bioprocess Biosyst. Eng. 2012, 35, 1651–1655. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.Y.; Laidlaw, R.D.; Marshall, J.; Keasling, J.D. Metabolic engineering of fungal secondary metabolic pathways. In Handbook of Industrial Mycology; An, Z.Q., Ed.; Marcel Dekker: New York, NY, USA, 2003; p. 10016. [Google Scholar]
- Miao, L.; Kwong, T.F.; Qian, P.-Y. Effect of culture conditions on mycelial growth, antibacterial activity, and metabolite profiles of the marine-derived fungus Arthrinium c.f. saccharicola. Appl. Microbiol. Biotechnol. 2006, 72, 1063–1073. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, R.; Molitoris, H.-P. High pressure cultivation of marine fungi: Apparatus and method. In High Pressure and Biotechnology, Proceedings of the First European Seminar on High Pressure and Biotechnology, a Joint Meeting with the Fifth Symposium on High Pressure and Food Science, La Grande Motte, France, 13–17 September 1992; Balny, C., Ed.; Libbey: London, UK, 1992; pp. 537–539. [Google Scholar]
- Ng, T.B.; Cheung, R.C.; Wong, J.H.; Bekhit, A.A.; Bekhit Ael, D. Antibacterial products of marine organisms. Appl. Microbiol. Biotechnol. 2015, 99, 4145–4173. [Google Scholar] [CrossRef] [PubMed]
- Monaghan, R.L.; Barrett, J.F. Antibacterial drug discovery—Then, now and the genomics future. Biochem. Pharmacol. 2006, 71, 901–909. [Google Scholar] [CrossRef] [PubMed]
- Linde, T.; Hansen, N.B.; Lübeck, M.; Lübeck, P.S. Fermentation in 24-well plates is an efficient screening platform for filamentous fungi. Lett. Appl. Microbiol. 2014, 59, 224–230. [Google Scholar] [CrossRef] [PubMed]
- Girarda, P.; Jordana, M.; Tsaob, M.; Wurma, F.M. Small-scale bioreactor system for process development and optimization. Biochem. Eng. J. 2001, 7, 117–119. [Google Scholar] [CrossRef]
- Kramer, A.; Paun, L.; Imhoff, J.F.; Kempken, F.; Labes, A. Development and validation of a fast and optimised screening method for enhanced production of secondary metabolites using the marine Scopulariopsis brevicaulis strain LF580 producing anti-cancer active scopularide A and B. PLoS ONE 2014, 9, e103320. [Google Scholar] [CrossRef] [PubMed]
- Bills, G.F.; Platas, G.; Fillola, A.; Jimenez, M.R.; Collado, J.; Vicente, F.; Martin, J.; Gonzalez, A.; Bur-Zimmermann, J.; Tormo, J.R.; et al. Enhancement of antibiotic and secondary metabolite detection from filamentous fungi by growth on nutritional arrays. J. Appl. Microbiol. 2008, 104, 1644–1658. [Google Scholar] [CrossRef] [PubMed]
- Duetz, W.A.; Ruedi, L.; Hermann, R.; O’Connor, K.; Buchs, J.; Witholt, B. Methods for intense aeration, growth, storage, and replication of bacterial strains in microtiter plates. Appl. Environ. Microbiol. 2000, 66, 2641–2646. [Google Scholar] [CrossRef] [PubMed]
- Samorski, M.; Müller-Newen, G.; Buchs, J. Quasi-continuous combined scattered light and fluorescence measurements: A novel measurement technique for shaken microtiter plates. Biotechnol. Bioeng. 2005, 92, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Kensy, F.; Zang, E.; Faulhammer, C.; Tan, R.K.; Buchs, J. Validation of a high-throughput fermentation system based on online monitoring of biomass and fluorescence in continuously shaken microtiter plates. Microb. Cell Fact. 2009. [Google Scholar] [CrossRef] [PubMed]
- Duetz, W.A. Microtiter plates as mini-bioreactors: Miniaturization of fermentation methods. Trends Microbiol. 2007, 15, 469–475. [Google Scholar] [CrossRef] [PubMed]
- Betts, J.I.; Baganz, F. Miniature bioreactors: Current practices and future opportunities. Microb. Cell Fact. 2006, 5, 21. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Posch, A.E.; Herwig, C.; Spadiut, O. Science-based bioprocess design for filamentous fungi. Trends Biotechnol. 2013, 31, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Joint, I.; Muhling, M.; Querellou, J. Culturing marine bacteria—An essential prerequisite for biodiscovery. Microb. Biotechnol. 2010, 3, 564–575. [Google Scholar] [CrossRef] [PubMed]
