Antimicrobial Activity of Monoramnholipids Produced by Bacterial Strains Isolated from the Ross Sea (Antarctica) †
Abstract
:1. Introduction
2. Results and Discussion
2.1. Isolation of Bacteria, Typing and Phylogenetic Analysis
- (i)
- As expected on the basis of the sharing of RAPD profiles, the six strains exhibiting the same RAPD profile (RAPD haplotype 1) share the same 16S rRNA gene sequence and were clustered together joining the species Pseudomonas azotoformans.
- (ii)
- Strain BTN4 was affiliated to the genus Arthrobacter.
- (iii)
- All the other strains were affiliated to the genus Psychrobacter and, according to the different RAPD profiles they exhibited, joined different Psychrobacter clades. The three strains (BTN20A, BTN24 and BTN 20B) sharing the same RAPD profile (RAPD haplotype 4), joined the same Psychrobacter cluster.
2.2. Cross-Streaking Experiments
2.3. Extracts’ Antimicrobial Assays
2.4. Bioassay-Guided Purification of BTN1 Extract
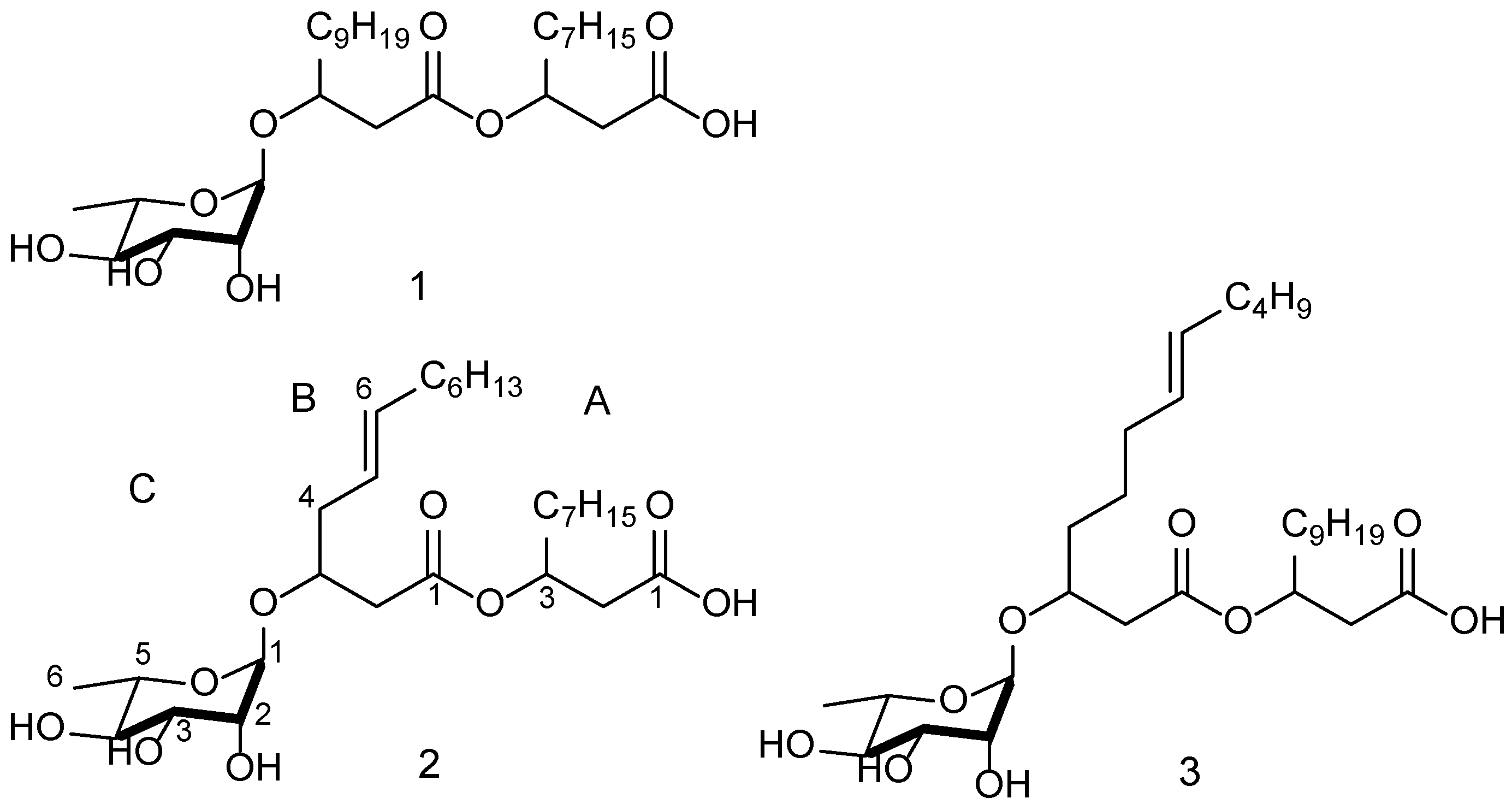
2.5. Compound Structure Elucidation
2.6. Antimicrobial Activity of BTN1 Pure Compounds
3. Experimental Section
3.1. Isolation of Bacterial Strains
3.2. Target Strains and Growth Conditions
3.3. RAPD Analysis
3.4 Phylogenetic Affiliation of BTN Strains
3.5. Cross-Streaking
3.6. Extract Preparation
3.7. Antimicrobial Assays
3.7.1. Minimal Inhibitory Concentration Assay (MIC)
3.7.2. Minimal Bactericidal Concentration (MBC) Assay
3.8. Purification of Ethyl-Acetate Crude Extract
3.9. NMR–LCMS Experiments
4. Conclusions
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Berdy, J. Thoughts and facts about antibiotics: Where we are now and where we are heading. J. Antibiot. 2012, 65, 385–395. [Google Scholar] [CrossRef] [PubMed]
- Tegos, G.P.; Hamblin, M.R. Disruptive innovations: New anti-infectives in the age of resistance. Curr. Opin. Pharmacol. 2013, 13, 673–677. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J. Nat. Prod. 2012, 75, 311–335. [Google Scholar] [CrossRef] [PubMed]
- Bologa, C.G.; Ursu, O.; Oprea, T.I.; Melancon, C.E., 3rd; Tegos, G.P. Emerging trends in the discovery of natural product antibacterials. Curr. Opin. Pharmacol. 2013, 13, 678–687. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Ma, X.H.; Qin, C.; Tao, L.; Liu, X.; Shi, Z.; Zhang, C.L.; Tan, C.Y.; Chen, Y.Z.; Jiang, Y.Y. Drug discovery prospect from untapped species: Indications from approved natural product drugs. PLoS ONE 2012, 7, e39782. [Google Scholar] [CrossRef] [PubMed]
- Jaspars, M.; Challis, G. Microbiology: A talented genus. Nature 2014, 506, 38–39. [Google Scholar] [CrossRef] [PubMed]
- Von Salm, J.L.; Wilson, N.G.; Vesely, B.A.; Kyle, D.E.; Cuce, J.; Baker, B.J. Shagenes A and B, new tricyclic sesquiterpenes produced by an undescribed Antarctic octocoral. Org. Lett. 2014, 16, 2630–2633. [Google Scholar] [CrossRef] [PubMed]
