Development of Synechocystis sp. PCC 6803 as a Phototrophic Cell Factory
Abstract
:1. Introduction
2. Influential Factors for Synechocystis 6803 Cultivation
2.1. CO2 Fixation
2.2. Organic Carbon Utilization
2.3. Light Harvesting
2.4. Nitrogen and Phosphorus Uptake
2.5. Cultivation Conditions
3. Genetic and Molecular Tools
3.1. Plasmid Vectors
3.2. Transformation and Segregation
3.3. Transcriptional Control Tools
3.4. Translational Control Tools
3.5. Posttranslational Control Tools
4. High Throughput System Biology for Synechocystis 6803
4.1. Transcriptomics
4.2. Proteomics
4.3. Metabolomics
5. Metabolic Modeling for Synechocystis 6803
5.1. Flux Balance Analysis (FBA)
5.2. Flux Coupling Finder (FCF)
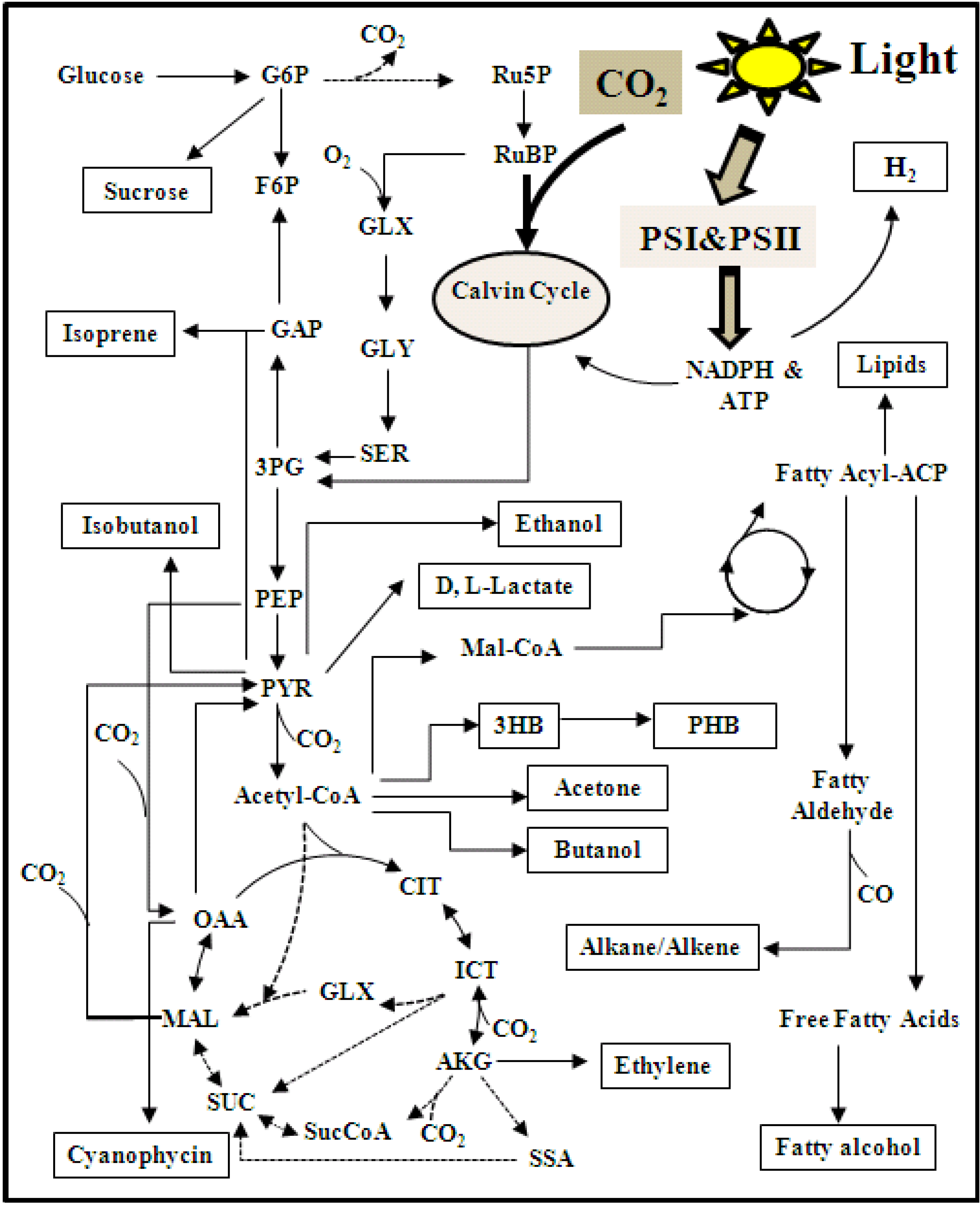
5.3. 13C Metabolic Flux Analysis (13C-MFA)
6. Applications of Synechocystis 6803 as a Cell Factory
| Strains | Chemicals | Genetic modification | Productivity | Growth conditions | References |
|---|---|---|---|---|---|
| Synechocystis 6803 | Ethanol | pdc and slr1192; ΔphaAB | 5.50 g/L | photoautotrophic; Sparging with 5% CO2-air | [92] |
| Promoter rbc | |||||
| Fatty acids | tesA, tesA137 (codon optimized), accBCDA, fatB1, fatB1, fatB2; ΔphaAB, Δsll1951, Δslr2001-slr2002, Δslr1710, Δslr2132 | 197 ± 14 mg/L | photoautotrophic; Bubbled with 1% CO2 | [44] | |
| Promoter psbA2, rbc, cpc, trc | |||||
| Isoprene | ispS (codon optimized) | 50 μg/g DCW/day | photoautotrophic | [93] | |
| Promoter psbA2 | |||||
| Alk(a/e)nes | sll0208 & sll0209 | 2.3 mg/L/OD730 | photoautotrophic | [94] | |
| Promoter rbc | |||||
| Fatty alcohols | far, at3g11980 Δagp, ΔphaAB | 761 ± 216 µg/g DCW | photoautotrophic | [95] | |
| Promoter psbD13, rbcL12 | |||||
| Sucrose | sps, spp, ugp ΔggpS | 35 mg/L/OD730 | photoautotrophic with 600 mM NaCl | [96] | |
| Promoter petE | |||||
| Hydrogen | ΔnarB, ΔnirA | 186 nmol/mg chl a/h | nitrogen-limiting in the dark | [97] | |
| Synechococcus7002 | Hydrogen | ΔldhA | 14.1 mol per day per 1017 cells | Anaerobic in the dark | [98] |
| Sucrose | ΔglgA-1, ΔglgA-II | 71 ± 3 mol per 1017 cells | Under hypersaline condition | [99] | |
| Synechococcus7942 | Ethanol | pdc and adhII | 0.23 g/L | photoautotrophic | [43] |
| Promoter rbcLS | |||||
| Isobutyraldehyde | kivd, alsS, ilvC, ilvD and rbclS | 1.1 g/L | photoautotrophic with NaHCO3 | [100] | |
| Promoter LlacO1, trc, tac | |||||
| Isobutanol | kivd, alsS, ilvCD and yqhD | 0.45 g/L | photoautotrophic with NaHCO3 | [100] | |
| Promoter LlacO1, trc | |||||
| Fatty acids | tesA and Δaas | 80 ± 10 mg/DCW | photoautotrophic; Bubbled with CO2 | [101] | |
| Promoter trc | |||||
| Hydrogen | hydEF, hydG, hydA | 2.8 µmol/h/mg Chl-a | Anaerobic in the dark | [102] | |
| Promoter psbA1, lac |
6.1. Biofuels
6.2. Commodity Chemical
6.3. Polyesters
7. Challenges and Perspectives
7.1. Low Product Titer and Rate
7.2. Product Loss and Process Failure during Long-Term Cultivation
7.3. Production Costs and Scale up
8. Conclusions
Acknowledgments
References
- Pisciotta, J.M.; Zou, Y.; Baskakov, I.V. Light-Dependent Electrogenic Activity of Cyanobacteria. PLoS One 2010, 5, e10821. [Google Scholar] [CrossRef]
- Wang, B.; Wang, J.; Zhang, W.; Meldrum, D.R. Application of synthetic biology in cyanobacteria and algae. Front.Microbiol. 2012, 3, 344. [Google Scholar]
- Ruffing, A.M. Engineered cyanobacteria: Teaching an old bug new tricks. Bioeng. Bugs 2011, 2, 136–149. [Google Scholar] [CrossRef]
- Kaneko, T.; Nakamura, Y.; Sasamoto, S.; Watanabe, A.; Kohara, M.; Matsumoto, M.; Shimpo, S.; Yamada, M.; Tabata, S. Structural analysis of four large plasmids harboring in a unicellular cyanobacterium, Synechocystis sp. PCC 6803. DNA Res. 2003, 10, 221–228. [Google Scholar] [CrossRef]
- Kaneko, T.; Sato, S.; Kotani, H.; Tanaka, A.; Asamizu, E.; Nakamura, Y.; Miyajima, N.; Hirosawa, M.; Sugiura, M.; Sasamoto, S.; et al. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996, 3, 109–136. [Google Scholar] [CrossRef]