- Kniemeyer, O. Proteomics of eukaryotic microorganisms: The medically and biotechnologically important fungal genus Aspergillus. Proteomics 2011, 11, 3232–3243. [Google Scholar] [CrossRef] [PubMed]
- Kramer, A.; Beck, H.C.; Kumar, A.; Kristensen, L.P.; Imhoff, J.F.; Labes, A. Proteomic analysis of anti-cancerous scopularide production by a marine Microascus brevicaulis strain and its UV mutant. PLoS ONE 2015, 10, e0140047. [Google Scholar] [CrossRef] [PubMed]
- Knuf, C.; Nielsen, J. Aspergilli: Systems biology and industrial applications. Biotechnol. J. 2012, 7, 1147–1155. [Google Scholar] [CrossRef] [PubMed]
- Kjer, J.; Debbab, A.; Aly, A.H.; Proksch, P. Methods for isolation of marine-derived endophytic fungi and their bioactive secondary products. Nature Protoc. 2010, 5, 479–490. [Google Scholar] [CrossRef] [PubMed]
- Lang, S.; Hüners, M.; Lurtz, V. Bioprocess engineering data on the cultivation of marine prokaryotes and fungi. Adv. Biochem. Eng. Biotechnol. 2005, 97, 29–62. [Google Scholar] [PubMed]
- Riley, G.L.; Tucker, K.G.; Paul, G.C.; Thomas, C.R. Effect of biomass concentration and mycelial morphology on fermentation broth rheology. Biotechnol. Bioeng. 2000, 68, 160–172. [Google Scholar] [CrossRef]
- Balakrishnan, K.; Pandey, A. Production of biologically active metabolites in solid-state fermentation. J. Sci. Ind. Res. 1996, 55, 365–372. [Google Scholar]
- Barrios-Gonzalez, J.; Meji´a, A. Production of secondary metabolites by solid-state fermentation. Biotechnol. Annu. Rev. 1996, 2, 85–88. [Google Scholar] [PubMed]
- Höllker, U.; Höfer, M.; Lenz, J. Biotechnological advantages of laboratory-scale solid state fermentation with fungi. Appl. Microbiol. Biotechnol. 2004, 64, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Robinson, T.; Singh, D.; Nigam, P. Solid-state fermentation: A promising microbial technology for secondary metabolite production. Appl. Microbiol. Biotechnol. 2001, 55, 284–289. [Google Scholar] [CrossRef] [PubMed]
- Papagianni, M. Fungal morphology and metabolite production in submerged mycelial processes. Biotechnol. Adv. 2004, 22, 189–259. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, P.A.; Seviour, R.J.; Schmid, F. Growth of filamentous fungi in submerged culture: Problems and possible solutions. Crit. Rev. Biotechnol. 2000, 20, 17–48. [Google Scholar] [CrossRef] [PubMed]
- Lara, A.R.; Galindo, E.; Ramírez, O.T.; Palomares, L.A. Living with heterogeneities in bioreactors: Understanding the effects of environmental gradients on cells. Mol. Biotechnol. 2006, 34, 355–382. [Google Scholar] [CrossRef]
- Neubauer, P.; Junne, S. Scale-down simulators for metabolic analysis of large-scale bioprocesses. Curr. Opin. Biotechnol. 2010, 21, 114–121. [Google Scholar] [CrossRef] [PubMed]
- Vardar, F.; Lilly, M.D. Effect of cycling oxygen concentrations on product formation in penicillin fermentations. Eur. J. Appl. Microbiol. Biotechnol. 1982, 14, 203–211. [Google Scholar] [CrossRef]
- Kaup, B.A.; Ehrich, K.; Pescheck, M.; Schrader, J. Microparticle-enhanced cultivation of filamentous microorganisms: Increased chloroperoxidase formation by Caldariomyces fumago as an example. Biotechnol. Bioeng. 2008, 99, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.Y.; Jin, B.; Kelly, J.M. Effects of cultivation parameters on the morphology of Rhizopus arrhizus and the lactic acid production in a bubble column reactor. Eng. Life Sci. 2007, 7, 490–496. [Google Scholar] [CrossRef]
- Krull, R.; Wucherpfennig, T.; Esfandabadi, M.E.; Walisko, R.; Melzer, G.; Hempel, D.C.; Kampen, I.; Kwade, A.; Wittmann, C. Characterization and control of fungal morphology for improved production performance in biotechnology. J. Biotechnol. 2013, 163, 112–123. [Google Scholar] [CrossRef] [PubMed]