- Godinho, V.M.; Goncalves, V.N.; Santiago, I.F.; Figueredo, H.M.; Vitoreli, G.A.; Schaefer, C.E.; Barbosa, E.C.; Oliveira, J.G.; Alves, T.M.; Zani, C.L.; et al. Diversity and bioprospection of fungal community present in oligotrophic soil of continental Antarctica. Extremophiles 2015, 19, 585–596. [Google Scholar] [CrossRef] [PubMed]
- Lee, L.H.; Cheah, Y.K.; Nurul Syakima, A.M.; Shiran, M.S.; Tang, Y.L.; Lin, H.P.; Hong, K. Analysis of Antarctic proteobacteria by PCR fingerprinting and screening for antimicrobial secondary metabolites. Genet. Mol. Res. 2012, 11, 1627–1641. [Google Scholar] [CrossRef] [PubMed]
- Papa, R.; Parrilli, E.; Sannino, F.; Barbato, G.; Tutino, M.L.; Artini, M.; Selan, L. Anti-biofilm activity of the Antarctic marine bacterium Pseudoalteromonas haloplanktis TAC125. Res. Microbiol. 2013, 164, 450–456. [Google Scholar] [CrossRef] [PubMed]
- Papaleo, M.C.; Fondi, M.; Maida, I.; Perrin, E.; Lo Giudice, A.; Michaud, L.; Mangano, S.; Bartolucci, G.; Romoli, R.; Fani, R. Sponge-associated microbial Antarctic communities exhibiting antimicrobial activity against Burkholderia cepacia complex bacteria. Biotechnol. Adv. 2012, 30, 272–293. [Google Scholar] [CrossRef] [PubMed]
- Papaleo, M.C.; Romoli, R.; Bartolucci, G.; Maida, I.; Perrin, E.; Fondi, M.; Orlandini, V.; Mengoni, A.; Emiliani, G.; Tutino, M.L.; et al. Bioactive volatile organic compounds from Antarctic (sponges) bacteria. N. Biotechnol. 2013, 30, 824–838. [Google Scholar] [CrossRef] [PubMed]
- Romoli, R.; Papaleo, M.C.; de Pascale, D.; Tutino, M.L.; Michaud, L.; LoGiudice, A.; Fani, R.; Bartolucci, G. Characterization of the volatile profile of Antarctic bacteria by using solid-phase microextraction-gas chromatography-mass spectrometry. J. Mass Spectrom. 2011, 46, 1051–1059. [Google Scholar] [CrossRef] [PubMed]
- Maida I., B.E.; Fondi, M.; Perrin, E.; Orlandini, V.; Papaleo, M.C.; Mengoni, A.; de Pascale, D.; Tutino, M.L.; Michaud, L.; Lo Giudice, A.; et al. Antimicrobial activity of Pseudoalteromonas strains isolated from the ross sea (Antarctica) vs. cystic fibrosis opportunistic pathogens. Hydrobiologia 2015, in press. [Google Scholar]
- Romoli, R.; Papaleo, M.C.; de Pascale, D.; Tutino, M.L.; Michaud, L.; Lo Giudice, A.; Fani, R.; Bartolucci, G. GC-MS volatolomic approach to study the antimicrobial activity of the Antarctic bacterium Pseudoalteromonas sp.Tb41. Metabolomics 2013, 10, 42–51. [Google Scholar]
- Coenye, T.; Vandamme, P. Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ. Microbiol. 2003, 5, 719–729. [Google Scholar] [CrossRef] [PubMed]
- Compant, S.; Nowak, J.; Coenye, T.; Clement, C.; Ait Barka, E. Diversity and occurrence of Burkholderia spp. In the natural environment. FEMS Microbiol. Rev. 2008, 32, 607–626. [Google Scholar] [CrossRef] [PubMed]
