Integrated (Meta) Genomic and Synthetic Biology Approaches to Develop New Biocatalysts
Abstract
:1. Introduction
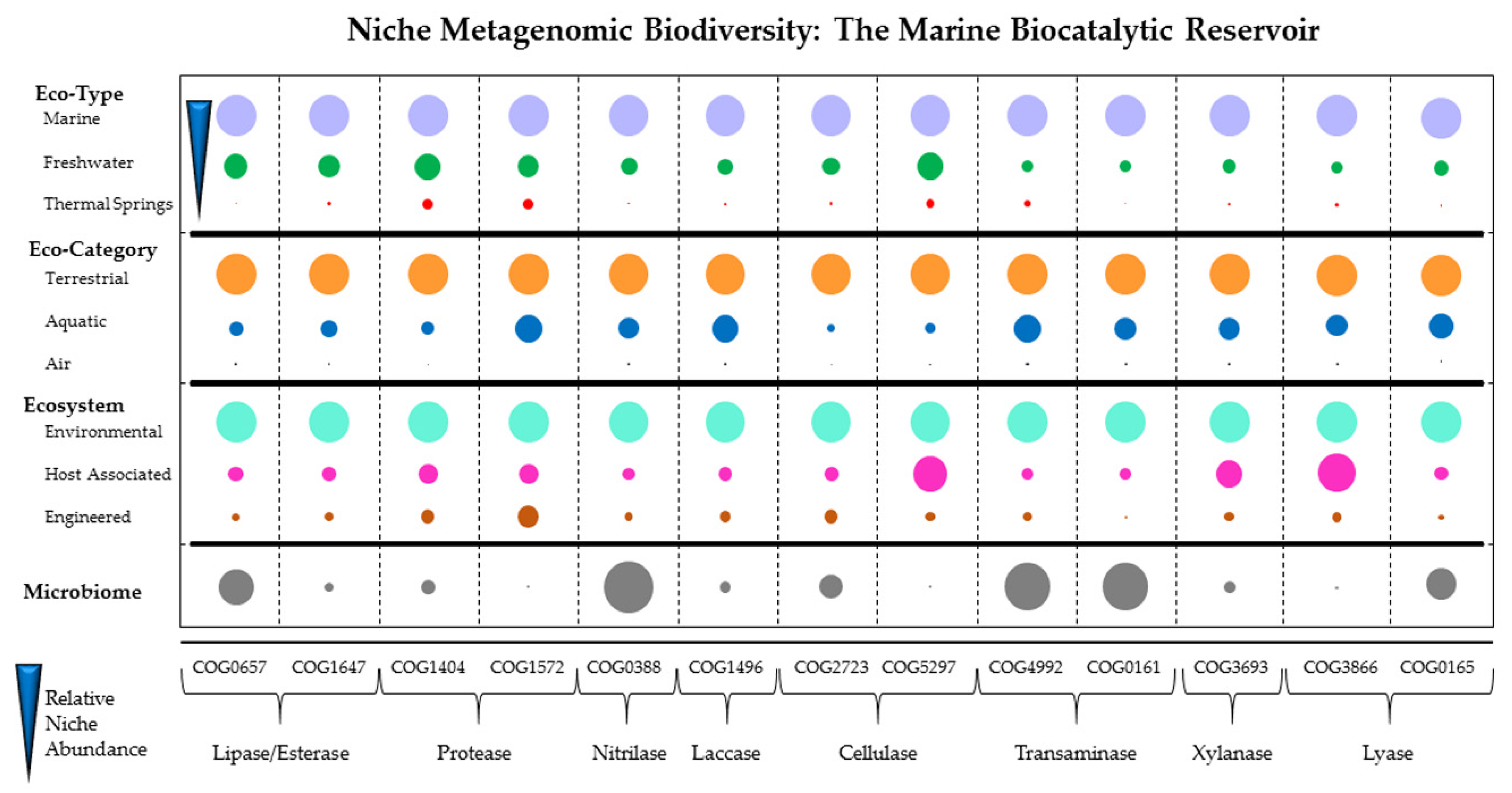
2. Marine Environment as a Biocatalytic Reservoir
3. Biocatalysts as a Valuable Alternative to Traditional Chiral Chemical Synthesis
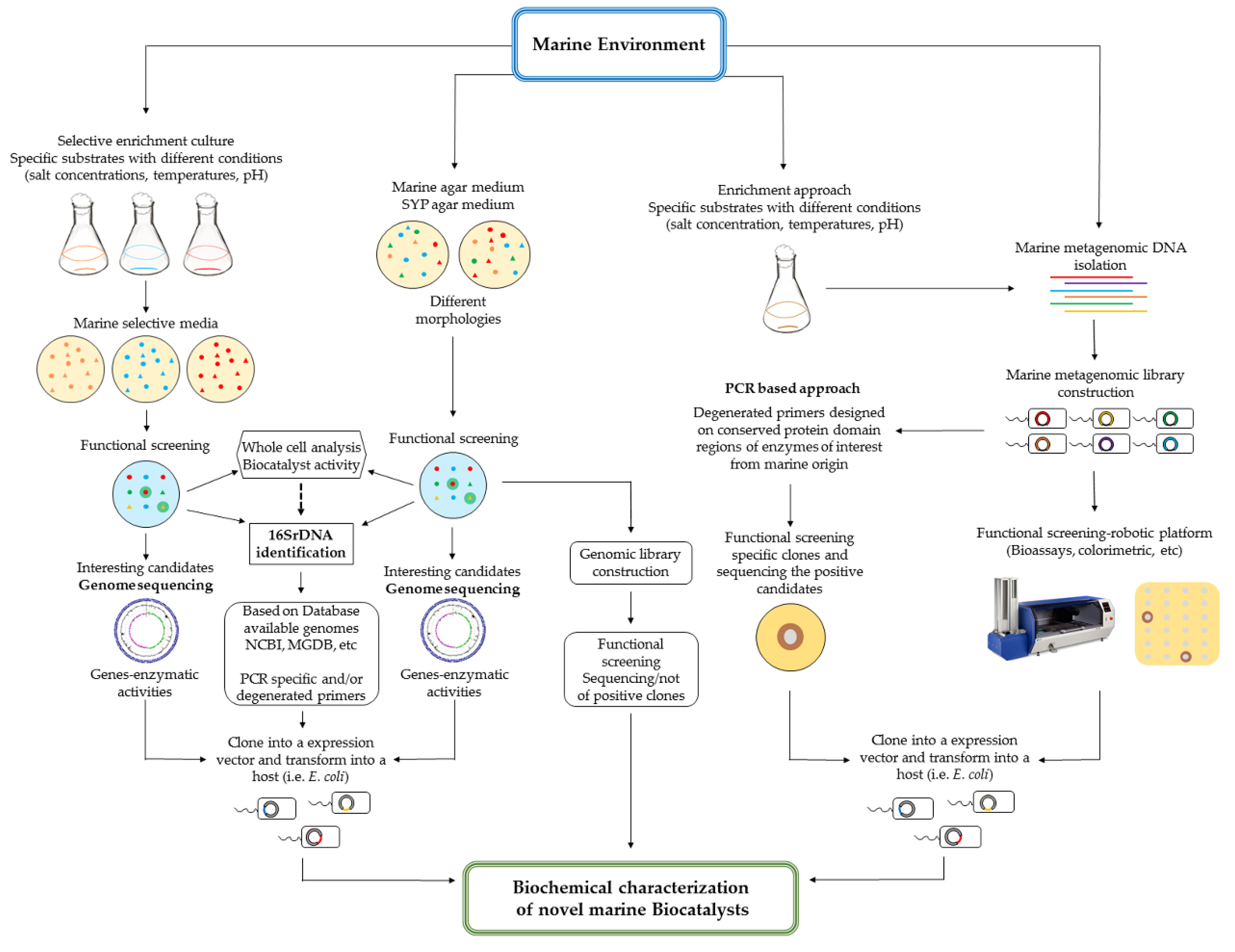
4. Culture Dependent and Independent Approaches to Unravel the Biocatalytic Potential of the Marine Environment
4.1. Culture Dependent Approach
4.1.1. Marine Bacteria as an Untapped Source of Novel Biocatalysts
4.1.2. Genome Mining: An Under Exploited Source of Biocatalyst Discovery
4.2. Culture Independent Approach: Metagenomic for Biocatalyst Discovery
4.2.1. Metagenomic Screening Strategies
Metagenomic Functional Screening
Metagenomic Sequence-Based Screening
5. Biocatalytic Improvement by Directed Evolution
5.1. Directed Evolution of Enzymes
5.1.1. Random Mutagenesis
5.1.2. Site-Directed Saturation Mutagenesis
5.1.3. Genetic Recombination
6. Synthetic Biology
7. Conclusions: The Future of the Biocatalysis Pipeline
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Barone, R.; de Santi, C.; Palma Esposito, F.; Tedesco, P.; Galati, F.; Visone, M.; di Scala, A.; de Pascale, D. Marine metagenomics, a valuable tool for enzymes and bioactive compounds discovery. Front. Mar. Sci. 2014, 1. [Google Scholar] [CrossRef]
- Felczykowska, A.; Bloch, S.K.; Nejman-faleńczyk, B.; Barańska, S. Metagenomic approach in the investigation of new bioactive compounds in the marine environment. Acta Biochim. Pol. 2012, 59, 501–505. [Google Scholar] [PubMed]
- Zhao, X. Genome-based studies of marine microorganisms to maximize the diversity of natural products discovery for medical treatments. Evid. Based Complement. Alternat. Med. 2011, 2011. [Google Scholar] [CrossRef] [PubMed]
- Trincone, A. Marine biocatalysts: Enzymatic features and applications. Mar. Drugs 2011, 9, 478–499. [Google Scholar] [CrossRef] [PubMed]
- Sunagawa, S.; Coelho, L.P.; Chaffron, S.; Kultima, J.R.; Labadie, K.; Salazar, G.; Djahanschiri, B.; Zeller, G.; Mende, D.R.; Alberti, A.; et al. Structure and function of the global ocean microbiome. Science 2015, 348. [Google Scholar] [CrossRef] [PubMed]
- Bérdy, J. Bioactive microbial metabolites. J. Antibiot. (Tokyo) 2005, 58, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Demain, A.L.; Sanchez, S. Microbial drug discovery: 80 years of progress. J. Antibiot. (Tokyo) 2009, 62, 5–16. [Google Scholar] [CrossRef] [PubMed]
- Ngo, D.-H.; Vo, T.-S.; Ngo, D.-N.; Wijesekara, I.; Kim, S.-K. Biological activities and potential health benefits of bioactive peptides derived from marine organisms. Int. J. Biol. Macromol. 2012, 51, 378–383. [Google Scholar] [CrossRef] [PubMed]
- Vinothkumar, S.; Parameswaran, P.S. Recent advances in marine drug research. Biotechnol. Adv. 2013, 31, 1826–1845. [Google Scholar] [CrossRef] [PubMed]
- Gardossi, L.; Molinari, F. Biocatalytic processes. In Catalysis; Centi, G., Ed.; Encyclopedia of Life Support Systems (Eolss): Paris, France, 2008. [Google Scholar]
- Herrera, S. Industrial biotechnology—A chance at redemption. Nat. Biotechnol. 2004, 22, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Bommarius, A.S.; Riebel-bommarius, B.R. Biocatalysis : Fundamentals and Applications, 1st ed.; Wiley-Blackwell: Hoboken, NJ, USA, 2004. [Google Scholar]
- Simon, R.C.; Mutti, F.G.; Kroutil, W. Biocatalytic synthesis of enantiopure building blocks for pharmaceuticals. Drug Discov. Today Technol. 2013, 10, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Gurung, N.; Ray, S.; Bose, S.; Rai, V. A broader view: Microbial enzymes and their relevance in industries, medicine, and beyond. Biomed Res. Int. 2013, 2013. [Google Scholar] [CrossRef] [PubMed]
- Guazzaroni, M.-E.; Silva-Rocha, R.; Ward, R.J. Synthetic biology approaches to improve biocatalyst identification in metagenomic library screening. Microb. Biotechnol. 2015, 8, 52–64. [Google Scholar] [CrossRef] [PubMed]
- Simon, C.; Wiezer, A.; Strittmatter, A.W.; Daniel, R. Phylogenetic diversity and metabolic potential revealed in a glacier ice metagenome. Appl. Environ. Microbiol. 2009, 75, 7519–7526. [Google Scholar] [CrossRef] [PubMed]
- Lipson, D.A.; Haggerty, J.M.; Srinivas, A.; Raab, T.K.; Sathe, S.; Dinsdale, E.A. Metagenomic insights into anaerobic metabolism along an arctic peat soil profile. PLoS ONE 2013, 8, e64659. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cao, H.; Zhang, G.; Bougouffa, S.; Lee, O.O.; Al-Suwailem, A.; Qian, P.-Y. Autotrophic microbe metagenomes and metabolic pathways differentiate adjacent Red Sea brine pools. Sci. Rep. 2013, 3. [Google Scholar] [CrossRef] [PubMed]