- Vermaas, W. Molecular genetics of the cyanobacterium Synechocystis sp. PCC 6803: Principles and possible biotechnology applications. J. Appl. Phycol. 1996, 8, 263–273. [Google Scholar] [CrossRef]
- Los, D.A.; Zorina, A.; Sinetova, M.; Kryazhov, S.; Mironov, K.; Zinchenko, V.V. Stress Sensors and Signal Transducers in Cyanobacteria. Sensors 2010, 10, 2386–2415. [Google Scholar] [CrossRef]
- Zang, X.N.; Liu, B.; Liu, S.M.; Arunakumara, K.K.I.U.; Zhang, X.C. Optimum conditions for transformation of Synechocystis sp PCC 6803. J. Microbiol. 2007, 45, 241–245. [Google Scholar]
- Kim, H.W.; Vannela, R.; Zhou, C.; Rittmann, B.E. Nutrient acquisition and limitation for the photoautotrophic growth of Synechocystis sp. PCC6803 as a renewable biomass source. Biotechnol. Bioeng. 2011, 108, 277–285. [Google Scholar] [CrossRef]
- Pierce, J.; Carlson, T.J.; Williams, J.G.K. A cyanobacterial mutant requiring the expression of ribulose bisphosphate carboxylase from a photosynthetic anaerobe. Proc. Natl. Acad. Sci. USA 1989, 86, 5753–5757. [Google Scholar] [CrossRef]
- Young, J.D.; Shastri, A.A.; Stephanopoulos, G.; Morgan, J.A. Mapping photoautotrophic metabolism with isotopically nonstationary 13C flux analysis. Metab. Eng. 2011, 13, 656–665. [Google Scholar] [CrossRef]
- Battchikova, N.; Vainonen, J.P.; Vorontsova, N.; Keranen, M.; Carmel, D.; Aro, E.M. Dynamic Changes in the Proteome of Synechocystis 6803 in Response to CO2 Limitation Revealed by Quantitative Proteomics. J. Proteome Res. 2010, 9, 5896–5912. [Google Scholar] [CrossRef]
- Kaplan, A.; Schwarz, R.; Lieman-Hurwitz, J.; Reinhold, L. Physiological and molecular aspects of the inorganic carbon-concentrating mechanism in cyanobacteria. Plant Physiol. 1991, 97, 851–855. [Google Scholar] [CrossRef]
- Maeda, S.; Price, G.D.; Badger, M.R.; Enomoto, C.; Omata, T. Bicarbonate binding activity of the CmpA protein of the cyanobacterium Synechococcus sp. strain PCC 7942 involved in active transport of bicarbonate. J. Biol. Chem. 2000, 275, 20551–20555. [Google Scholar]
- Xu, M.; Bernat, G.; Singh, A.; Mi, H.L.; Rogner, M.; Pakrasi, H.B.; Ogawa, T. Properties of mutants of Synechocystis sp. Strain PCC 6803 lacking inorganic carbon sequestration systems. Plant Cell Physiol. 2008, 49, 1672–1677. [Google Scholar] [CrossRef]
- Williams, J.G.K. Construction of specific mutations in photosystem-II photosynthetic reaction center by genetic-engineering methods in Synechocystis-6803. Methods Enzymol. 1988, 167, 766–778. [Google Scholar] [CrossRef]
- Bartsevich, V.V.; Pakrasi, H.B. Molecular-identification of an abc transporter complex for manganese—Analysis of a cyanobacterial mutant strain impaired in the photosynthetic oxygen evolution process. EMBO J. 1995, 14, 1845–1853. [Google Scholar]
- Ikeuchi, M.; Tabata, S. Synechocystis sp. PCC 6803—A useful tool in the study of the genetics of cyanobacteria. Photosynth. Res. 2001, 70, 73–83. [Google Scholar] [CrossRef]
- Yoshikawa, K.; Hirasawa, T.; Ogawa, K.; Hidaka, Y.; Nakajima, T.; Furusawa, C.; Shimizu, H. Integrated transcriptomic and metabolomic analysis of the central metabolism of Synechocystis sp. PCC 6803 under different trophic conditions. Biotechnol. J. 2013, 8, 571–580. [Google Scholar] [CrossRef]
- Anderson, S.L.; Mcintosh, L. Light-activated heterotrophic growth of the cyanobacterium Synechocystis sp. strain PCC 6803: A blue-light-requiring process. J. Bact. 1991, 173, 2761–2767. [Google Scholar]
- You, L.; He, L.; Tang, Y.J. 13C-metabolism Analysis of Photoheterotrophic Cyanobacterium Synechocystis sp. Strain PCC 6803. Appl. Environ. Microbiol. 2013. submitted for publication. [Google Scholar]
- Marcus, Y.; Altman-Gueta, H.; Wolff, Y.; Gurevitz, M. Rubisco mutagenesis provides new insight into limitations on photosynthesis and growth in Synechocystis PCC6803. J. Exp. Bot. 2011, 62, 4173–4182. [Google Scholar] [CrossRef]