- De Smet, B.; Mayo, M.; Peeters, C.; Zlosnik, J.E.; Spilker, T.; Hird, T.J.; LiPuma, J.J.; Kidd, T.J.; Kaestli, M.; Ginther, J.L.; et al. Burkholderia stagnalis sp. Nov. And Burkholderia territorii sp. Nov., two novel Burkholderia cepacia complex species from environmental and human sources. Int. J. Syst. Evol. Microbiol. 2015, 65, 2265–2271. [Google Scholar] [CrossRef] [PubMed]
- Drevinek, P.; Mahenthiralingam, E. Burkholderia cenocepacia in cystic fibrosis: Epidemiology and molecular mechanisms of virulence. Clin. Microbiol. Infect. 2010, 16, 821–830. [Google Scholar] [CrossRef] [PubMed]
- Ramsay, K.A.; Butler, C.A.; Paynter, S.; Ware, R.S.; Kidd, T.J.; Wainwright, C.E.; Bell, S.C. Factors influencing acquisition of Burkholderia cepacia complex organisms in patients with cystic fibrosis. J. Clin. Microbiol. 2013, 51, 3975–3980. [Google Scholar] [CrossRef] [PubMed]
- Rose, H.; Baldwin, A.; Dowson, C.G.; Mahenthiralingam, E. Biocide susceptibility of the Burkholderia cepacia complex. J. Antimicrob. Chemother. 2009, 63, 502–510. [Google Scholar] [CrossRef] [PubMed]
- Nash, E.F.; Thomas, A.; Whitmill, R.; Rashid, R.; Barker, B.; Rayner, R.J.; Whitehouse, J.L.; Honeybourne, D. “Cepacia syndrome” associated with Burkholderia cepacia (Genomovar I) infection in an adolescent with cystic fibrosis. Pediatr. Pulmonol. 2010, 46, 512–514. [Google Scholar] [CrossRef] [PubMed]
- Tegos, G.P.; Haynes, M.K.; Schweizer, H.P. Dissecting novel virulent determinants in the Burkholderia cepacia complex. Virulence 2012, 3, 234–237. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Jansen, R.; Nimtz, M.; Johri, B.N.; Wray, V. Rhamnolipids from the rhizosphere bacterium Pseudomonas sp. Grp(3) that reduces damping-off disease in chilli and tomato nurseries. J. Nat. Prod. 2007, 70, 941–947. [Google Scholar] [CrossRef] [PubMed]
- De Bruyn, A.; Anteunis, M. 1H-N.m.r. study of l-rhamnose, methyl alpha-l-rhamnopyranoside, and 4-O-beta-d-galactopranosyl-l-rhamnose in deuterium oxide. Carbohydr. Res. 1976, 47, 158–163. [Google Scholar] [CrossRef]
- Das, P.; Mukherjee, S.; Sen, R. Antimicrobial potential of a lipopeptide biosurfactant derived from a marine Bacillus circulans. J. Appl. Microbiol. 2008, 104, 1675–1684. [Google Scholar] [CrossRef] [PubMed]
- Das, P.; Yang, X.P.; Ma, L.Z. Analysis of biosurfactants from industrially viable pseudomonas strain isolated from crude oil suggests how rhamnolipids congeners affect emulsification property and antimicrobial activity. Front. Microbiol. 2014, 5, 696. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Mawgoud, A.M.; Lepine, F.; Deziel, E. Rhamnolipids: Diversity of structures, microbial origins and roles. Appl. Microbiol. Biotechnol. 2010, 86, 1323–1336. [Google Scholar] [CrossRef] [PubMed]