- Daniel, R. The metagenomics of soil. Nat. Rev. Microbiol. 2005, 3, 470–478. [Google Scholar] [CrossRef] [PubMed]
- Simon, C.; Daniel, R. Metagenomic analyses: Past and future trends. Appl. Environ. Microbiol. 2011, 77, 1153–1161. [Google Scholar] [CrossRef] [PubMed]
- Behzad, H.; Ibarra, M.A.; Mineta, K.; Gojobori, T. Metagenomic studies of the Red Sea. Gene 2016, 576, 717–723. [Google Scholar] [CrossRef] [PubMed]
- Burton, S.G.; Cowan, D.A.; Woodley, J.M. The search for the ideal biocatalyst. Nat. Biotechnol. 2002, 20, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Cowan, D.A.; Arslanoglu, A.; Burton, S.G.; Baker, G.C.; Cameron, R.A.; Smith, J.J.; Meyer, Q. Metagenomics, gene discovery and the ideal biocatalyst. Biochem. Soc. Trans. 2004, 32, 298–302. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, P.; Eck, J. Metagenomics and industrial applications. Nature 2005, 3, 510–516. [Google Scholar]
- Singhania, R.R.; Patel, A.K.; Sukumaran, R.K.; Larroche, C.; Pandey, A. Role and significance of beta-glucosidases in the hydrolysis of cellulose for bioethanol production. Bioresour. Technol. 2013, 127, 500–507. [Google Scholar] [CrossRef] [PubMed]
- Ward, R.J. Cellulase Engineering for Biomass Saccharification. In Routes to Cellulosic Ethanol; Buckeridge, M.S., Goldman, G.H., Eds.; Springer: New York, NY, USA, 2011; pp. 135–151. [Google Scholar]
- Brum, J.R.; Ignacio-espinoza, J.C.; Roux, S.; Doulcier, G.; Acinas, S.G.; Alberti, A.; Chaffron, S.; Cruaud, C.; de Vargas, C.; Gasol, J.M.; et al. Ocean plankton. Patterns and ecological driveres of ocean viral communities. Science 2015, 348. [Google Scholar] [CrossRef] [PubMed]
- Armbrust, E.V.; Palumbi, S.R. Uncovering hidden worlds of ocean biodiversity. Science 2015, 348, 865–867. [Google Scholar] [CrossRef] [PubMed]
- Dhakar, K.; Pandey, A. Phialophora Melinii ( NFCCI 3617 ): A newly isolated psychrotolerant fungus that produces enhanced laccase under the influence of organic solvents. Adv. Nat. Sci. 2014, 8, 14–20. [Google Scholar]
- Sarmiento, F.; Peralta, R.; Blamey, J.M. Cold and hot extremozymes: Industrial relevance and current trends. Front. Bioeng. Biotechnol. 2015, 3. [Google Scholar] [CrossRef] [PubMed]
- Huston, A.L. Biotechnological aspects of cold-adapted enzymes. In Psychrophiles: From Biodiversity to Biotechnology; Margesin, R., Schinner, F., Marx, J.-C., Gerday, C., Eds.; Springer: Heidelberg, Germany, 2008; pp. 347–363. [Google Scholar]
- De Santi, C.; Altermark, B.; de Pascale, D.; Willassen, N.-P. Bioprospecting around Arctic islands : Marine bacteria as rich source of biocatalysts. J. Basic Microbiol. 2015, 55, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Gifford, S.M.; Sharma, S.; Booth, M.; Moran, M.A. Expression patterns reveal niche diversification in a marine microbial assemblage. ISME J. 2012, 7, 281–298. [Google Scholar] [CrossRef] [PubMed]
- Arrieta, J.M.; Arnaud-Haond, S.; Duarte, C.M. What lies underneath: Conserving the oceans’ genetic resources. Proc. Natl. Acad. Sci. USA 2010, 107, 18318–18324. [Google Scholar] [CrossRef] [PubMed]
- Venugopal, V. Marine Products for Healthcare: Functional and Bioactive Nutraceutical Compounds from the Ocean, 1st ed.; CRC Press: Florida, FL, USA, 2008. [Google Scholar]
- Desantis, G. Enzymes in organic synthesis. Mod. Drug Discov. 2002, 5, 43–47. [Google Scholar]
- Kisukuri, C.M.; Andrade, L.H. Production of chiral compounds using immobilized cells as a source of biocatalysts. Org. Biomol. Chem. 2015, 13, 10086–10107. [Google Scholar] [CrossRef] [PubMed]
- Skarydova, L.; Skarka, A.; Solich, P.; Wsol, V. Enzyme stereospecificity as a powerful tool in searching for new enzymes. Curr. Drug Metab. 2010, 11, 547–559. [Google Scholar] [CrossRef] [PubMed]
- García-vilas, J.A.; Martínez-Poveda, B.; Quesada, A.R.; Medina, M.Á. Aeroplysinin-1, a sponge-derived multi-targeted bioactive. Mar. Drugs. 2016, 14. [Google Scholar] [CrossRef]
- Sekhon, B.S. Exploiting the power of stereochemistry in drugs: An overview of racemic and enantiopure drugs. J. Mod. Med. Chem. 2013, 1, 10–36. [Google Scholar] [CrossRef]
- Rat’ko, A.A.; Stefan-van Staden, R.-I. Determination of baclofen enantiomers in pharmaceutical formulations using maltodextrin-based enantioselective, potentiometric membrane electrodes. Farmaco 2004, 59, 993–997. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.W. Chiral toxicology: It’s the same thing...only different. Toxicol. Sci. 2009, 110, 4–30. [Google Scholar] [CrossRef] [PubMed]
