Preparation and Antitumor Activity of CS5931, A Novel Polypeptide from Sea Squirt Ciona Savignyi
Abstract
:1. Introduction
2. Results
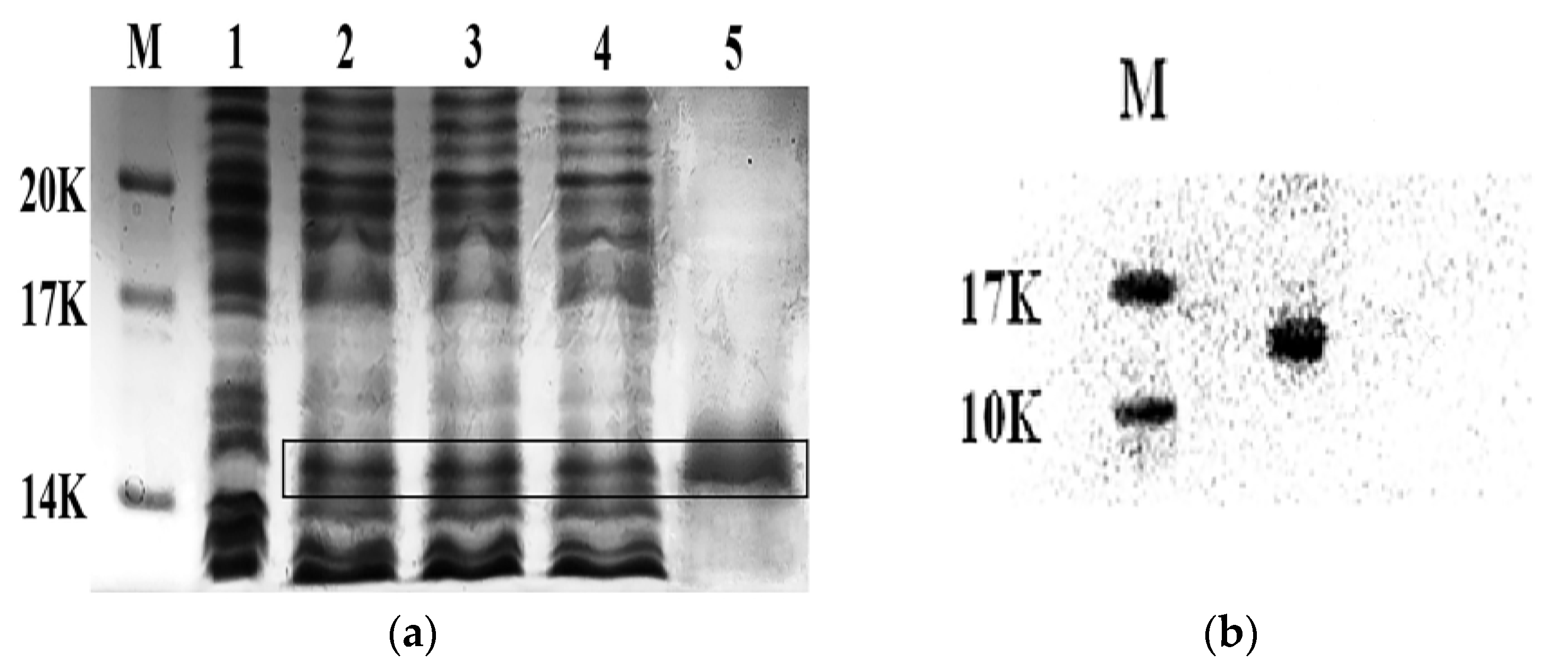
2.1. Expression and Purification of the His-Tag CS5931