- Beckmann, J.; Lehr, F.; Finazzi, G.; Hankamer, B.; Posten, C.; Wobbe, L.; Kruse, O. Improvement of light to biomass conversion by de-regulation of light-harvesting protein translation in Chlamydomonas reinhardtii. J. Biotechnol. 2009, 142, 70–77. [Google Scholar] [CrossRef]
- Page, L.E.; Liberton, M.; Pakrasi, H.B. Reduction of photoautotrophic productivity in the cyanobacterium Synechocystis sp. strain PCC 6803 by phycobilisome antenna truncation. Appl. Environ. Microbiol. 2012, 78, 6349–6351. [Google Scholar] [CrossRef]
- Collins, A.M.; Liberton, M.; Jones, H.D.T.; Garcia, O.F.; Pakrasi, H.B.; Timlin, J.A. Photosynthetic pigment localization and thylakoid membrane morphology are altered in Synechocystis 6803 phycobilisome mutants. Plant Physiol. 2012, 158, 1600–1609. [Google Scholar] [CrossRef]
- Krasikov, V.; Aguirre von Wobeser, E.; Dekker, H.L.; Huisman, J.; Matthijs, H.C. Time-series resolution of gradual nitrogen starvation and its impact on photosynthesis in the cyanobacterium Synechocystis PCC 6803. Physiol. Plant 2012, 145, 426–439. [Google Scholar] [CrossRef]
- Flores, E.; Herrero, A. Nitrogen assimilation and nitrogen control in cyanobacteria. Biochem. Soc. Trans. 2005, 33, 164–167. [Google Scholar] [CrossRef]
- Richter, R.; Hejazi, M.; Kraft, R.; Ziegler, K.; Lockau, W. Cyanophycinase, a peptidase degrading the cyanobacterial reserve material multi-l-arginyl-poly-l-aspartic acid (cyanophycin). Eur. J. Biochem. 1999, 263, 163–169. [Google Scholar] [CrossRef]
- Osanai, T.; Imamura, S.; Asayama, M.; Shirai, M.; Suzuki, I.; Murata, N.; Tanaka, K. Nitrogen induction of sugar catabolic gene expression in Synechocystis sp PCC 6803. DNA Res. 2006, 13, 185–195. [Google Scholar] [CrossRef]
- Burut-Archanai, S.; Eaton-Rye, J.J.; Incharoensakdi, A.; Powtongsook, S. Phosphorus removal in a closed recirculating aquaculture system using the cyanobacterium Synechocystis sp. PCC 6803 strain lacking the SphU regulator of the Pho regulon. Biochem. Eng. J. 2013, 74, 69–75. [Google Scholar] [CrossRef]
- Fuszard, M.A.; Ow, S.Y.; Gan, C.S.; Noirel, J.; Ternan, N.G.; McMullan, G.; Biggs, C.A.; Reardon, K.F.; Wright, P.C. The quantitative proteomic response of Synechocystis sp. PCC6803 to phosphate acclimation. Aquat. Biosyst. 2013, 9, 5. [Google Scholar] [CrossRef]
- Collier, J.L.; Grossman, A.R. Chlorosis induced by nutrient deprivation in Synechococcus sp. strain PCC 7942: Not all bleaching is the same. J. Bact. 1992, 174, 4718–4726. [Google Scholar]
- Heidorn, T.; Camsund, D.; Huang, H.H.; Lindberg, P.; Oliveira, P.; Stensjo, K.; Lindblad, P. Synthetic biology in cyanobacteria: Engineering and analyzing novel functions. Method Enzymol. 2011, 497, 539–579. [Google Scholar] [CrossRef]
- Muramatsu, M.; Sonoike, K.; Hihara, Y. Mechanism of downregulation of photosystem I content under high-light conditions in the cyanobacterium Synechocystis sp. PCC 6803. Microbiology (SGM) 2009, 155, 989–996. [Google Scholar] [CrossRef]
- Mohamed, A.; Jansson, C. Influence of light on accumulation of photosynthesis-specific transcripts in the cyanobacterium Synechocystis-6803. Plant Mol. Biol. 1989, 13, 693–700. [Google Scholar] [CrossRef]
- Mermet-bouvier, P.; Cassier-chauvat, C.; Marraccini, P.; Chauvat, F. Transfer and replication of Rsf1010-derived plasmids in several cyanobacteria of the general Synechocystis and Synechococcus. Curr. Microbiol. 1993, 27, 323–327. [Google Scholar]
- Mermet-bouvier, P.; Chauvat, F. A Conditional expression vector for the cyanobacteria Synechocystis sp. strains PCC6803 and PCC6714 or Synechococcus sp. strains PCC7942 and PCC6301. Curr. Microbiol. 1994, 28, 145–148. [Google Scholar] [CrossRef]