- Patel, R.N. Biocatalytic synthesis of chiral alcohols and amino acids for development of pharmaceuticals. Biomolecules 2013, 3, 741–777. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.A.; He, H.; Pham-Huy, C. Chiral drugs: An overview. Int. J. Biomed. Sci. 2006, 2, 85–100. [Google Scholar] [PubMed]
- Darwish, M.; Kirby, M.; Hellriegel, E.T.; Robertson, P.J. Armodafinil and modafinil have substantially different pharmacokinetic profiles despite having the same terminal half-lives: Analysis of data from three randomized, single-dose, pharmacokinetic studied pharmacokinetic studies. Clin. Drug Investig. 2009, 29, 613–623. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Pan, J.; Ma, B.; Xu, J. Efficient biocatalytic synthesis of chiral chemicals. In Advances in Biochemical Engineering/Biotechnology; Scheper, T.H., Belkin, S., Bley, T.H., Bohlmann, J., Doran, P.M., Gu, M.B., Hu, W., Mattiasson, B., Nielsen, J., Seitz, H., et al., Eds.; Springer: New York, NY, USA, 2014; pp. 1–52. [Google Scholar]
- FDA: U.S. Food and Drug Administration. Development of New Stereoisomeric Drugs. Available online: http://www.fda.gov/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/ucm122883.htm (accessed on 12 November 2015).
- Ishihara, K.; Fujita, A.; Sakiyama, A.; Kobayashi, Y.; Hori, K.; Maruike, K.; Masuoka, N.; Nakajima, N.; Hamada, H. Preparation of chiral hydroxy esters using Actinobacteria : Biocatalyst activity of marine-derived Micromonospora and Streptomyces strains. Open J. Appl. Sci. 2013, 3, 116–122. [Google Scholar] [CrossRef]
- Hughes, A.J.; Detelich, J.F.; Keatinge-Clay, A.T. Employing a polyketide synthase module and thioesterase in the semipreparative biocatalysis of diverse triketide pyrones. MedChemComm 2012, 3, 956–959. [Google Scholar] [CrossRef]
- Piasecki, S.K.; Taylor, C.A.; Detelich, J.F.; Liu, J.; Zheng, J.; Komsoukaniants, A.; Siegel, D.R.; Keatinge-clay, A.T. Employing modular polyketide synthase ketoreductases as biocatalysts in the preparative chemoenzymatic syntheses of diketide chiral building blocks. Chem. Biol. 2011, 18, 1331–1340. [Google Scholar] [CrossRef] [PubMed]
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Munro, M.H.G.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2012, 29, 144–222. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.K.; Wijesekara, I. Development and biological activities of marine-derived bioactive peptides: A review. J. Funct. Foods 2010, 2, 1–9. [Google Scholar] [CrossRef]
- Montaser, R.; Luesch, H. Marine natural products: A new wave of drugs? Future Med. Chem. 2011, 3, 1475–1489. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J. Nat. Prod. 2012, 75, 311–335. [Google Scholar] [CrossRef] [PubMed]
- Rocha-Martin, J.; Harrington, C.; Dobson, A.D.W.; O’Gara, F. Emerging strategies and integrated systems microbiology technologies for biodiscovery of marine bioactive compounds. Mar. Drugs 2014, 12, 3516–3559. [Google Scholar] [CrossRef] [PubMed]
- Reen, F.J.; Gutiérrez-Barranquero, J.A.; Dobson, A.D.W.; Adams, C.; O’Gara, F. Emerging concepts promising new horizons for marine biodiscovery and synthetic biology. Mar. Drugs 2015, 13, 2924–2954. [Google Scholar] [CrossRef] [PubMed]
- Reen, F.J.; Romano, S.; Dobson, A.D.W.; O’Gara, F. The sound of silence: Activating silent biosynthetic gene clusters in marine microorganisms. Mar. Drugs 2015, 13, 4754–4783. [Google Scholar] [CrossRef] [PubMed]
- Suarez-Jimenez, G.-M.; Burgos-Hernandez, A.; Ezquerra-Brauer, J.-M. Bioactive peptides and depsipeptides with anticancer potential: Sources from marine animals. Mar. Drugs 2012, 10, 963–986. [Google Scholar] [CrossRef] [PubMed]
- Ran, N.; Zhao, L.; Chen, Z.; Tao, J. Recent applications of biocatalysis in developing green chemistry for chemical synthesis at the industrial scale. Green Chem. 2008, 10, 361–372. [Google Scholar] [CrossRef]
- Trincone, A. Biocatalytic processes using marine biocatalysts : Ten cases in point. Curr. Org. Chem. 2013, 17, 1058–1066. [Google Scholar] [CrossRef]
- Bernard, L.; Schäfer, H.; Joux, F.; Courties, C.; Muyzer, G.; Lebaron, P. Genetic diversity of total, active and culturable marine bacteria in coastal seawater. Aquat. Microb. Ecol. 2000, 23, 1–11. [Google Scholar] [CrossRef]
- Tyson, G.W.; Banfield, J.F. Cultivating the uncultivated: A community genomics perspective. Trends Microbiol. 2005, 13, 411–415. [Google Scholar] [CrossRef] [PubMed]
- MaCuMBA-Marine Microorganimsms: Cultivation Methods for Improving Their Biotechnological Applications. Available online: http://www.macumbaproject.eu (accessed on 12 November 2015).