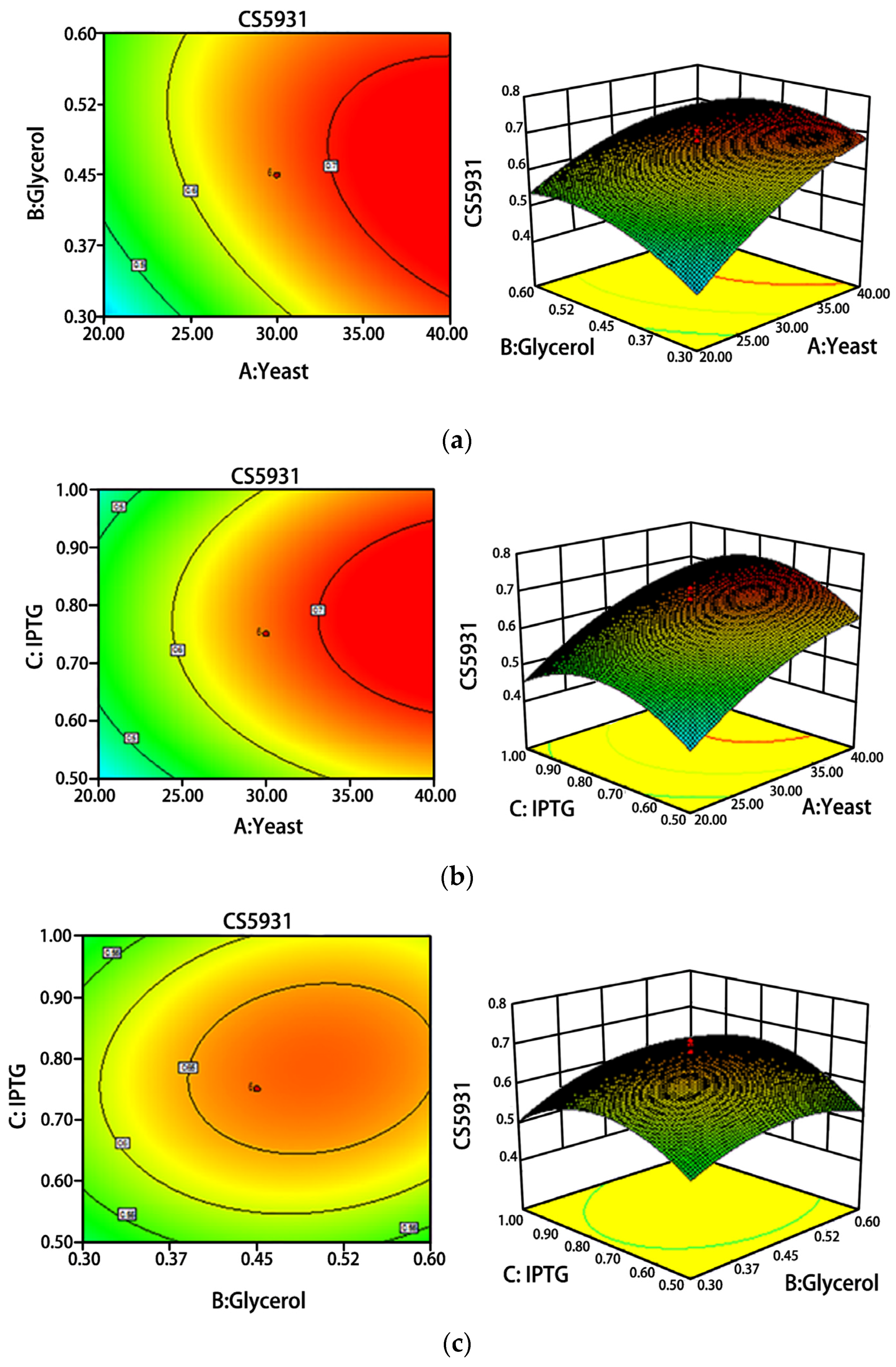
2.2. Optimization of Fermentation Conditions
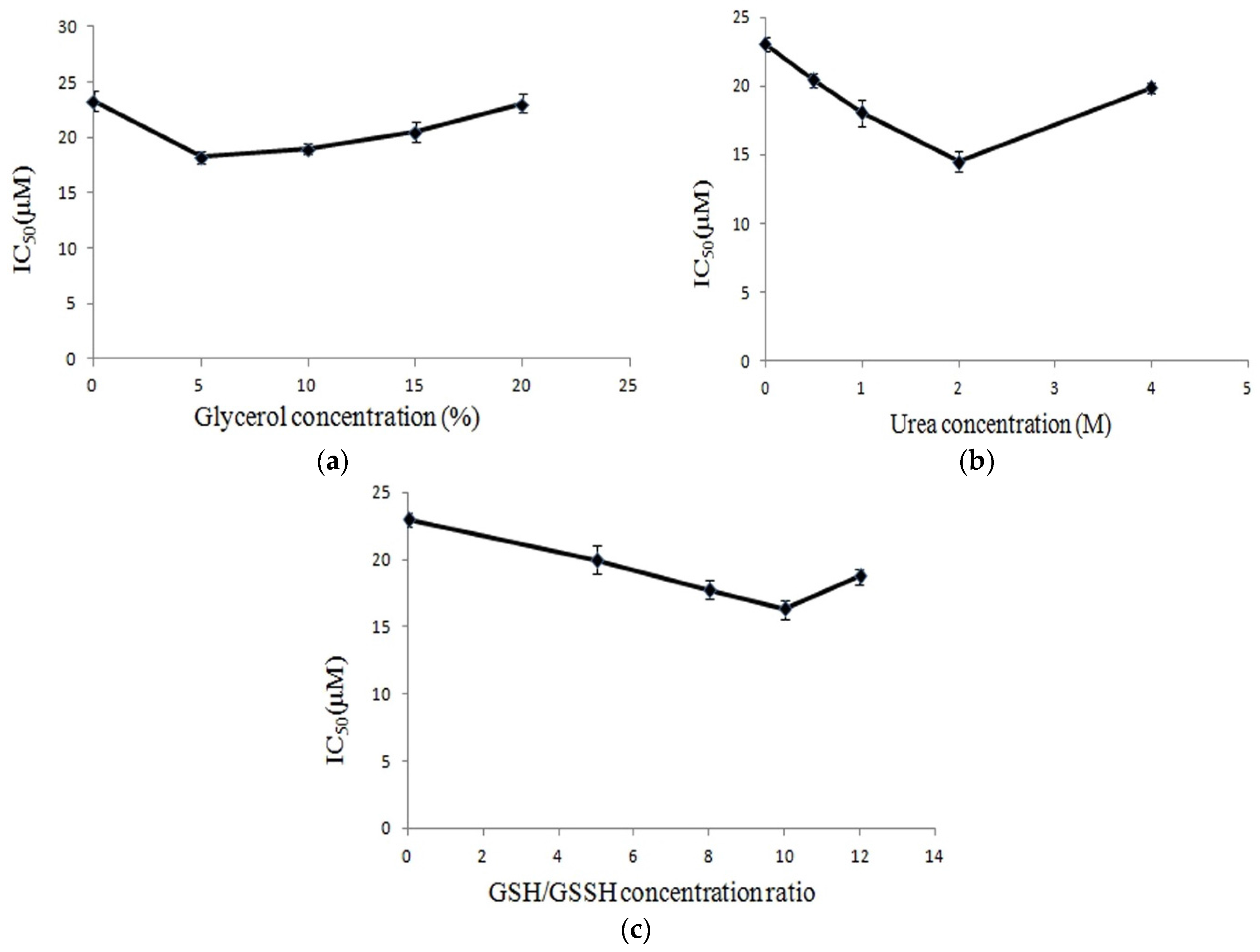
2.3. Renaturation of CS5931
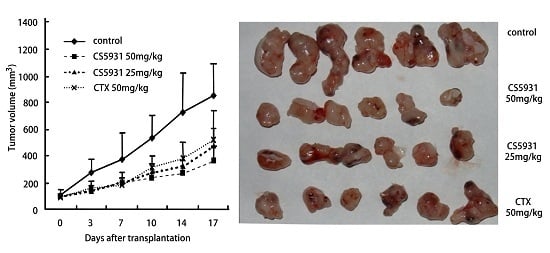
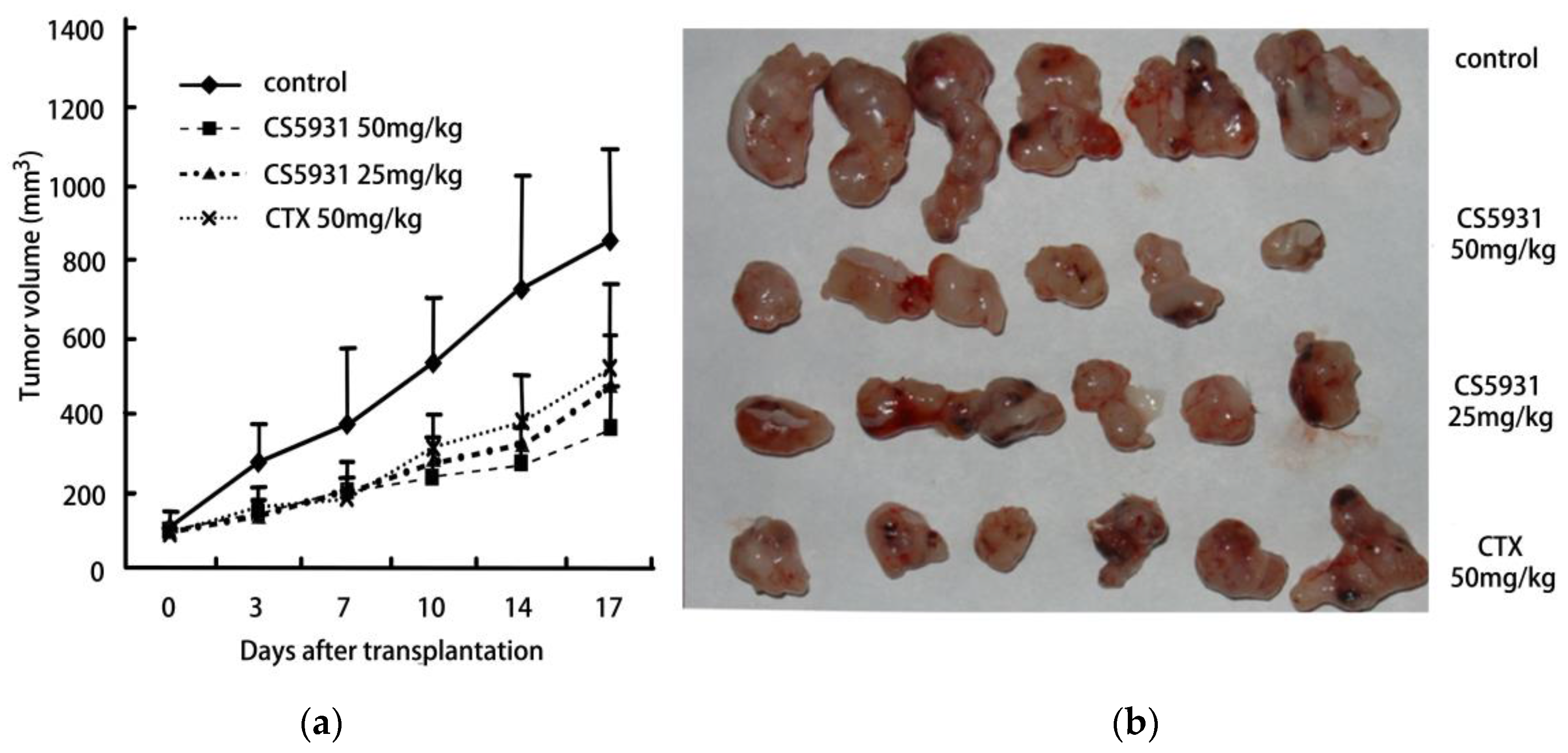
2.4. CS5931 Inhibits the Growth of HCT116 Xenograft in Athymic Mice
3. Discussion
4. Materials and Methods
4.1. Recombinant Expression of CS5931 from E. coli BL21 (DE3)
4.2. Purification of the His-Tag CS5931 Polypeptide
4.3. Optimization of Fermentation Conditions of CS5931 by Response Surface Methodology
4.4. Denaturing and Renaturing of the His-Tag CS5931 Polypeptide
4.5. Western Blotting Analysis
4.6. Cell Proliferation Assay
- Relative inhibition rate (%) = (ODcontrol group − ODexperimental group)/ODcontrol group × 100%.