- Ozcengiz, G.; Demain, A.L. Recent advances in the biosynthesis of penicillins, cephalosporins and clavams and its regulation. Biotechnol. Adv. 2013, 31, 287–311. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, S.; Pramanik, A.; Mitra, A.; Mukherjee, J. Bioprocessing data for the production of marine enzymes. Mar. Drugs 2010, 8, 1323–1372. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, J. It is all about metabolic fluxes. J. Bacteriol. 2003, 185, 7031–7035. [Google Scholar] [CrossRef] [PubMed]
- Schneider, P.; Misiek, M.; Hoffmeister, D. In vivo and in vitro production options for fungal secondary metabolites. Mol. Pharm. 2008, 5, 234–242. [Google Scholar] [CrossRef] [PubMed]
- Brotzu, G. Ricerche su di un nuovo antibiotico. Lavori dell'Istituto di Igiene di Cagliari 1948. Available online: http://medicina.unica.it/pacs/brotzu.pdf (accessed on 13 July 2016). [Google Scholar]
- Kück, U.; Bloemendal, S.; Teichert, I. Putting fungi to work: Harvesting a cornucopia of drugs, toxins, and antibiotics. PLoS ONE 2014, 10, e1003950. [Google Scholar] [CrossRef] [PubMed]
- Abraham, E.P.; Loder, P.B. Cephalosporin C. In Cephalosporins and Penicillins; Flynn, E.H., Ed.; Elsevier: Amsterdam, The Netherlands, 1972; pp. 1–26. [Google Scholar]
- Wright, P.M.; Seiple, I.B.; Myers, A.G. The evolving role of chemical synthesis in antibacterial drug discovery. Angew. Chem. Int. Ed. Engl. 2014, 53, 8840–8869. [Google Scholar] [CrossRef] [PubMed]
- Mancini, I.; Defant, A.; Guella, G. Recent synthesis of marine natural products with antibacterial activities. Anti Infect. Agents Med. Chem. 2007, 6, 17–48. [Google Scholar] [CrossRef]
- Bugni, T.S.; Abbanat, D.; Bernan, V.S.; Maiese, W.M.; Greenstein, M.; van Wagoner, R.M.; Ireland, C.M. Yanuthones: Novel metabolites from a marine isolate of Aspergillus niger. J. Org. Chem. 2000, 65, 7195–7200. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Z.Q.; Wang, J.F.; Hao, Y.Y.; Wang, Y. Recent advances in the discovery and development of marine microbial natural products. Mar. Drugs 2013, 11, 700–717. [Google Scholar] [CrossRef] [PubMed]
- Meyer, V.; Wu, B.; Ram, A.F. Aspergillus as a multi-purpose cell factory: Current status and perspectives. Biotechnol. Lett. 2011, 33, 469–476. [Google Scholar] [CrossRef] [PubMed]
- Wanka, F.; Cairns, T.; Boecker, S.; Berens, C.; Happel, A.; Zheng, X.; Sun, J.; Krappmann, S.; Meyer, V. Tet-on, or Tet-off, that is the question: Advanced conditional gene expression in Aspergillus. Fungal Genet. Biol. 2016, 89, 72–83. [Google Scholar] [CrossRef] [PubMed]
- Helmschrott, C.; Sasse, A.; Samantaray, S.; Krappmann, S.; Wagener, J. Upgrading fungal gene expression on demand: Improved systems for doxycycline-dependent silencing in Aspergillus fumigatus. Appl. Environ. Microbiol. 2013, 79, 1751–1754. [Google Scholar] [CrossRef] [PubMed]
- Anyaogu, D.C.; Mortensen, U.H. Heterologous production of fungal secondary metabolites in Aspergilli. Front. Microbiol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Lukassen, M.B.; Saei, W.; Sondergaard, T.E.; Tamminen, A.; Kumar, A.; Kempken, F.; Wiebe, M.G.; Sorensen, J.L. Identification of the scopularide biosynthetic gene cluster in Scopulariopsis brevicaulis. Mar. Drugs 2015, 13, 4331–4343. [Google Scholar] [CrossRef] [PubMed]
- Muffler, K.; Ulber, R. Downstream processing in marine biotechnology. Adv. Biochem. Eng. Biotechnol. 2005, 97, 63–103. [Google Scholar] [PubMed]
- Jungbauer, A. Continuous downstream processing of biopharmaceuticals. Trends Biotechnol. 2013, 31, 479–492. [Google Scholar] [CrossRef] [PubMed]
- Zydney, A.L. Continuous downstream processing for high value biological products: A review. Biotechnol. Bioeng. 2016, 113, 465–475. [Google Scholar] [CrossRef] [PubMed]