- Bachmann, B.O.; van Lanen, S.G.; Baltz, R.H. Microbial genome mining for accelerated natural products discovery: Is a renaissance in the making? J. Ind. Microbiol. Biotechnol. 2014, 41, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Harvey, A.L.; Edrada-Ebel, R.; Quinn, R.J. The re-emergence of natural products for drug discovery in the genomics era. Nat. Rev. Drug Discov. 2015, 14, 111–129. [Google Scholar] [CrossRef] [PubMed]
- Handelsman, J. Metagenomics: Application of genomics to uncultured microorganisms metagenomics. Microbiol. Mol. Biol. Rev. 2004, 68, 669–685. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Ren, J.; Wu, Q.; Feng, J.; Zhu, D.; Ma, Y. Identification of a marine NADPH-dependent aldehyde reductase for chemoselective reduction of aldehydes. J. Mol. Catal. B Enzym. 2013, 90, 17–22. [Google Scholar] [CrossRef]
- Novak, H.R.; Sayer, C.; Panning, J.; Littlechild, J.A. Characterisation of an l-haloacid dehalogenase from the marine psychrophile Psychromonas ingrahamii with potential industrial application. Mar. Biotechnol. (N. Y.) 2013, 15, 695–705. [Google Scholar] [CrossRef] [PubMed]
- De Pascale, D.; Cusano, A.M.; Autore, F.; Parrilli, E.; di Prisco, G.; Marino, G.; Tutino, M.L. The cold-active Lip1 lipase from the Antarctic bacterium Pseudoalteromonas haloplanktis TAC125 is a member of a new bacterial lipolytic enzyme family. Extremophiles 2008, 12, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Xiao, X.; Wang, F. Isolation and characterization of alkane hydroxylases from a metagenomic library of Pacific deep-sea sediment. Extremophiles 2008, 12, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Zemin, F.; Fang, W.; Liu, J.; Hong, Y.; Peng, H.; Zhang, X.; Sun, B.; Xiao, Y. Cloning and characterization of a β-glucosidase from marine microbial metagenome with excellent glucose tolerance. J. Microbiol. Biotechnol. 2010, 20, 1351–1358. [Google Scholar]
- Song, J.S.; Jeon, J.H.; Lee, J.H.; Jeong, S.H.; Jeong, B.C.; Kim, S.J.; Lee, J.H.; Lee, S.H. Molecular characterization of TEM-type beta-lactamases identified in cold-seep sediments of Edison Seamount (south of Lihir Island, Papua New Guinea). J. Microbiol. 2005, 43, 172–178. [Google Scholar] [PubMed]
- LeCleir, G.R.; Buchan, A.; Hollibaugh, J.T. Chitinase gene sequences retrieved from diverse aquatic habitats reveal environment-specific distributions. Appl. Environ. Microbiol. 2004, 70, 6977–6983. [Google Scholar] [CrossRef] [PubMed]
- Cottrell, M.T.; Moore, J.A.; Kirchman, D.L. Chitinases from uncultured marine microorganisms. Appl. Environ. Microbiol. 1999, 65, 2553–2557. [Google Scholar] [PubMed]
- Martin, M.; Biver, S.; Steels, S.; Barbeyron, T.; Jam, M.; Portetelle, D.; Michel, G.; Vandenbol, M. Identification and characterization of a halotolerant, cold-active marine Endo-β-1,4-Glucanase by using functional metagenomics of seaweed-associated microbiota. Appl. Environ. Microbiol. 2014, 80, 4958–4967. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, M.; Golyshina, O.V.; Chernikova, T.N.; Khachane, A.N.; Martins Dos Santos, V.A.P.; Yakimov, M.M.; Timmis, K.N.; Golyshin, P.N. Microbial enzymes mined from the Urania deep-sea hypersaline anoxic basin. Chem. Biol. 2005, 12, 895–904. [Google Scholar] [CrossRef] [PubMed]
- Chu, X.; He, H.; Guo, C.; Sun, B. Identification of two novel esterases from a marine metagenomic library derived from South China Sea. Appl. Microbiol. Biotechnol. 2008, 80, 615–625. [Google Scholar] [CrossRef] [PubMed]
- Jeon, J.H.; Kim, J.-T.; Kang, S.G.; Lee, J.-H.; Kim, S.-J. Characterization and its potential application of two esterases derived from the arctic sediment metagenome. Mar. Biotechnol. 2009, 11, 307–316. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Fu, C.; Huang, Y.; Yin, Y.; Cheng, G.; Lei, F.; Lu, N.; Li, J.; Ashforth, E.J.; Zhang, L.; Zhu, B. Novel lipolytic genes from the microbial metagenomic library of the South China Sea marine sediment. FEMS Microbiol. Ecol. 2010, 72, 228–237. [Google Scholar] [CrossRef] [PubMed]
- Okamura, Y.; Kimura, T.; Yokouchi, H.; Meneses-Osorio, M.; Katoh, M.; Matsunaga, T.; Takeyama, H. Isolation and characterization of a GDSL esterase from the metagenome of a marine sponge-associated bacteria. Mar. Biotechnol. 2010, 12, 395–402. [Google Scholar] [CrossRef] [PubMed]
- Fu, C.; Hu, Y.; Xie, F.; Guo, H.; Ashforth, E.J.; Polyak, S.W.; Zhu, B.; Zhang, L. Molecular cloning and characterization of a new cold-active esterase from a deep-sea metagenomic library. Appl. Microbiol. Biotechnol. 2011, 90, 961–970. [Google Scholar] [CrossRef] [PubMed]
- Jeon, J.H.; Lee, H.S.; Kim, J.T.; Kim, S.-J.; Choi, S.H.; Kang, S.G.; Lee, J.-H. Identification of a new subfamily of salt-tolerant esterases from a metagenomic library of tidal flat sediment. Appl. Microbiol. Biotechnol. 2012, 93, 623–631. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Xu, X.; Huo, Y.; Wu, Y.; Zhu, X.; Zhang, X.; Wu, M. Identification and characterization of novel esterases from a deep-sea sediment metagenome. Arch. Microbiol. 2012, 194, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, Y.M.; Ghazy, M.A.; Sayed, A.; Ouf, A.; El-Dorry, H.; Siam, R. Isolation and characterization of a heavy metal-resistant, thermophilic esterase from a Red Sea brine pool. Sci. Rep. 2013, 3. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Leiros, H.-K.; de Pascale, D.; Johnson, K.A.; Blencke, H.-M.; Landfald, B. Functional and structural studies of a novel cold-adapted esterase from an Arctic intertidal metagenomic library. Appl. Microbiol. Biotechnol. 2013, 97, 3965–3978. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Li, J.; Cai, H.; Ni, H.; Xiao, A.; Hou, L. Characterization of a new and thermostable esterase from a metagenomic library. Microbiol. Res. 2013, 168, 589–597. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.; Li, J.; Wang, Q.; Fang, W.; Peng, H.; Zhang, X.; Xiao, Y. A novel esterase from a marine metagenomic library exhibiting salt tolerance ability. J. Microbiol. Biotechnol. 2014, 24, 771–780. [Google Scholar] [PubMed]
- Jiang, C.; Wu, L.-L.; Zhao, G.-C.; Shen, P.-H.; Jin, K.; Hao, Z.-Y.; Li, S.-X.; Ma, G.-F.; Luo, F.-F.; Hu, G.-Q.; et al. Identification and characterization of a novel fumarase gene by metagenome expression cloning from marine microorganisms. Microb. Cell Fact. 2010, 9. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Gong, Y.; Xie, W.; Xiao, W.; Wang, J.; Zheng, Y.; Hu, J.; Liu, Z. Identification and characterization of a novel thermostable gh-57 gene from metagenomic fosmid library of the Juan de Fuca Ridge hydrothemal vent. Appl. Biochem. Biotechnol. 2011, 164, 1323–1338. [Google Scholar] [CrossRef] [PubMed]
- Wierzbicka-Woś, A.; Bartasun, P.; Cieśliński, H.; Kur, J. Cloning and characterization of a novel cold-active glycoside hydrolase family 1 enzyme with β-glucosidase, β-fucosidase and β-galactosidase activities. BMC Biotechnol. 2013, 13. [Google Scholar] [CrossRef] [PubMed]
- Cottrell, M.T.; Yu, L.; Kirchman, D.L. Sequence and expression analyses of Cytophaga-like hydrolases in a western Arctic metagenomic library and the Sargasso Sea. Appl. Environ. Microbiol. 2005, 71, 8506–8513. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.; Li, T.; Wang, Q.; Zhang, X.; Peng, H.; Fang, W.; Hong, Y.; Ge, H.; Xiao, Y. A bacterial laccase from marine microbial metagenome exhibiting chloride tolerance and dye decolorization ability. Appl. Microbiol. Biotechnol. 2011, 89, 1103–1110. [Google Scholar] [CrossRef] [PubMed]
- Hårdeman, F.; Sjöling, S. Metagenomic approach for the isolation of a novel low-temperature-active lipase from uncultured bacteria of marine sediment. FEMS Microbiol. Ecol. 2007, 59, 524–534. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.-H.; Lee, C.-H.; Oh, T.-K.; Song, J.K.; Yoon, J.-H. Isolation and characterization of a novel lipase from a metagenomic library of tidal flat sediments: Evidence for a new family of bacterial lipases. Appl. Environ. Microbiol. 2006, 72, 7406–7409. [Google Scholar] [CrossRef] [PubMed]