- Haba, E.; Pinazo, A.; Jauregui, O.; Espuny, M.J.; Infante, M.R.; Manresa, A. Physicochemical characterization and antimicrobial properties of rhamnolipids produced by Pseudomonas aeruginosa 47T2 NCBIM 40044. Biotechnol. Bioeng. 2003, 81, 316–322. [Google Scholar] [CrossRef] [PubMed]
- Benincasa, M.; Abalos, A.; Oliveira, I.; Manresa, A. Chemical structure, surface properties and biological activities of the biosurfactant produced by Pseudomonas aeruginosa lBI from soapstock. Antonie van Leeuwenhoek 2004, 85, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Haba, E.; Bouhdid, S.; Torrego-Solana, N.; Marques, A.M.; Espuny, M.J.; Garcia-Celma, M.J.; Manresa, A. Rhamnolipids as emulsifying agents for essential oil formulations: Antimicrobial effect against Candidaalbicans and methicillin-resistant Staphylococcus aureus. Int. J. Pharm. 2014, 476, 134–141. [Google Scholar] [CrossRef] [PubMed]
- Sotirova, A.V.; Spasova, D.I.; Galabova, D.N.; Karpenko, E.; Shulga, A. Rhamnolipid-biosurfactant permeabilizing effects on gram-positive and gram-negative bacterial strains. Curr. Microbiol. 2008, 56, 639–644. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.G.; Kubelik, A.R.; Livak, K.J.; Rafalski, J.A.; Tingey, S.V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990, 18, 6531–6535. [Google Scholar] [CrossRef] [PubMed]
- Welsh, J.; McClelland, M. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 1990, 18, 7213–7218. [Google Scholar] [CrossRef] [PubMed]
- Mori, E.; Lio, P.; Daly, S.; Damiani, G.; Perito, B.; Fani, R. Molecular nature of RAPD markers from Haemophilus influenzae Rd genome. Res. Microbiol. 1999, 150, 83–93. [Google Scholar] [CrossRef]
- Grifoni, A.; Bazzicalupo, M.; Di Serio, C.; Fancelli, S.; Fani, R. Identification of Azospirillum strains by restriction fragment length polymorphism of the 16s rDNA and of the histidine operon. FEMS Microbiol. Lett. 1995, 127, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. Clustal W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Bharali, P.; Saikia, J.P.; Ray, A.; Konwar, B.K. Rhamnolipid (RL) from Pseudomonas aeruginosa OBP1: A novel chemotaxis and antibacterial agent. Colloids Surf. B Biointerfaces 2013, 103, 502–509. [Google Scholar] [CrossRef] [PubMed]
- Dobler, L.; Vilela, L.F.; Almeida, R.V.; Neves, B.C. Rhamnolipids in perspective: Gene regulatory pathways, metabolic engineering, production and technological forecasting. N. Biotechnol. 2016, 33, 123–135. [Google Scholar] [CrossRef] [PubMed]
| Genus | Strains | RAPD Profile | Accession Number |
|---|---|---|---|
| Pseudomonas | BTN1 | 1 | KT989002 |
| BTN6 | KT989003 | ||
| BTN7 | KT989004 | ||
| BTN8 | KT989005 | ||
| BTN9 | KT989006 | ||
| BTN10 | KT989007 | ||
| Psychrobacter | BTN3 | 2 | KT989009 |
| BTN19 | 3 | KT989019 | |
| BTN20B | 4 | KT989021 | |
| BTN24 | KT989022 | ||
| BTN21 | 5 | KT989025 | |
| BTN23 | 6 | KT989024 | |
| BTN2 | 7 | KT989008 | |
| BTN11 | 8 | KT989011 | |