- Lightfoot, E.N.; Moscariello, J.S. Bioseparations. Biotechnol. Bioeng. 2004, 87, 259–273. [Google Scholar] [CrossRef] [PubMed]
- Weissman, S.A.; Anderson, N.G. Design of experiments (DoE) and process optimization. A review of recent publications. Org. Process Res. Dev. 2015, 19, 1605–1633. [Google Scholar] [CrossRef]
- Barber, M.S.; Giesecke, U.; Reichert, A.; Minas, W. Industrial enzymatic production of cephalosporin-based beta-lactams. Adv. Biochem. Eng. Biotechnol. 2004, 88, 179–215. [Google Scholar] [PubMed]
- Takimoto, A.; Mitsushima, K.; Yagi, S.; Sonoyama, T. Purification, characterization and partial amino-acid-sequences of a novel cephalosporin-C deacetylase from Bacillus subtilis. J. Ferment. Bioeng. 1994, 77, 17–22. [Google Scholar] [CrossRef]
- Sonawane, V.C. Enzymatic modifications of cephalosporins by cephalosporin acylase and other enzymes. Crit. Rev. Biotechnol. 2006, 26, 95–120. [Google Scholar] [CrossRef] [PubMed]
- Abbott, B.J. Preparation of pharmaceutical compounds by immobilized enzymes and cells. Adv. Appl. Microbiol. 1976, 20, 203–257. [Google Scholar] [PubMed]
- US House of Representatives. Antibiotic Development to Advance Patient Treatment; Press: Washington, DC, USA, 2013. [Google Scholar]
 |  |  |
| 3-Chloro-2,5-dihydroxy benzyl alcohol [37] | 15G265α [38] | Ascochytatin [39] |
 |  |  |
| Ascosetin [40] | Bis(2-ethylhexyl)phthalate [41] | Calcaride A [42] |
 |  | 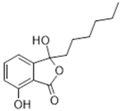 |
| Cephalosporin C [43] | Chrysogenazine [44] | Corollosporine [45,46] |
 |  |  |
| Cyclo-(Pro-Phe) [37] | Enniatin B [47] | Exophilin A [48] |
 |  | 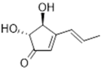 |
| Lindgomycin x [40] | Obioninene [49] | (+)-Terrein [50] |
| Compound, Chemical Class | Producer, Origin | Biotechnological Approach | Antibiotic Activity Against | Ref. |
|---|---|---|---|---|
| 15G265α,β,γ macrocyclic polylactones and lipodepsipeptide | Hypoxylon oceanicum LL‑15G256, mangrove | Optimised medium to increase titres Effect of seawater (negative at low temperature) Transfer to Fernbach flasks and 300-L fermenter | Staphylococcus epidermidis, Xanthomonas campestris Propionibacterium acnes | [38,42] |
| Ascochytatin, spirodioxynaphthalene | Ascochyta sp. NGB4, floating scrap of festering rope collected at a fishing port | Optimisation of medium at small scale | Bacterial two-component regulatory system | [39] |
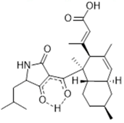
| Ascosetin, tetramic acid | Lindgomycetaceae, Halichondria panicea, (sponge from Baltic Sea) | Transfer from EMF to STR (10 L): adaptation of medium, increase of yield (factor 100) and decrease of cultivation time | S. epidermidis, S. aureus, MR S. aureus, P. acnes, X. campestris, Septoria tritici | [40] |
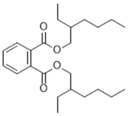
| Bis(2-ethylhexyl)phthalate, phthalate * | Cladosporium sp., sea water in mangrove area | Transfer from EMF to STR Record of conditions Scaling (2-L fermenter) | Loktanella hongkongensis, M. luteus, Rhodovulum sp., Ruegeria sp., Pseudoalteromonas piscida, Vibrio harveyi | [41,51] |
| Calcarides A–E, macrocyclic and linear polyesters | Calcarisporium sp., Wadden sea water | Biosynthesis study for strain characterisation Biological derivatisation For calcaride A: Adaptation of medium in flasks (13‑fold improvement) STR: 200-fold improvement by pH adaptation, C/N ratio, nature of mycelial growth | Macrocyclic compounds: S. epidermidis, X. campestris linear polyesters: no antibiotic activity | [42,52] |
| Cephalosporin, β-lactam | Aspergillus chrysogenum, sewage water | Full fermentative optimised process, titres up to 25 g/L Semi-synthesis from 7‑aminocephalosporanic acid (enzymatic) Genetic engineering to reduce by‑products Enzymatic treatment in DSP Immobilised cells in a repeated batch tower reactor | Broad spectrum | [43] |
| Cephalosporium chrysogenum, sea water | DNA modified by mutagenesis | Broad spectrum | [53] | |