- Ferino, F.; Chauvat, F. A Promoter-probe vector-host system for the cyanobacterium, Synechocystis PCC6803. Gene 1989, 84, 257–266. [Google Scholar] [CrossRef]
- Chauvat, F.; Devries, L.; Vanderende, A.; Vanarkel, G.A. A host-vector system for gene cloning in the cyanobacterium Synechocystis PCC6803. Mol. Gen. Genet. 1986, 204, 185–191. [Google Scholar] [CrossRef]
- Becker, E.C.; Meyer, R.J. Acquisition of resista nce genes by the IncQ plasmid R1162 is limited by its high copy number and lack of a partitioning mechanism. J. Bact. 1997, 179, 5947–5950. [Google Scholar]
- Kufryk, G.I.; Sachet, M.; Schmetterer, G.; Vermaas, W.F. Transformation of the cyanobacterium Synechocystis sp. PCC6803 as a tool for genetic mapping: Optimization of efficiency. FEMS Microbiol. Lett. 2002, 206, 215–219. [Google Scholar] [CrossRef]
- Tajima, N.; Sato, S.; Maruyama, F.; Kaneko, T.; Sasaki, N.V.; Kurokawa, K.; Ohta, H.; Kanesaki, Y.; Yoshikawa, H.; Tabata, S.; et al. Genomic structure of the cyanobacterium Synechocystis sp. PCC6803 strain GT-S. DNA Res. 2011, 18, 393–399. [Google Scholar] [CrossRef]
- Deng, M.D.; Coleman, J.R. Ethanol synthesis by genetic engineering in cyanobacteria. Appl. Environ. Microbiol. 1999, 65, 523–528. [Google Scholar]
- Liu, X.; Sheng, J.; Curtiss, R., III. Fatty acid production in genetically modified cyanobacteria. Proc. Natl. Acad. Sci. USA 2011, 108, 6899–6904. [Google Scholar] [CrossRef]
- Aoki, S.; Kondo, T.; Ishiura, M. A promoter-trap vector for clock-controlled genes in the cyanobacterium Synechocystis sp. PCC6803. J. Microbiol. Methods 2002, 49, 265–274. [Google Scholar] [CrossRef]
- Liu, X.; Curtiss, R., III. Nickel-inducible lysis system in Synechocystis sp. PCC 6803. Proc. Natl. Acad. Sci. USA 2009, 106, 21550–21554. [Google Scholar] [CrossRef]
- Huang, H.H.; Camsund, D.; Lindblad, P.; Heidorn, T. Design and characterization of molecular tools for a synthetic biology approach towards developing cyanobacterial biotechnology. Nucleic Acids Res. 2010, 38, 2577–2593. [Google Scholar] [CrossRef]
- Imamura, S.; Asayama, M. Sigma factors for cyanobacterial transcription. Gene Regul. Syst. Biol. 2009, 3, 65–87. [Google Scholar]
- Osanai, T.; Oikawa, A.; Azuma, M.; Tanaka, K.; Saito, K.; Hirai, M.Y.; Ikeuchi, M. Genetic engineering of group 2 sigma factor SigE widely activates expressions of sugar catabolic genes in Synechocystis species PCC 6803. J. Biol. Chem. 2011, 286, 30962–30971. [Google Scholar]
- Zhang, F.; Keasling, J. Biosensors and their applications in microbial metabolic engineering. Trends Microbiol. 2011, 19, 323–329. [Google Scholar] [CrossRef]
- Zhang, F.; Carothers, J.M.; Keasling, J.D. Design of a dynamic sensor-regulator system for production of chemicals and fuels derived from fatty acids. Nat. Biotechnol. 2012, 30, 354–359. [Google Scholar] [CrossRef]
- Ma, J.; Campbell, A.; Karlin, S. Correlations between Shine-Dalgarno sequences and gene features such as predicted expression levels and operon structures. J. Bact. 2002, 184, 5733–5745. [Google Scholar] [CrossRef]
- Salis, H.M.; Mirsky, E.A.; Voigt, C.A. Automated design of synthetic ribosome binding sites to control protein expression. Nat. Biotechnol. 2009, 27, 946–950. [Google Scholar] [CrossRef]
- Beiter, T.; Reich, E.; Williams, R.W.; Simon, P. Antisense transcription: A critical look in both directions. Cell Mol. Life Sci. 2009, 66, 94–112. [Google Scholar] [CrossRef]
- Georg, J.; Hess, W.R. cis-Antisense RNA, another level of gene regulation in bacteria. Microbiol. Mol. Biol. Rev. 2011, 75, 286–300. [Google Scholar] [CrossRef]
- Duhring, U.; Axmann, I.M.; Hess, W.R.; Wilde, A. An internal antisense RNA regulates expression of the photosynthesis gene isiA. Proc. Natl. Acad. Sci. USA 2006, 103, 7054–7058. [Google Scholar]
- Georg, J.; Voss, B.; Scholz, I.; Mitschke, J.; Wilde, A.; Hess, W.R. Evidence for a major role of antisense RNAs in cyanobacterial gene regulation. Mol. Syst. Biol. 2009, 5, 305. [Google Scholar]