| BTN5 | 9 | KT989010 | |
| BTN20A | 4 | KT989020 | |
| BTN15 | 10 | KT989015 | |
| BTN13 | 11 | KT989012 | |
| BTN14 | 12 | KT989013 | |
| BTN17 | 13 | KT989017 | |
| BTN16 | 14 | KT989016 | |
| BTN18 | 15 | KT989018 | |
| BTN12 | 16 | KT989014 | |
| BTN22 | 17 | KT989023 | |
| Arthrobacter | BTN4 | 18 | KT989001 |
| Bcc Strain | S | BTN Strain | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 5 | 11 | 13 | 14 | 4 | 12 | 15 | 16 | 17 | 18 | 19 | 20 a | 20 b | 21 | 22 | 23 | C+ | ||
| B. ambifaria LMG 19182 | W | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | + |
| N | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | + | |
| B. anthina LMG 20980 | W | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | + |
| N | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | + | |
| B. vietnamensis LMG10929 | W | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | ± | - | - | - | + |
| N | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | + | |
| B. cenocepacia LMG 16656 | W | ± | - | ± | ± | - | ± | - | - | - | - | - | ± | ± | - | ± | ± | - | - | - | + |
| N | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | + | |
| B. cepacia LMG 1222 | W | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | + |
| N | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | + | |
| B. contaminas LMG 23361 | W | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | + |
| N | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | + | |
| B. diffusa LMG 24065 | W | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | + |
| N | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | + | |
| B. dolosa LMG 18943 | W | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | + |
| N | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | + | |
| B. lata LMG 22485 | W | ± | ± | ± | ± | ± | ± | ± | - | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | + |
| N | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | + | |
| B. latens LMG 24064 | W | - | - | ± | ± | ± | - | - | - | - | ± | ± | ± | ± | - | ± | ± | - | - | ± | + |
| N | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | + | |
| B. metallica LMG 24068 | W | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | + |
| N | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | + | |
| B. multivorans LMG 13010 | W | - | ± | ± | ± | ± | ± | ± | - | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | + |
| N | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | + | |
| B. pseudomultivorans LMG 26883 | W | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | + |
| N | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | + | |
| B. pyrrocinia LMG 14191 | W | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | + |
| N | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | + | |
| B. seminalis LMG 24067 | W | - | ± | ± | ± | ± | ± | ± | - | ± | - | ± | ± | ± | ± | ± | - | ± | ± | ± | + |
| N | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | + | |
| B. stabilis LMG 14294 | W | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± | - | ± | ± | ± | + |
| N | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | + | |
| B. uborrensis LMG 20358 | W | - | - | ± | ± | ± | ± | ± | - | ± | ± | ± | ± | ± | - | ± | ± | ± | ± | ± | + |
| N | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | + | |
| Pseudomonas | Psychrobacter | Arthrobacter | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Species | Strain | BTN 1 | BTN 2 | BTN 15 | BTN 3 | BTN 19 | BTN 21 | BTN 5 | BTN 4 |
| B. diffusa | LMG 24065 | 100 ± 0 | 75 ± 3 | 77 ± 3 | 43 ± 7 | 45 ± 11 | 70 ± 4 | 77 ± 9 | 63 ± 3 |
| B. metallica | LMG 24068 | 92 ± 4 | 70 ± 5 | 71 ± 3 | 32 ± 2 | 30 ± 3 | 53 ± 5 | 77 ± 4 | 64 ± 9 |
| B. cenocepacia | LMG 16656 | 100 ± 0 | 78 ± 2 | 87 ± 1 | 84 ± 6 | 64 ± 4 | 45 ± 1 | 84 ± 2 | 57 ± 1 |
| B. latens | LMG 24064 | 100 ± 0 | 53 ± 11 | 75 ± 2 | 55 ± 6 | 43 ± 3 | 65 ± 2 | 56 ± 3 | 41 ± 2 |
| B. seminalis | LMG 24067 | 100 ± 0 | 43 ± 6 | 67 ± 5 | 73 ± 8 | 45 ± 6 | 78 ± 11 | 40 ± 3 | 56 ± 3 |
| 2 | 3 | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Position | ™C/ppm a, m | ™H/ppm (m, J in Hz) b | COSY 1H–1H | HMBC H→C | ™C/ppm a, m | ™H/ppm (m, J in Hz) b | COSY 1H–1H | HMBC H→C | |
| A | 1 | 173.4, C | 175.5, C | ||||||
| 2 | 38.9, CH2 | 2.58, m | A3 | A1 | 40.9, CH2 | 2.54, m | A3 | A1 | |
| 3 | 71.1, CH | 5.27, pentet, 6.4 | A2, A3 | A1, A2 | 72.7, CH | 5.29, pentet, 6.5 | A2, A4 | A1, A2 | |
| 4 | 33.8, CH2 | 1.64, m | A3 | A3 | 34.9, CH2 | 1.63, bm | A3 | A3 | |
| 5 | 24.9, CH2 | 1.35, overlap | 26.0, CH2 | 1.35, overlap | |||||
| 6 | 29.3, CH2 | 1.31, overlap | 30.5, CH2 | 1.37, overlap | |||||
| 7 | 29.3, CH2 | 1.31, overlap | 30.1, CH2 | 1.32, overlap | |||||
| 8 | 31. 6, CH2 | 1.31, overlap | 29.8, CH2 | 1.33, overlap | |||||
| 9 | 22.3, CH2 | 1.33, overlap | A10 | A10 | 30.2, CH2 | 1.36, overlap | A10 | A10 | |
| 10 | 13.1, CH3 | 0.92, m | A9 | A9 | 32.7, CH2 | 1.31, overlap | A9 | A9 | |
| 11 | 23.4, CH2 | 1.33, overlap | |||||||
| 12 | 14.1, CH3 | 0.92, m | |||||||
| B | 1 | 171.4, C | 172.3, CH | ||||||
| 2 | 39.5, CH2 | 2.53, m | B3 | B1 | 41.0, CH2 | A: 2.60, m B: 2.50, m | B3 | B1 | |
| 3 | 72.9, CH | 4.16, pentet, 5.8 | B2, B4 | B1, B5 | 74.8, CH | 4.10, pentet, 5.9 | B2, B4 | B1, B5 | |
| 4 | 30.4, CH2 | A: 2.39, m B: 2.33, m | B3, B5 | B3, B5 | 33.5, CH2 | 1.58, bm | B3,B5 | B3, B5 | |
| 5 | 123.7, CH | 5.40, m | B4, B6 | B3, B4, B6, B7 | 25.7, CH2 | 1.43, overlap | B4, B6 | ||
| 6 | 132.8, CH | 5.55, m | B5, B7 | B5, B8 | 27.8, CH2 | 2.08, overlap | B5, B7 | ||
| 7 | 27.1, CH2 | 2.08, m | B6 | B5, B6 | 130.0, CH | 5.37, m | B6, B8 | B8, B6, B9 | |
| 8 | 29.3, CH2 | 1.31, overlap | 131.2, CH | 5.39, m | B7 | B7 | |||