- The relative viability rate (%) = 1 − relative inhibition rate × 100%.
- The IC50 value was expressed as the concentration of drugs causing 50% inhibition.
4.7. Antitumor Activity in Vivo
- Group A received normal saline;
- Group B received CTX 50 mg/kg as positive drug;
- Groups C and D received CS5931 with 25.0 and 50.0 mg/kg, respectively.
5. Conclusions
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Vera, M.D.; Joullié, M.M. Natural products as probes of cell biology: 20 years of didemnin research. Med. Res. Rev. 2007, 22, 102–145. [Google Scholar] [CrossRef] [PubMed]
- Mirian, F.D.; Hayashi, A.F. Katsuhiro Konno, natural peptides with potential applications in drug development, diagnosis, and/or biotechnology. Int. J. Pept. 2012, 2, 1–2. [Google Scholar]
- Zhou, Z.; Wang, X.; Zhang, H.; Guo, Y. Chromopeptide A, a highly cytotoxic depsipeptide from the marine sediment-derived bacterium Chromobacterium sp. HS-13-94. Acta Pharm. Sin. B 2015, 5, 62–66. [Google Scholar] [CrossRef] [PubMed]
- Depenbrock, H.; Peter, R.; Faircloth, G.T.; Manzanares, I.; Jimeno, J.; Hanauske, A.R. In vitro activity of aplidine, a new marine-derived anti-cancer compound, on freshly explanted clonogenic human tumour cells and haematopoietic precursor cells. Br. J. Cancer 1998, 78, 739–744. [Google Scholar] [CrossRef] [PubMed]
- Urdiales, J.L.; Morata, P.; Nunez De Castro, I.; Sanchez-Jimenez, F. Antiproliferative effect of dehydrodidemnin B (DDB), a depsipeptide isolated from Mediterranean tunicates. Cancer Lett. 1996, 102, 31–37. [Google Scholar] [CrossRef]
- Luesch, H.; Moore, R.E.; Paul, V.J.; Mooberry, S.L.; Corbett, T.H. Isolation of Dolastatin 10 from the marine cyanobacterium Symploca species VP642 and total stereochemistry and biological evaluation of its analogue Symplostatin 1. J. Nat. Prod. 2001, 64, 907–910. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.H.; Lin, X.K. Targeting cellular apoptotic pathway with peptides from marine organisms. Biochim. Biophys. Acta 2013, 1836, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Van de Donk, N.W.; Dhimolea, E. Brentuximab vedotin. MAbs 2012, 4, 458–465. [Google Scholar] [CrossRef] [PubMed]
- Deutsch, Y.E.; Lossos, I.S.; Rosenblatt, J.D. Brentuximab vedotin for Hodgkin lymphoma and systemic anaplastic large-cell lymphoma: A review of clinical experience and future directions. Int. J. Hematol. Oncol. 2013, 2, 455–465. [Google Scholar] [CrossRef]
- Minich, S.S. Brentuximab vedotin: A new age in the treatment of Hodgkin lymphoma and anaplastic large cell lymphoma. Ann. Pharmacother. 2012, 46, 377–383. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.Y.; Wang, C.C. A novel polypeptide extracted from Ciona savignyi induces apoptosis through a mitochondrial-mediated pathway in human colorectal carcinoma cells. Clin. Colorectal Cancer 2012, 11, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Wei, J. Cloning, characterization and expression of a cDNA encoding a granulin-like polypeptide in Ciona savignyi. Biochimie 2013, 95, 1611–1619. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Luana, W. A novel tumor necrosis factor ligand superfamily member (CsTL) from Ciona savignyi: Molecular identification and expression analysis. Dev. Comp. Immunol. 2008, 32, 1362–1373. [Google Scholar] [CrossRef] [PubMed]