- Jeon, J.H.; Kim, J.-T.; Kim, Y.J.; Kim, H.-K.; Lee, H.S.; Kang, S.G.; Kim, S.-J.; Lee, J.-H. Cloning and characterization of a new cold-active lipase from a deep-sea sediment metagenome. Appl. Microbiol. Biotechnol. 2009, 81, 865–874. [Google Scholar] [CrossRef] [PubMed]
- Selvin, J.; Kennedy, J.; Lejon, D.P.H.; Kiran, G.S.; Dobson, A.D.W. Isolation identification and biochemical characterization of a novel halo-tolerant lipase from the metagenome of the marine sponge Haliclona simulans. Microb. Cell Fact. 2012, 11. [Google Scholar] [CrossRef] [PubMed]
- Su, J.; Zhang, F.; Sun, W.; Karuppiah, V.; Zhang, G.; Li, Z.; Jiang, Q. A new alkaline lipase obtained from the metagenome of marine sponge Ircinia sp. World J. Microbiol. Biotechnol. 2015, 31, 1093–1102. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.-G.; Jeon, J.H.; Jang, M.K.; Kim, N.Y.; Lee, J.H.; Lee, J.-H.; Kim, S.-J.; Kim, G.-D.; Lee, S.-H. Screening and characterization of a novel fibrinolytic metalloprotease from a metagenomic library. Biotechnol. Lett. 2007, 29, 465–472. [Google Scholar] [CrossRef] [PubMed]
- Prabavathi, R.; Mathivanan, V.; Ambika, A. Screening of protease enzyme by construction of metagenomic library from marine soil sediments. Int. J. Pharma Sci. Res. 2012, 3, 396–399. [Google Scholar]
- Bull, A.T.; Goodfellow, M.; Slater, J.H. Biodiversity as a source of innovation in biotechnology. Annu. Rev. Microbiol. 1992, 46, 219–252. [Google Scholar] [CrossRef] [PubMed]
- Short, J.M. Recombinant approaches for accessing biodiversity. Nat. Biotechnol. 1997, 15, 1322–1323. [Google Scholar] [CrossRef] [PubMed]
- Shigematsu, T.; Hayashi, M.; Kikuchi, I.; Ueno, S.; Masaki, H.; Fujii, T. A culture-dependent bacterial community structure analysis based on liquid cultivation and its application to a marine environment. FEMS Microbiol. Lett. 2009, 293, 240–247. [Google Scholar] [CrossRef] [PubMed]
- Simu, K.; Holmfeldt, K.; Zweifel, U.L.; Hagström, Å. Culturability and coexistence of colony-forming and single-cell marine bacterioplankton. Appl. Environ. Microbiol. 2005, 71, 4793–4800. [Google Scholar] [CrossRef] [PubMed]
- Ivars-Martínez, E.; D’Auria, G.; Rodríguez-Valera, F.; Sánchez-Porro, C.; Ventosa, A.; Joint, I.; Mühling, M. Biogeography of the ubiquitous marine bacterium Alteromonas macleodii determined by multilocus sequence analysis. Mol. Ecol. 2008, 17, 4092–4106. [Google Scholar] [CrossRef] [PubMed]
- Karl, D.M. Microbial oceanography: Paradigms, processes and promise. Nat. Rev. Microbiol. 2007, 5, 759–769. [Google Scholar] [CrossRef] [PubMed]
- Kolter, R.; Greenberg, E.P. The superficial life of microbes. Nature 2006, 18, 300–302. [Google Scholar] [CrossRef] [PubMed]
- Mikkelsen, H.; Duck, Z.; Lilley, K.S.; Welch, M. Interrelationships between colonies, biofilms and planktonic cells of Pseudomonas aeruginosa. J. Bacteriol. 2007, 189, 2411–2416. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, J.; Marchesi, J.R.; Dobson, A.D.W. Metagenomic approaches to exploit the biotechnological potential of the microbial consortia of marine sponges. Appl. Microbiol. Biotechnol. 2007, 75, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Mehbub, M.F.; Lei, J.; Franco, C.; Zhang, W. Marine sponge derived natural products between 2001 and 2010: Trends and opportunities for discovery of bioactives. Mar. Drugs 2014, 12, 4539–4577. [Google Scholar] [CrossRef] [PubMed]
- Perdicaris, S.; Vlachogianni, T.; Valavanidis, A. Bioactive natural substances from marine sponges: New developments and prospects for future pharmaceuticals. Nat. Prod. Chem. Res. 2013, 1. [Google Scholar] [CrossRef]
- Van Pée, K.H.; Patallo, E.P. Flavin-dependent halogenases involved in secondary metabolism in bacteria. Appl. Microbiol. Biotechnol. 2006, 70, 631–641. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Cao, X.; Xin, Y.; Xue, S.; Zhang, W. Purification and characterization of a dehalogenase from Pseudomonas stutzeri DEH130 isolated from the marine sponge Hymeniacidon perlevis. World J. Microbiol. Biotechnol. 2013, 29, 1791–1799. [Google Scholar] [CrossRef] [PubMed]
- Aurilia, V.; Parracino, A.; D’Auria, S. Microbial carbohydrate esterases in cold adapted environments. Gene 2008, 410, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Collins, T.; Meuwis, M.A.; Stals, I.; Claeyssens, M.; Feller, G.; Gerday, C. A novel family 8 xylanase, functional and physicochemical characterization. J. Biol. Chem. 2002, 277, 35133–35139. [Google Scholar] [CrossRef] [PubMed]
- Hobel, C.F.V.; Hreggvidsson, G.Ò.; Marteinsson, V.T.; Bahrani-Mougeot, F.; Einarsson, J.M.; Kristjánsson, J.K. Cloning, expression, and characterization of a highly thermostable family 18 chitinase from Rhodothermus marinus. Extremophiles 2005, 9, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Irwin, J.A.; Lynch, S.V.; Coughlan, S.; Baker, P.J.; Gudmundsson, H.M.; Alfredsson, G.A.; Rice, D.W.; Engel, P.C. Alanine dehydrogenase from the psychrophilic bacterium strain PA-43: Overexpression, molecular characterization, and sequence analysis. Extremophiles 2003, 7, 135–143. [Google Scholar] [PubMed]
- Maki, S.; Yoneta, M.; Takada, Y. Two isocitrate dehydrogenases from a psychrophilic bacterium, Colwellia psychrerythraea. Extremophiles 2006, 10, 237–249. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Watanabe, S.; Yamaoka, N.; Takada, Y. Gene cloning of cold-adapted isocitrate lyase from a psychrophilic bacterium, Colwellia psychrerythraea, and analysis of amino acid residues involved in cold adaptation of this enzyme. Extremophiles 2008, 12, 107–117. [Google Scholar] [CrossRef] [PubMed]
- Woo, J.H.; Hwang, Y.O.; Kang, S.G.; Lee, H.S.; Cho, J.C.; Kim, S.J. Cloning and characterization of three epoxide hydrolases from a marine bacterium, Erythrobacter litoralis HTCC2594. Appl. Microbiol. Biotechnol. 2007, 76, 365–375. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.-W.; Zeng, R.-Y. Purification and characterization of a cold-adapted alpha-amylase produced by Nocardiopsis sp. 7326 isolated from Prydz Bay, Antarctic. Mar. Biotechnol. 2008, 10, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Guo, L.Z.; Lu, W.D. Extracellular production of novel halotolerant, thermostable, and alkali-stable carboxymethyl cellulase by marine bacterium Marinimicrobium sp. LS-A18. Appl. Biochem. Biotechnol. 2012, 168, 550–567. [Google Scholar] [CrossRef] [PubMed]
- Lailaja, V.P.; Chandrasekaran, M. Detergent compatible alkaline lipase produced by marine Bacillus smithii BTMS 11. World J. Microbiol. Biotechnol. 2013, 29, 1349–1360. [Google Scholar] [CrossRef] [PubMed]
- Garsoux, G.; Lamotte, J.; Gerday, C.; Feller, G. Kinetic and structural optimization to catalysis at low temperatures in a psychrophilic cellulase from the Antarctic bacterium Pseudoalteromonas haloplanktis. Biochem. J. 2004, 384, 247–253. [Google Scholar] [CrossRef] [PubMed]
- Truong, L.V.; Tuyen, H.; Helmke, E.; Binh, L.T.; Schweder, T. Cloning of two pectate lyase genes from the marine Antarctic bacterium Pseudoalteromonas haloplanktis strain ANT/505 and characterization of the enzymes. Extremophiles 2001, 5, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Land, M.; Hauser, L.; Jun, S.-R.; Nookaew, I.; Leuze, M.R.; Ahn, T.-H.; Karpinets, T.; Lund, O.; Kora, G.; Wassenaar, T.; Poudel, S.; Ussery, D.W. Insights from 20 years of bacterial genome sequencing. Funct. Integr. Genomics 2015, 15, 141–161. [Google Scholar] [CrossRef] [PubMed]
- Mineta, K.; Gojobori, T. Databases of the marine metagenomics. Gene 2015, 576, 724–728. [Google Scholar] [CrossRef] [PubMed]
- The Micro B3 Project, Ocean Sampling Day. Available online: http://www.microb3.eu/osd (accessed on 12 November 2015).