- Horvath, P.; Barrangou, R. CRISPR/Cas, the immune system of bacteria and archaea. Science 2010, 327, 167–170. [Google Scholar] [CrossRef]
- Makarova, K.S.; Haft, D.H.; Barrangou, R.; Brouns, S.J.; Charpentier, E.; Horvath, P.; Moineau, S.; Mojica, F.J.; Wolf, Y.I.; Yakunin, A.F.; et al. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 2011, 9, 467–477. [Google Scholar]
- Scholz, I.; Lange, S.J.; Hein, S.; Hess, W.R.; Backofen, R. CRISPR-Cas systems in the cyanobacterium Synechocystis sp. PCC6803 exhibit distinct processing pathways involving at least two Cas6 and a Cmr2 protein. PLoS One 2013, 8, e56470. [Google Scholar]
- van Kasteren, S. Synthesis of post-translationally modified proteins. Biochem. Soc. Trans. 2012, 40, 929–944. [Google Scholar] [CrossRef]
- Landry, B.P.; Stockel, J.; Pakrasi, H.B. Use of degradation tags to control protein levels in the cyanobacterium Synechocystis sp. Strain PCC6803. Appl. Environ. Microbiol. 2013, 79, 2833–2835. [Google Scholar] [CrossRef]
- Hagemann, M. Molecular biology of cyanobacterial salt acclimation. FEMS Microbiol. Rev. 2011, 35, 87–123. [Google Scholar] [CrossRef]
- Singh, A.K.; Elvitigala, T.; Cameron, J.C.; Ghosh, B.K.; Bhattacharyya-Pakrasi, M.; Pakrasi, H.B. Integrative analysis of large scale expression profiles reveals core transcriptional response and coordination between multiple cellular processes in a cyanobacterium. Bmc. Syst. Biol. 2010, 4, 105. [Google Scholar]
- Wang, J.; Chen, L.; Huang, S.; Liu, J.; Ren, X.; Tian, X.; Qiao, J.; Zhang, W. RNA-seq based identification and mutant validation of gene targets related to ethanol resistance in cyanobacterial Synechocystis sp. PCC6803. Biotechnol. Biofuels 2012, 5, 89. [Google Scholar] [CrossRef]
- Tian, X.; Chen, L.; Wang, J.; Qiao, J.; Zhang, W. Quantitative proteomics reveals dynamic responses of Synechocystis sp. PCC6803 to next-generation biofuel butanol. J. Proteomics 2013, 78, 326–345. [Google Scholar]
- Qiao, J.; Wang, J.; Chen, L.; Tian, X.; Huang, S.; Ren, X.; Zhang, W. Quantitative iTRAQ LC-MS/MS proteomics reveals metabolic responses to biofuel ethanol in cyanobacterial Synechocystis sp. PCC6803. J. Proteome Res. 2012, 11, 5286–5300. [Google Scholar] [CrossRef]
- Liu, J.; Chen, L.; Wang, J.; Qiao, J.; Zhang, W. Proteomic analysis reveals resistance mechanism against biofuel hexane in Synechocystis sp. PCC6803. Biotechnol. Biofuels 2012, 5, 68. [Google Scholar] [CrossRef]
- Krall, L.; Huege, J.; Catchpole, G.; Steinhauser, D.; Willmitzer, L. Assessment of sampling strategies for gas chromatography-mass spectrometry (GC-MS) based metabolomics of cyanobacteria. J. Chromatogr. B 2009, 877, 2952–2960. [Google Scholar] [CrossRef]
- Pearce, J.; Leach, C.K.; Carr, N.G. The incomplete tricarboxylic acid cycle in the blue-green alga Anabaena variabilis. J. Gen. Microbiol. 1969, 55, 371–378. [Google Scholar] [CrossRef]
- Zhang, S.; Bryant, D.A. The tricarboxylic acid cycle in Cyanobacteria. Science 2011, 334, 1551–1553. [Google Scholar] [CrossRef]
- Steinhauser, D.; Fernie, A.R.; Araújo, W.L. Unusual cyanobacterial TCA cycles: Not broken just different. Trends Plant Sci. 2012, 17, 503–509. [Google Scholar] [CrossRef]
- Pelroy, R.A.; Rippka, R.; Stanier, R.Y. Metabolism of glucose by unicellular blue-green algae. Arch. Microbiol. 1972, 87, 303–322. [Google Scholar]
- Yang, C.Y.; Hua, Q.H.; Shimizu, K.S. Integration of the information from gene expression and metabolic fluxes for the analysis of the regulatory mechanisms in Synechocystis. Appl. Microbiol. Biotechnol. 2002, 58, 813–822. [Google Scholar] [CrossRef]
- Kahlon, S.; Beeri, K.; Ohkawa, H.; Hihara, Y.; Murik, O.; Suzuki, I.; Ogawa, T.; Kaplan, A. A putative sensor kinase, Hik31, is involved in the response of Synechocystis sp. strain PCC6803 to the presence of glucose. Microbiology 2006, 152, 647–655. [Google Scholar] [CrossRef]