- Morowvat, M.H.; Babaeipour, V. Optimization of fermentation conditions for recombinant human interferon Beta production by Escherichia coli using the response surface methology. Jundishapur J. Microbiol. 2015, 8, e16236. [Google Scholar] [CrossRef]
- Tsumoto, K.; Umetsu, M.; Kumagai, I.; Ejima, D.; Philo, J.S.; Arakawa, T. Role of arginine in protein refolding, solubilization, and purification. Biotechnol. Prog. 2004, 20, 1301–1308. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, I.; Mastui, H.; Ito, T. L-Cysteine-enhanced renaturation of bioactive soluble tumor necrosis factor ligand family member LIGHT from inclusion bodies in Escherichia coli. Protein Expr. Purif. 2011, 80, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Clark, E.D.; Schwarz, E.; Rudolph, R. Inhibition of aggregation side reactions during in vitro protein folding. Methods Enzymol. 1999, 309, 217–236. [Google Scholar]
- Berndt, C.; Lillig, C.H. Thioredoxins and glutaredoxins as facilitators of protein folding. Biochim. Biophys. Acta 2008, 1783, 641–650. [Google Scholar] [CrossRef] [PubMed]
- Clark, E.D. Refolding of recombinant proteins. Curr. Opin. Biotechnol. 1998, 9, 157–163. [Google Scholar] [CrossRef]
- Burgess, R.R. Refolding solubilized inclusion body proteins. Methods Enzymol. 2009, 463, 259–282. [Google Scholar] [PubMed]
- Maharjan, S.; Singh, B. Exploring codon optimization and Response Surface Methodology to express biologically active transmembrane RANKL in E. coli. PLoS ONE 2014, 9, 1–15. [Google Scholar]
- Mohd Salim, R.J.; Adenan, M.I. Statistical analysis of metal chelating activity of centella asiatica and erythroxylum cuneatum using Response Surface Methodology. Biotechnol. Res. Int. 2013, 5, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Bose, A.; Wong, T.W. Formulation development and optimization of sustained release matrix tablet of itopride HCl by response surface methodology and its evaluation of release kinetics. Saudi Pharm. J. 2013, 21, 201–213. [Google Scholar] [CrossRef] [PubMed]
- Pirzadah, T.; Garg, S. Characterization of actinomycetes and trichoderma spp. for cellulase production utilizing crude substrates by response surface methodology. SpringerPlus 2014, 3, 622–628. [Google Scholar] [CrossRef] [PubMed]
- Chambers, S.P.; Swalley, S.E. Designing experiments for high-throughput protein expression. Methods Mol. Biol. 2009, 498, 19–29. [Google Scholar] [PubMed]
- Du, C.; Niu, R.; Chu, E.; Zhang, P.; Lin, X. Sequence analysis and functional study of thymidylate synthase from zebrafish, Danio rerio. J. Biochem. 2006, 139, 913–920. [Google Scholar] [CrossRef] [PubMed]