- Médigue, C.; Krin, E.; Pascal, G.; Barbe, V.; Bernsel, A.; Bertin, P.N.; Cheung, F.; Cruveiller, S.; D’Amico, S.; Duilio, A.; et al. Coping with cold: The genome of the versatile marine Antarctica bacterium Pseudoalteromonas haloplanktis TAC125. Genome Res. 15, 1325–1335.
- Riley, M.; Staley, J.T.; Danchin, A.; Wang, T.Z.; Brettin, T.S.; Hauser, L.J.; Land, M.L.; Thompson, L.S. Genomics of an extreme psychrophile, Psychromonas ingrahamii. BMC Genomics 2008, 9. [Google Scholar] [CrossRef] [PubMed]
- Arahal, D.R.; Lekunberri, I.; González, J.M.; Pascual, J.; Pujalte, M.J.; Pedrós-Alió, C.; Pinhassi, J. Neptuniibacter caesariensis gen. nov., sp. nov., a novel marine genome-sequenced gammaproteobacterium. Int. J. Syst. Evol. Microbiol. 2007, 57, 1000–1006. [Google Scholar] [CrossRef] [PubMed]
- Glöckner, F.O.; Stal, L.J.; Sandaa, R.-A.; Gasol, J.M.; O’Gara, F.; Hernandez, F.; Labrenz, M.; Stoica, E.; Varela, M.M.; Bordalo, A.; Pitta, P. Marine microbial diversity and its role in ecosystem functioning and environmental change. In Marine Board-ESF; Calewaert, J.B., McDonough, N., Eds.; European Science Foundation: Ostend, Belgium, 2012. [Google Scholar]
- Handelsman, J.; Rondon, M.R.; Brady, S.F.; Clardy, J.; Goodman, R.M. Molecular biological access to the chemistry of unknown soil microbes: A new frontier for natural products. Chem. Biol. 1998, 5, 245–249. [Google Scholar] [CrossRef]
- Schloss, P.D.; Handelsman, J. Biotechnological prospects from metagenomics. Curr. Opin. Biotechnol. 2003, 14, 303–310. [Google Scholar] [CrossRef]
- Steele, H.L.; Streit, W.R. Metagenomics: Advances in ecology and biotechnology. FEMS Microbiol. Lett. 2005, 247, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Streit, W.R.; Schmitz, R.A. Metagenomics—The key to the uncultured microbes. Curr. Opin. Microbiol. 2004, 7, 492–498. [Google Scholar] [CrossRef] [PubMed]
- Venter, J.C.; Remington, K.; Heidelberg, J.F.; Halpern, A.L.; Rusch, D.; Eisen, J.A.; Wu, D.; Paulsen, I.; Nelson, K.E.; Nelson, W.; et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science 2004, 304, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Chistoserdova, L. Is metagenomics resolving identification of functions in microbial communities? Microbiol. Biotechnol. 2013, 7, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Bonugli-santos, R.C.; dos Santos Vasconcelos, M.R.; Passarini, M.R.Z.; Vieira, G.A.L.; Lopes, V.C.P.; Mainardi, P.H.; dos Santos, J.A.; de Azevedo Duarte, L.; Otero, I.V.R.; da Silva Yoshida, A.M.; et al. Marine-derived fungi: Diversity of enzymes and biotechnological applications. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Wong, D.W.S. Applications of metagenomics for industrial bioproducts. In Metagenomics: Theory, Methods and Applications, 1st ed.; Marco, D., Ed.; Caister Academic Press: Norfolk, UK, 2010; pp. 141–158. [Google Scholar]
- Imhoff, J.F.; Labes, A.; Wiese, J. Bio-mining the microbial treasures of the ocean: New natural products. Biotechnol. Adv. 2011, 29, 468–482. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, J.; O’Leary, N.D.; Kiran, G.S.; Morrissey, J.P.; O’Gara, F.; Selvin, J.; Dobson, A.D.W. Functional metagenomic strategies for the discovery of novel enzymes and biosurfactants with biotechnological applications from marine ecosystems. J. Appl. Microbiol. 2011, 111, 787–799. [Google Scholar] [CrossRef] [PubMed]
- Piel, J. Approaches to capturing and designing biologically active small molecules produced by uncultured microbes. Annu. Rev. Microbiol. 2011, 65, 431–453. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.C.; Piel, J. Metagenomic approaches for exploiting uncultivated bacteria as a resource for novel biosynthetic enzymology. Chem. Biol. 2013, 20, 636–647. [Google Scholar] [CrossRef] [PubMed]
- Gurgui, C.; Piel, J. Metagenomic approaches to identify and isolate bioactive natural products from microbiota of marine sponges. Methods Mol. Biol. 2010, 668, 247–264. [Google Scholar] [PubMed]
- Liles, M.R.; Williamson, L.L.; Rodbumrer, J.; Torsvik, V.; Goodman, R.M.; Handelsman, J. Recovery, purification, and cloning of high-molecular-weight DNA from soil microorganisms. Appl. Environ. Microbiol. 2008, 74, 3302–3305. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, Y.; Dai, S.; Xie, L.; Kumar, M.S.R.; Sun, W.; Sun, H.; Tang, D.; Li, X. Isolation of high molecular weight DNA from marine sponge bacteria for BAC library construction. Mar. Biotechnol. 2010, 12, 318–325. [Google Scholar] [CrossRef] [PubMed]
- Abe, T.; Sahin, F.P.; Akiyama, K.; Naito, T.; Kishigami, M.; Miyamoto, K.; Sakakibara, Y.; Uemura, D. Construction of a metagenomic library for the marine sponge Halichondria okadai. Biosci. Biotechnol. Biochem. 2012, 76, 633–639. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, J.; Marchesi, J.R.; Dobson, A.D.W. Marine metagenomics: Strategies for the discovery of novel enzymes with biotechnological applications from marine environments. Microb. Cell Fact. 2008, 7. [Google Scholar] [CrossRef] [PubMed]
- Uchiyama, T.; Miyazaki, K. Functional metagenomics for enzyme discovery: Challenges to efficient screening. Curr. Opin. Biotechnol. 2009, 20, 616–622. [Google Scholar] [CrossRef] [PubMed]
- Ufarté, L.; Laville, É.; Duquesne, S.; Potocki-Veronese, G. Metagenomics for the discovery of pollutant degrading enzymes. Biotechnol. Adv. 2015, 33, 1845–1854. [Google Scholar] [CrossRef] [PubMed]
- Rashamuse, K.; Magomani, V.; Ronneburg, T.; Brady, D. A novel family VIII carboxylesterase derived from a leachate metagenome library exhibits promiscuous beta-lactamase activity on nitrocefin. Appl. Microbiol. Biotechnol. 2009, 83, 491–500. [Google Scholar] [CrossRef] [PubMed]
- GOLD: Genomes Online Database. Available online: http://www.genomesonline.org (accessed on 21 November 2015).