- Herranen, M.; Battchikova, N.; Zhang, P.; Graf, A.; Sirpiö, S.; Paakkarinen, V.; Aro, E.-M. Towards Functional Proteomics of Membrane Protein Complexes in Synechocystis sp. PCC6803. Plant Physiol. 2004, 134, 470–481. [Google Scholar] [CrossRef]
- Orth, J.D.; Thiele, I.; Palsson, B.O. What is flux balance analysis? Nat. Biotechnol. 2010, 28, 245–248. [Google Scholar]
- Varma, A.; Palsson, B.O. Metabolic Flux Balancing: Basic Concepts, Scientific and Practical Use. Nat. Biotechnol. 1994, 12, 994–998. [Google Scholar] [CrossRef]
- Shastri, A.A.; Morgan, J.A. Flux balance analysis of photoautotrophic metabolism. Biotechnol. Prog. 2005, 21, 1617–1626. [Google Scholar] [CrossRef]
- Nogales, J.; Gudmundsson, S.; Knight, E.M.; Palsson, B.O.; Thiele, I. Detailing the optimality of photosynthesis in cyanobacteria through systems biology analysis. Proc. Natl. Acad. Sci. USA 2012, 109, 2678–2683. [Google Scholar]
- Fischer, E.; Sauer, U. Large-scale in vivo flux analysis shows rigidity and suboptimal performance of Bacillus subtilis metabolism. Nat. Genet. 2005, 37, 636–640. [Google Scholar] [CrossRef]
- Mahadevan, R.; Schilling, C.H. The effects of alternate optimal solutions in constraint-based genome-scale metabolic models. Metab. Eng. 2003, 5, 264–276. [Google Scholar] [CrossRef]
- Price, N.D.; Reed, J.L.; Palsson, B.O. Genome-scale models of microbial cells: Evaluating the consequences of constraints. Nat. Rev. Microbiol. 2004, 2, 886–897. [Google Scholar] [CrossRef]
- Chen, X.; Alonso, A.P.; Allen, D.K.; Reed, J.L.; Shachar-Hill, Y. Synergy between 13C-metabolic flux analysis and flux balance analysis for understanding metabolic adaption to anaerobiosis in E. coli. Metab. Eng. 2011, 13, 38–48. [Google Scholar] [CrossRef]
- Burgard, A.P.; Nikolaev, E.V.; Schilling, C.H.; Maranas, C.D. Flux coupling analysis of genome-scale metabolic network reconstructions. Genome Res. 2004, 14, 301–312. [Google Scholar] [CrossRef]
- Montagud, A.; Zelezniak, A.; Navarro, E.; de Córdoba, P.F.; Urchueguía, J.F.; Patil, K.R. Flux coupling and transcriptional regulation within the metabolic network of the photosynthetic bacterium Synechocystis sp. PCC6803. Biotechnol. J. 2011, 6, 330–342. [Google Scholar]
- Wiechert, W. 13C metabolic flux analysis. Metab. Eng. 2001, 3, 195–206. [Google Scholar] [CrossRef]
- You, L.; Page, L.; Feng, X.; Berla, B.; Pakrasi, H.B.; Tang, Y.J. Metabolic pathway confirmation and discovery through 13C-labeling of proteinogenic amino acids. J. Vis. Exp. 2012, 59, e3583. [Google Scholar]
- Tang, J.K.-H.; You, L.; Blankenship, R.E.; Tang, Y.J. Recent advances in mapping environmental microbial metabolisms through 13C isotopic fingerprints. J. Royal Soc. Interface 2012, 9, 2767–2780. [Google Scholar] [CrossRef]
- Yang, C.; Hua, Q.; Shimizu, K. Metabolic flux analysis in Synechocystis using isotope distribution from 13C-labeled glucose. Metab. Eng. 2002, 4, 202–216. [Google Scholar] [CrossRef]
- Huege, J.; Goetze, J.; Schwarz, D.; Bauwe, H.; Hagemann, M.; Kopka, J. Modulation of the major paths of carbon in photorespiratory mutants of Synechocystis. PLoS One 2011, 6, e16278. [Google Scholar]
- Gao, Z.; Zhao, H.; Li, Z.; Tan, X.; Lu, X. Photosynthetic production of ethanol from carbon dioxide in genetically engineered cyanobacteria. Energy Environ. Sci. 2012, 5, 9857–9865. [Google Scholar] [CrossRef]
- Lindberg, P.; Park, S.; Melis, A. Engineering a platform for photosynthetic isoprene production in cyanobacteria, using Synechocystis as the model organism. Metab. Eng. 2010, 12, 70–79. [Google Scholar] [CrossRef]
- Wang, W.; Liu, X.; Lu, X. Engineering cyanobacteria to improve photosynthetic production of alka(e)nes. Biotechnol. Biofuels 2013, 6, 69. [Google Scholar] [CrossRef]