| Purification Steps | Total Protein (mg) | CS5931 (mg) | Yield (%) | Purity (%) | IC50 (μM) |
|---|---|---|---|---|---|
| Cell lysate | 539 | 54 | - | 10 | - |
| Ni-NTA column | 48.5 | 47 | 9 | 97 | 23.2 |
| Protein renaturation | 24.3 | 23.8 | 50 | 98 | - |
| Protein frozen-dried | 7.65 | 7.5 | 32 | 98 | 11.6 |
| Group | Dosage (mg/kg) | No. of Mice (Begin/End) | Body Weight Change (g) | Tumor Weight (mean ± SD, g) | %TGI |
|---|---|---|---|---|---|
| Control | - | 6/6 | +4.2 | 1.35 ± 0.20 | - |
| CTX | 50 | 6/6 | +0.9 | 0.63 ± 0.13 | 54 ** |
| CS5931 | 25 | 6/6 | +1.2 | 0.69 ± 0.13 | 48 ** |
| CS5931 | 50 | 6/6 | +0.4 | 0.45 ± 0.14 | 67 ** |
| Independent Variables | Symbol | Code Levels | |
|---|---|---|---|
| −1 | 1 | ||
| Yeast extract concentration, g·L−1 | X1 | 20 | 40 |
| Glycerol concentration, % | X2 | 0.3 | 0.6 |
| IPTG concentration, mM | X3 | 0.5 | 1.0 |
| Experiment | Factor A, g·L−1 | Factor B, % | Factor C, mM | CS5931, mg |
|---|---|---|---|---|
| X1 | X2 | X3 | ||
| 1 | 40.00 (1) | 0.30 (−1) | 1.00 (1) | 0.708 |
| 2 | 20.00 (−1) | 0.60 (1) | 1.00 (1) | 0.474 |
| 3 | 20.00 (−1) | 0.30 (−1) | 1.00 (1) | 0.370 |
| 4 | 40.00 (1) | 0.60 (1) | 0.50 (−1) | 0.538 |
| 5 | 30.00 (0) | 0.45 (0) | 0.75 (0) | 0.680 |
| 6 | 30.00 (0) | 0.45 (0) | 0.75 (0) | 0.659 |
| 7 | 46.82 (1.682) | 0.45 (0) | 0.75 (0) | 0.700 |
| 8 | 30.00 (0) | 0.45 (0) | 0.75 (0) | 0.681 |
| 9 | 40.00 (1) | 0.60 (1) | 1.00 (1) | 0.610 |
| 10 | 30.00 (0) | 0.45 (0) | 0.33 (−1.682) | 0.423 |
| 11 | 30.00 (0) | 0.45 (0) | 1.17 (1.682) | 0.401 |
| 12 | 30.00 (0) | 0.45 (0) | 0.75 (0) | 0.597 |
| 13 | 40.00 (1) | 0.30 (−1) | 0.50 (−1) | 0.627 |
| 14 | 20.00 (−1) | 0.30 (−1) | 0.50 (−1) | 0.378 |
| 15 | 30.00 (0) | 0.45 (0) | 0.75 (0) | 0.700 |
| 16 | 30.00 (0) | 0.45 (0) | 0.75 (0) | 0.709 |
| 17 | 30.00 (0) | 0.20 (−1.682) | 0.75 (0) | 0.348 |
| 18 | 20.00 (−1) | 0.60 (1) | 0.50 (−1) | 0.340 |
| 19 | 13.18 (−1.682) | 0.45 (0) | 0.75 (0) | 0.392 |
| 20 | 30.00 (0) | 0.70 (1.682) | 0.75 (0) | 0.671 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, X.; Xu, H.; Li, B.; Wang, F.; Chen, X.; Kong, D.; Lin, X. Preparation and Antitumor Activity of CS5931, A Novel Polypeptide from Sea Squirt Ciona Savignyi. Mar. Drugs 2016, 14, 47. https://doi.org/10.3390/md14030047
Chen X, Xu H, Li B, Wang F, Chen X, Kong D, Lin X. Preparation and Antitumor Activity of CS5931, A Novel Polypeptide from Sea Squirt Ciona Savignyi. Marine Drugs. 2016; 14(3):47. https://doi.org/10.3390/md14030047
Chicago/Turabian StyleChen, Xiaoshuang, Huanli Xu, Bo Li, Feng Wang, Xiaoliang Chen, Dexin Kong, and Xiukun Lin. 2016. "Preparation and Antitumor Activity of CS5931, A Novel Polypeptide from Sea Squirt Ciona Savignyi" Marine Drugs 14, no. 3: 47. https://doi.org/10.3390/md14030047
APA StyleChen, X., Xu, H., Li, B., Wang, F., Chen, X., Kong, D., & Lin, X. (2016). Preparation and Antitumor Activity of CS5931, A Novel Polypeptide from Sea Squirt Ciona Savignyi. Marine Drugs, 14(3), 47. https://doi.org/10.3390/md14030047