- Bell, P.J.L.; Sunna, A.; Gibbs, M.D.; Curach, N.C.; Nevalainen, H.; Bergquist, P.L. Prospecting for novel lipase genes using PCR. Microbiology 2002, 148, 2283–2291. [Google Scholar] [CrossRef] [PubMed]
- Tuffin, M.; Anderson, D.; Heath, C.; Cowan, D.A. Metagenomic gene discovery: How far have we moved into novel sequence space? Biotechnol. J. 2009, 4, 1671–1683. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Arrojo, L.; Guazzaroni, M.-E.; López-Cortés, N.; Beloqui, A.; Ferrer, M. Metagenomic era for biocatalyst identification. Curr. Opin. Biotechnol. 2010, 21, 725–733. [Google Scholar] [CrossRef] [PubMed]
- Turner, N.J. Directed evolution drives the next generation of biocatalysts. Nat. Chem. Biol. 2009, 5, 567–573. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Si, T.; Zhao, H. Biocatalyst development by directed evolution. Bioresour. Technol. 2012, 115, 117–125. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Singh, S. Directed evolution: Tailoring biocatalysts for industrial applications. Crit. Rev. Biotechnol. 2013, 33, 365–378. [Google Scholar] [CrossRef] [PubMed]
- Currin, A.; Swainston, N.; Day, P.J.; Kell, D.B. Synthetic biology for the directed evolution of protein biocatalysts: Navigating sequence space intelligently. Chem. Soc. Rev. 2015, 44, 1172–1239. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; An, J.; Ye, W.; Yang, G.; Qian, Z.-G.; Chen, H.-F.; Cui, L.; Feng, Y. Enhancing the promiscuous phosphotriesterase activity of a thermostable Lactonase (GkaP) for the efficient degradation of organophosphate pesticides. Appl. Environ. Microbiol. 2012, 78, 6647–6655. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Pereira, J.H.; Liu, H.; Tran, H.M.; Hsu, N.S.Y.; Dibble, D.; Singh, S.; Adams, P.D.; Sapra, R.; Hadi, M.Z.; Simmons, B.A.; Sale, K.L. Improved activity of a thermophilic cellulase, Cel5A, from Thermotoga maritima on ionic liquid pretreated switchgrass. PLoS ONE 2013, 8, e79725. [Google Scholar] [CrossRef] [PubMed]
- Zhan, T.; Zhang, K.; Chen, Y.; Lin, Y.; Wu, G.; Zhang, L.; Yao, P.; Shao, Z.; Liu, Z. Improving glyphosate oxidation activity of glycine oxidase from Bacillus cereus by directed evolution. PLoS ONE 2013, 8, e79175. [Google Scholar] [CrossRef] [PubMed]
- Suribabu, K.; Govardhan, L.; Hemalatha, K.P.J. Strain improvement of Brevibacillus borostelensis R1 for optimization of α-amylase production by mutagens. J. Microb. Biochem. Technol. 2014, 6, 123–127. [Google Scholar] [CrossRef]
- Zhou, C.; Xue, Y.; Ma, Y. Evaluation and directed evolution for thermostability improvement of a GH13 thermostable α-glucosidase from Thermus thermophilus TC11. BMC Biotechnol. 2015, 15. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; An, J.; Yang, G.-Y.; Bai, A.; Zheng, B.; Lou, Z.; Wu, G.; Ye, W.; Chen, H.-F.; Feng, Y.; et al. Active site loop conformation regulates promiscuous activity in a lactonase from Geobacillus kaustophilus HTA426. PLoS ONE 2015, 10, e0115130. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Zhang, S.; Gao, H.; Hu, N. Characterization of a cold-active esterase from Serratia sp and improvement of thermostability by directed evolution. BMC Biotechnol. 2016, 16. [Google Scholar] [CrossRef] [PubMed]
- Packer, M.S.; Liu, D.R. Methods for the directed evolution of proteins. Nat. Rev. Genet. 2015, 16, 379–394. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.P.; Huang, J.; Wang, L.F.; Li, J.; Wu, Z.R. A new approach to random mutagenesis in vitro. Biotechnol. Bioeng. 2004, 86, 622–627. [Google Scholar] [CrossRef] [PubMed]
- Mohan, U.; Banerjee, U.C. Molecular evolution of a defined DNA sequence with accumulation of mutations in a single round by a dual approach to random chemical mutagenesis (DuARCheM). ChemBioChem 2008, 9, 2238–2243. [Google Scholar] [CrossRef] [PubMed]
- Selifonova, O.; Valle, F.; Schellenberger, V. Rapid evolution of novel traits in microorganisms. Appl. Environ. Microbiol. 2001, 67, 3645–3649. [Google Scholar] [CrossRef] [PubMed]
- Stefan, A.; Radeghieri, A.; Rodriguez, A.G.V.Y.; Hochkoeppler, A. Directed evolution of beta-galactosidase from Escherichia coli by mutator strains defective in the 3’→5’ exonuclease activity of DNA polymerase III. FEBS Lett. 2001, 493, 139–143. [Google Scholar] [CrossRef]
- Leung, D.W.; Chen, E.; Goeddel, D.V. A method for random mutagenesis of a defined DNA segment using a modified polymerase chain reaction. Technique 1989, 1, 11–15. [Google Scholar]
- Shivange, A.V.; Marienhagen, J.; Mundhada, H.; Schenk, A.; Schwaneberg, U. Advances in generating functional diversity for directed protein evolution. Curr. Opin. Chem. Biol. 2009, 13, 19–25. [Google Scholar] [CrossRef] [PubMed]
- DeSantis, G.; Wong, K.; Farwell, B.; Chatman, K.; Zhu, Z.; Tomlinson, G.; Huang, H.; Tan, X.; Bibbs, L.; Chen, P.; et al. Creation of a productive, highly enantioselective nitrilase through gene site saturation mutagenesis (GSSM). J. Am. Chem. Soc. 2003, 125, 11476–11477. [Google Scholar] [CrossRef] [PubMed]
- Valetti, F.; Gilardi, G. Improvement of biocatalysts for industrial and environmental purposes by saturation mutagenesis. Biomolecules 2013, 3, 778–811. [Google Scholar] [CrossRef] [PubMed]
- Reetz, M.T.; Carballeira, J.D. Iterative saturation mutagenesis (ISM) for rapid directed evolution of functional enzymes. Nat. Protoc. 2007, 2, 891–903. [Google Scholar] [CrossRef] [PubMed]
- Reetz, M.T.; Prasad, S.; Carballeira, J.D.; Gumulya, Y.; Bocola, M. Iterative saturation mutagenesis accelerates laboratory evolution of enzyme stereoselectivity: Rigorous comparison with traditional methods. J. Am. Chem. Soc. 2010, 132, 9144–9152. [Google Scholar] [CrossRef] [PubMed]
- Nobili, A.; Gall, M.G.; Pavlidis, I.V.; Thompson, M.L.; Schmidt, M.; Bornscheuer, U.T. Use of “small but smart” libraries to enhance the enantioselectivity of an esterase from Bacillus stearothermophilus towards tetrahydrofuran-3-yl acetate. FEBS J. 2013, 280, 3084–3093. [Google Scholar] [CrossRef] [PubMed]
- Parra, L.P.; Agudo, R.; Reetz, M.T. Directed evolution by using iterative saturation mutagenesis based on multiresidue sites. ChemBioChem 2013, 14, 2301–2309. [Google Scholar] [CrossRef] [PubMed]