- Qi, F.; Yao, L.; Tan, X.; Lu, X. Construction, characterization and application of molecular tools for metabolic engineering of Synechocystis sp. Biotechnol. Lett. 2013, in press. [Google Scholar]
- Du, W.; Liang, F.; Duan, Y.; Tan, X.; Lu, X. Exploring the photosynthetic production capacity of sucrose by cyanobacteria. Metab. Eng. 2013, 19, 17–25. [Google Scholar] [CrossRef]
- Baebprasert, W.; Jantaro, S.; Khetkorn, W.; Lindblad, P.; Incharoensakdi, A. Increased H2 production in the cyanobacterium Synechocystis sp. strain PCC6803 by redirecting the electron supply via genetic engineering of the nitrate assimilation pathway. Metab. Eng. 2011, 13, 610–616. [Google Scholar] [CrossRef]
- McNeely, K.; Xu, Y.; Bennette, N.; Bryant, D.A.; Dismukes, G.C. Redirecting reductant flux into hydrogen production via metabolic engineering of fermentative carbon metabolism in a cyanobacterium. Appl. Environ. Microbiol. 2010, 76, 5032–5038. [Google Scholar] [CrossRef]
- Xu, Y.; Guerra, L.T.; Li, Z.; Ludwig, M.; Dismukes, G.C.; Bryant, D.A. Altered carbohydrate metabolism in glycogen synthase mutants of Synechococcus sp. strain PCC7002: Cell factories for soluble sugars. Metab. Eng. 2013, 16, 56–67. [Google Scholar] [CrossRef]
- Atsumi, S.; Higashide, W.; Liao, J.C. Direct photosynthetic recycling of carbon dioxide to isobutyraldehyde. Nat. Biotechnol. 2009, 27, 1177–1180. [Google Scholar]
- Ruffing, A.M.; Jones, H.D.T. Physiological effects of free fatty acid production in genetically engineered Synechococcus elongatus PCC7942. Biotechnol. Bioeng. 2012, 109, 2190–2199. [Google Scholar] [CrossRef]
- Ducat, D.C.; Sachdeva, G.; Silver, P.A. Rewiring hydrogenase-dependent redox circuits in cyanobacteria. Proc. Natl. Acad. Sci. USA 2011, 108, 3941–3946. [Google Scholar]
- Chisti, Y. Biodiesel from microalgae beats bioethanol. Trends Biotechnol. 2008, 26, 126–131. [Google Scholar] [CrossRef]
- Lan, E.I.; Liao, J.C. Metabolic engineering of cyanobacteria for 1-butanol production from carbon dioxide. Metab. Eng. 2011, 13, 353–363. [Google Scholar] [CrossRef]
- Varman, A.M.; Xiao, Y.; Pakrasi, H.B.; Tang, Y.J. Metabolic engineering of Synechocystis sp. strain PCC6803 for isobutanol production. Appl. Environ. Microbiol. 2013, 79, 908–914. [Google Scholar] [CrossRef]
- Tan, X.; Yao, L.; Gao, Q.; Wang, W.; Qi, F.; Lu, X. Photosynthesis driven conversion of carbon dioxide to fatty alcohols and hydrocarbons in cyanobacteria. Metab. Eng. 2011, 13, 169–176. [Google Scholar] [CrossRef]
- Nakao, M.; Okamoto, S.; Kohara, M.; Fujishiro, T.; Fujisawa, T.; Sato, S.; Tabata, S.; Kaneko, T.; Nakamura, Y. CyanoBase: The cyanobacteria genome database update 2010. Nucleic Acids Res. 2010, 38, 379–381. [Google Scholar] [CrossRef]
- Cournac, L.; Guedeney, G.; Peltier, G.; Vignais, P.M. Sustained photoevolution of molecular hydrogen in a mutant of Synechocystis sp. strain PCC6803 deficient in the type I NADPH-dehydrogenase complex. J. Bacteriol. 2004, 186, 1737–1746. [Google Scholar] [CrossRef]
- Bermejo, L.L.; Welker, N.E.; Papoutsakis, E.T. Expression of Clostridium acetobutylicum ATCC 824 genes in Escherichia coli for acetone production and acetate detoxification. Appl. Environ. Microbiol. 1998, 64, 1079–1085. [Google Scholar]
- Zhou, J.; Zhang, H.F.; Zhang, Y.P.; Li, Y.; Ma, Y.H. Designing and creating a modularized synthetic pathway in cyanobacterium Synechocystis enables production of acetone from carbon dioxide. Metab. Eng. 2012, 14, 394–400. [Google Scholar] [CrossRef]
- Guerrero, F.; Carbonell, V.; Cossu, M.; Correddu, D.; Jones, P.R. Ethylene synthesis and regulated expression of recombinant protein in Synechocystis sp. PCC6803. PLoS One 2012, 7, e50470. [Google Scholar] [CrossRef]
- Ungerer, J.; Tao, L.; Davis, M.; Ghirardi, M.; Maness, P.C.; Yu, J.P. Sustained photosynthetic conversion of CO2 to ethylene in recombinant cyanobacterium Synechocystis 6803. Energy Environ. Sci. 2012, 5, 8998–9006. [Google Scholar] [CrossRef]