| 3-Chloro-2,5-dihydroxy benzyl alcohol, benzene derivative | Ampelomyces sp., marine biofilm | Scaling in EMF | Micrococcus sp., Vibrio sp., Pseudoalteromonas sp., S. aureus, S. haemolyticus | [37] |
| Chrysogenazine, diketopiperazine | Penicillium chrysogenum, Porteresia coarctata (mangrove plant, leaves) | Scaling from 1-L to 5-L flasks Yield of the compound enhanced by modifying the carbon and nitrogen source | Vibrio cholera | [44] |
| Corollosporin and derivates, phthalide derivatives | Corollospora maritima, marine driftwood | Biological derivatives by enzymatic treatment Salt dependency of fermentation | Candida maltosa, Escherichia coli, Pseudomonas aeruginosa, Bacillus subtilis, S. aureus, S. aureus North German epidemic strain, S. epidermidis, S. haemolyticus | [45,54] |
| Cyclo-(Pro-Phe), diketopiperazine | Unidentified marine fungus UST030110-009, marine biofilm | Scaling in EMF | Antibacterial antibiofilm: Micrococcus sp., Vibrio sp., Pseudoalteromonas, S. aureus, S. haemolyticus | [37] |
| Enniatins, cyclodepsipeptides | Halosarpheia sp., mangrove | Heterologous reprogramming of biosynthetic pathways | E. coli, Enterococcus faecium, Salmonella enterica, Shigella dysenteriae, Listeria monocytogenes, Yersinia enterocolitica, Clostridium perfringens, P. aeruginosa, S. aureus | [47] |
| Exophilin A, 3,5-dihydroxy-decanoic polyester | Exophiala pisciphila, Mycale adhaerens (sponge) | Transfer from EMF to STR (glass bottle fermenter, 20 L) | E. facium, E. faecalis, S. aureus, MR S. aureus | [48] |
| Lindgomycin, tetramic acid | Lindgomycetaceae, Halichondria panicea (sponge from Baltic Sea) | Adaptation of medium Transfer from EMF to STR (10 L): increase of yield (factor 100) and decrease of cultivation time (from 14 to 7 days) | MR S. aureus, S. epidermidis, P. acnes, X. campestris, S. tritici | [40] |
| Obioninene, ortho-quinone | Leptosphaeria oraemaris, marine driftwood | Effect of salinity on antibiotic production (in EMF) | Fucus-associated not identified bacterium | [49] |
| (+)-Terrein, cyclopentenone | Aspergillus terreus PF-26, Phakellia fusca (sponge) | Optimisation of operating factors (5-L STR) such as inoculation, agitation speed, aeration rate, pH control and nutrient feeding | B. subtilis | [55] |
| Not determined, sesterterpenoid | Fusarium heterosporum and Aspergillus versicolor, driftwood and alga | Metabolic engineering | Broad spectrum | [56] |
| Not determined | Arthrinium c.f. saccharicola, seawater from a mangrove habitat | Co-culture Stimulation with bacterial elucidators Systematic manipulation of culture conditions: salinity, temperature, pH, and culture medium composition | Pseudoalteromonas spongiae, Vibrio vulnificus | [57] |
| Not determined | Obligate fungi, marine deep-sea habitats | High pressure cultivation Scaling 20–100 L | Broad spectrum | [58] |
© 2016 by the author; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silber, J.; Kramer, A.; Labes, A.; Tasdemir, D. From Discovery to Production: Biotechnology of Marine Fungi for the Production of New Antibiotics. Mar. Drugs 2016, 14, 137. https://doi.org/10.3390/md14070137
Silber J, Kramer A, Labes A, Tasdemir D. From Discovery to Production: Biotechnology of Marine Fungi for the Production of New Antibiotics. Marine Drugs. 2016; 14(7):137. https://doi.org/10.3390/md14070137
Chicago/Turabian StyleSilber, Johanna, Annemarie Kramer, Antje Labes, and Deniz Tasdemir. 2016. "From Discovery to Production: Biotechnology of Marine Fungi for the Production of New Antibiotics" Marine Drugs 14, no. 7: 137. https://doi.org/10.3390/md14070137
APA StyleSilber, J., Kramer, A., Labes, A., & Tasdemir, D. (2016). From Discovery to Production: Biotechnology of Marine Fungi for the Production of New Antibiotics. Marine Drugs, 14(7), 137. https://doi.org/10.3390/md14070137