- Reetz, M.T.; Bocola, M.; Carballeira, J.D.; Zha, D.; Vogel, A. Expanding the range of substrate acceptance of enzymes: Combinatorial active-site saturation test. Angew. Chem. Int. Ed. Engl. 2005, 44, 4192–4196. [Google Scholar] [CrossRef] [PubMed]
- Sandström, A.G.; Wikmark, Y.; Engström, K.; Nyhlén, J.; Bäckvall, J-E. Combinatorial reshaping of the Candida antarctica lipase A substrate pocket for enantioselectivity using an extremely condensed library. Proc. Natl. Acad. Sci. USA 2012, 109, 78–83. [Google Scholar] [PubMed]
- Stemmer, W.P. Rapid evolution of a protein in vitro by DNA shuffling. Nature 1994, 370, 389–391. [Google Scholar] [CrossRef] [PubMed]
- Yano, T.; Oue, S.; Kagamiyama, H. Directed evolution of an aspartate aminotransferase with new substrate specificities. Proc. Natl. Acad. Sci. USA 1998, 95, 5511–5515. [Google Scholar] [CrossRef] [PubMed]
- Coco, W.M.; Levinson, W.E.; Crist, M.J.; Hektor, H.J.; Darzins, A.; Pienkos, P.T.; Squires, C.H.; Monticello, D.J. DNA shuffling method for generating highly recombined genes and evolved enzymes. Nat. Biotechnol. 2001, 19, 354–359. [Google Scholar] [CrossRef] [PubMed]
- Gillam, E.M.J.; Copp, J.N.; Ackerley, D.F. Directed Evolution Library Creation, 2nd ed.; Springer: New York, NY, USA, 2014. [Google Scholar]
- Kelley, N.J.; Whelan, D.J.; Kerr, E.; Apel, A.; Beliveau, R.; Scanlon, R. Engineering biology to address global problems: Synthetic biology markets, needs, and applications. Ind. Biotechnol. 2014, 10, 140–149. [Google Scholar] [CrossRef]
- Craig, J.W.; Chang, F.-Y.; Kim, J.H.; Obiajulu, S.C.; Brady, S.F. Expanding small-molecule functional metagenomics through parallel screening of broad-host-range cosmid cnvironmental DNA libraries in diverse Proteobacteria. Appl. Environ. Microbiol. 2010, 76, 1633–1641. [Google Scholar] [CrossRef] [PubMed]
- Wexler, M.; Bond, P.L.; Richardson, D.J.; Johnston, A.W.B. A wide host-range metagenomic library from a waste water treatment plant yields a novel alcohol/aldehyde dehydrogenase. Environ. Microbiol. 2005, 7, 1917–1926. [Google Scholar] [CrossRef] [PubMed]
- Reen, F.J.; Barret, M.; Fargier, E.; O’Muinneacháin, M.; O’Gara, F. Molecular evolution of LysR-type transcriptional regulation in Pseudomonas aeruginosa. Mol. Phylogenet. Evol. 2013, 66, 1041–1049. [Google Scholar] [CrossRef] [PubMed]
- Bayer, T.S.; Widmaier, D.M.; Temme, K.; Mirsky, E.A.; Santi, D.V.; Voigt, C.A. Synthesis of methyl halides from biomass using engineered microbes. J. Am. Chem. Soc. 2009, 131, 6508–6515. [Google Scholar] [CrossRef] [PubMed]
- The Nagoya Protocol. Available online: https://www.cbd.int/abs/ (accessed on 25 November 2015).


| Marine microbial enzymes | Screening method | Environmental DNA source (G or M)a | Reference |
|---|---|---|---|
| Aldehyde reductase | Genome-based | G-Oceanospirillum sp. MED92 | [67] |
| Dehalogenase | Genome-based | G-Psychromonas ingrahamii | [68] |
| Lipase (Lip 1) | Genome-based | G-Pseudoalteromonas haloplanktis | [69] |
| Alkane hydroxylase (AlkB) | Function-based | M-Deep sea sediment | [70] |
| β-Glucosidase (Bgl1A) | Function-based | M-Surface seawater | [71] |
| β-Lactamase | Function-based | M-Cold seep sediments | [72] |
| Chitinase | Sequence-based | M-Aquatic habitats | [73] |
| Chitinase | Function-based | M-Coastal and estuarine waters | [74] |
| Endo-1,4-Glucanase | Function-based | M-Brown algae | [75] |
| Esterase (5 different Est) | Function-based | M-Brine:seawater interface | [76] |
| Esterase (EstA and B) | Function-based | M-Surface seawater | [77] |
| Esterase (EstAT1 and AT11) | Function-based | M-Seashore sediment | [78] |
| Esterase/Lipase | Function-based | M-Deep-sea sediment | [79] |
| Esterase (EstEH1) | Function-based | M-Marine sponge | [80] |
| Esterase (EstF) | Function-based | M-Sea sediment | [81] |
| Esterase (EstKT4, T7 and T9) | Function-based | M-Tidal flat sediment | [82] |
| Esterase (Est6) | Function-based | M-Sea sediment | [83] |
| Esterase (EstATII) | Function-based | M-Red Sea brine pool | [84] |
| Esterase (Est97) | Function-based | M-Intertidal zone | [85] |
| Esterase (EstEP16) | Function-based | M-Deep sea sediment | [86] |
| Esterase (Est9X) | Function-based | M-Surface seawater | [87] |
| Fumarase (FumF) | Sequence-based | M-Sea water | [88] |
| Glycoside hydrolase (GH-57) | Sequence-based | M-Hydrothermal vent | [89] |
| Glycoside hydrolase (BglMKg) | Function-based | M-Sea water | [90] |
| Hydrolase (CelM) | Sequence-based | M-Artic ocean | [91] |
| Laccase (Lac15) | Sequence-based | M-Surface seawater | [92] |
| Lipase (h1Lip1) | Function-based | M-Sea sediment | [93] |
| Lipase (LipG) | Function-based | M-Tidal flat sediment | [94] |
| Lipase (EML1) | Function-based | M-Deep-sea sediment | [95] |
| Lipase (Lpc53E1) | Function-based | M-Marine sponge | [96] |
| Lipase (LipA) | Function-based | M-Marine sponge | [97] |
| Protease | Function-based | M-Sea sediment | [98] |
| Protease | Function-based | M-Sea sediment | [99] |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parages, M.L.; Gutiérrez-Barranquero, J.A.; Reen, F.J.; Dobson, A.D.W.; O’Gara, F. Integrated (Meta) Genomic and Synthetic Biology Approaches to Develop New Biocatalysts. Mar. Drugs 2016, 14, 62. https://doi.org/10.3390/md14030062
Parages ML, Gutiérrez-Barranquero JA, Reen FJ, Dobson ADW, O’Gara F. Integrated (Meta) Genomic and Synthetic Biology Approaches to Develop New Biocatalysts. Marine Drugs. 2016; 14(3):62. https://doi.org/10.3390/md14030062
Chicago/Turabian StyleParages, María L., José A. Gutiérrez-Barranquero, F. Jerry Reen, Alan D.W. Dobson, and Fergal O’Gara. 2016. "Integrated (Meta) Genomic and Synthetic Biology Approaches to Develop New Biocatalysts" Marine Drugs 14, no. 3: 62. https://doi.org/10.3390/md14030062
APA StyleParages, M. L., Gutiérrez-Barranquero, J. A., Reen, F. J., Dobson, A. D. W., & O’Gara, F. (2016). Integrated (Meta) Genomic and Synthetic Biology Approaches to Develop New Biocatalysts. Marine Drugs, 14(3), 62. https://doi.org/10.3390/md14030062