- Wee, Y.J.; Kim, J.N.; Ryu, H.W. Biotechnological production of lactic acid and its recent applications. Food Technol. Biotechnol. 2006, 44, 163–172. [Google Scholar]
- Angermayr, S.A.; Paszota, M.; Hellingwerf, K.J. Engineering a Cyanobacterial Cell Factory for Production of Lactic Acid. Appl. Environ. Microbiol. 2012, 78, 7098–7106. [Google Scholar] [CrossRef]
- Luengo, J.M.; Garcia, B.; Sandoval, A.; Naharro, G.; Olivera, E.R. Bioplastics from microorganisms. Curr. Opin. Microbiol. 2003, 6, 251–260. [Google Scholar]
- Sudesh, K.; Taguchi, K.; Doi, Y. Effect of increased PHA synthase activity on polyhydroxyalkanoates biosynthesis in Synechocystis sp. PCC6803. Int. J. Biol. Macromol. 2002, 30, 97–104. [Google Scholar] [CrossRef]
- Wang, B.; Pugh, S.; Nielsen, D.R.; Zhang, W.W.; Meldrum, D.R. Engineering cyanobacteria for photosynthetic production of 3-hydroxybutyrate directly from CO2. Metab. Eng. 2013, 16, 68–77. [Google Scholar] [CrossRef]
- Wu, G.F.; Wu, Q.Y.; Shen, Z.Y. Accumulation of poly-β-hydroxybutyrate in cyanobacterium Synechocystis sp. PCC6803. Bioresource Technol. 2001, 76, 85–90. [Google Scholar] [CrossRef]
- Tyo, K.E.J.; Jin, Y.-S.; Espinoza, F.A.; Stephanopoulos, G. Identification of gene disruptions for increased poly-3-hydroxybutyrate accumulation in Synechocystis PCC6803. Biotechnol. Prog. 2009, 25, 1236–1243. [Google Scholar] [CrossRef]
- Shin, J.H.; Kim, H.U.; Kim, D.I.; Lee, S.Y. Production of bulk chemicals via novel metabolic pathways in microorganisms. Biotechnol. Adv. 2012. [Google Scholar] [CrossRef]
- Zhang, F.; Ouellet, M.; Batth, T.S.; Adams, P.D.; Petzold, C.J.; Mukhopadhyay, A.; Keasling, J.D. Enhancing fatty acid production by the expression of the regulatory transcription factor FadR. Metab. Eng. 2012, 14, 653–660. [Google Scholar] [CrossRef]
- Zheng, Y.N.; Li, L.L.; Liu, Q.; Yang, J.M.; Wang, X.W.; Liu, W.; Xu, X.; Liu, H.; Zhao, G.; Xian, M. Optimization of fatty alcohol biosynthesis pathway for selectively enhanced production of C12/14 and C16/18 fatty alcohols in engineered E. coli. Microb. Cell Fact. 2012, 11, 65. [Google Scholar] [CrossRef]
- Wijffels, R.H.; Kruse, O.; Hellingwerf, K.J. Potential of industrial biotechnology with cyanobacteria and eukaryotic microalgae. Curr. Opin. Biotechnol. 2013, 24, 405–413. [Google Scholar] [CrossRef]
- Oncel, S.; Sabankay, M. Microalgal biohydrogen production considering light energy and mixing time as the two key features for scale-up. Bioresour. Technol. 2012, 121, 228–234. [Google Scholar] [CrossRef]
- Chance, R.; McCool, B.; Coleman, J. A Cyanobacteria-Based Photosynthetic Process for the Production of Ethanol. Presented at the National Research Council Committee on Sustainable Development of Algal Biofuels, Washington, DC, USA, 13 June 2011; Available online: http://khlaw.com/Files/10645_Chance_corrected.pdf (accessed on 2 August 2011).
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Yu, Y.; You, L.; Liu, D.; Hollinshead, W.; Tang, Y.J.; Zhang, F. Development of Synechocystis sp. PCC 6803 as a Phototrophic Cell Factory. Mar. Drugs 2013, 11, 2894-2916. https://doi.org/10.3390/md11082894
Yu Y, You L, Liu D, Hollinshead W, Tang YJ, Zhang F. Development of Synechocystis sp. PCC 6803 as a Phototrophic Cell Factory. Marine Drugs. 2013; 11(8):2894-2916. https://doi.org/10.3390/md11082894
Chicago/Turabian StyleYu, Yi, Le You, Dianyi Liu, Whitney Hollinshead, Yinjie J. Tang, and Fuzhong Zhang. 2013. "Development of Synechocystis sp. PCC 6803 as a Phototrophic Cell Factory" Marine Drugs 11, no. 8: 2894-2916. https://doi.org/10.3390/md11082894
APA StyleYu, Y., You, L., Liu, D., Hollinshead, W., Tang, Y. J., & Zhang, F. (2013). Development of Synechocystis sp. PCC 6803 as a Phototrophic Cell Factory. Marine Drugs, 11(8), 2894-2916. https://doi.org/10.3390/md11082894