| 9 | 28.9, CH2 | 1.33, overlap | B7 | 32. 7, CH2 | 1.31, overlap | B8 | |||
| 10 | 31.6, CH2 | 1.31, overlap | 32.7, CH2 | 1.31, overlap | |||||
| 11 | 22.3, CH2 | 1.33, overlap | B12 | 23.4, CH2 | 1.33, overlap | B12 | |||
| 12 | 13.1, CH3 | 0.92, m | B11 | 14.1, CH3 | 0.92, m | B11 | |||
| C | 1 | 98.5, CH | 4.86, overlap | C2 | B3, C2 | 100.0, CH | 4.80, d, 1.4 | C2 | B3, C2 |
| 2 | 71.2, CH | 3.77, dd, 3.5, 1.4 | C1, C3 | C3, C4 | 72.4, CH | 3.76, dd, 3.4, 1.4 | C1, C3 | C3, C4 | |
| 3 | 70.9, CH | 3.64, dd, 9.5, 3.5 | C2, C4 | C5 | 71.9, CH | 3.66, dd, 9.7, 3.4 | C2, C4 | C5 | |
| 4 | 72.7, CH | 3.38, dd, 9.5, 9.8 | C3,C5 | C3 | 73.8, CH | 3.35, dd, 9.7, 9.8 | C3, C5 | C3 | |
| 5 | 68.7, CH | 3.67, m | C4, C6 | C4, C6 | 69.8, CH | 3.68, m | C4, C6 | C4, C6 | |
| 6 | 16.6, CH3 | 1.27, d, 6.2 | C5 | C5 | 17.6, CH3 | 1.27, d, 6.3 | C5 | C5 |
| Antimicrobial Activity (μg/mL) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B. cenocepacia LMG 16656 | B. metallica LMG 24068 | B. seminalis LMG 24067 | B. diffusa LMG 24065 | B. latens LMG 24064 | S. aureus 6538P | |||||||
| MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | |
| C1 | 3.12 | 3.12 | 50 | 50 | 12.5 | 12.5 | >200 | >200 | 12.5 | 12.5 | 1.56 | 1.56 |
| C2 | 3.12 | 3.12 | 25 | 25 | 3.12 | 3.12 | 200 | 200 | 12.5 | 12.5 | 3.12 | 3.12 |
| C3 | 200 | 200 | >200 | >200 | >200 | >200 | >200 | >200 | >200 | >200 | 100 | 100 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tedesco, P.; Maida, I.; Palma Esposito, F.; Tortorella, E.; Subko, K.; Ezeofor, C.C.; Zhang, Y.; Tabudravu, J.; Jaspars, M.; Fani, R.; et al. Antimicrobial Activity of Monoramnholipids Produced by Bacterial Strains Isolated from the Ross Sea (Antarctica). Mar. Drugs 2016, 14, 83. https://doi.org/10.3390/md14050083
Tedesco P, Maida I, Palma Esposito F, Tortorella E, Subko K, Ezeofor CC, Zhang Y, Tabudravu J, Jaspars M, Fani R, et al. Antimicrobial Activity of Monoramnholipids Produced by Bacterial Strains Isolated from the Ross Sea (Antarctica). Marine Drugs. 2016; 14(5):83. https://doi.org/10.3390/md14050083
Chicago/Turabian StyleTedesco, Pietro, Isabel Maida, Fortunato Palma Esposito, Emiliana Tortorella, Karolina Subko, Chidinma Christiana Ezeofor, Ying Zhang, Jioji Tabudravu, Marcel Jaspars, Renato Fani, and et al. 2016. "Antimicrobial Activity of Monoramnholipids Produced by Bacterial Strains Isolated from the Ross Sea (Antarctica)" Marine Drugs 14, no. 5: 83. https://doi.org/10.3390/md14050083
APA StyleTedesco, P., Maida, I., Palma Esposito, F., Tortorella, E., Subko, K., Ezeofor, C. C., Zhang, Y., Tabudravu, J., Jaspars, M., Fani, R., & De Pascale, D. (2016). Antimicrobial Activity of Monoramnholipids Produced by Bacterial Strains Isolated from the Ross Sea (Antarctica). Marine Drugs, 14(5), 83. https://doi.org/10.3390/md14050083








