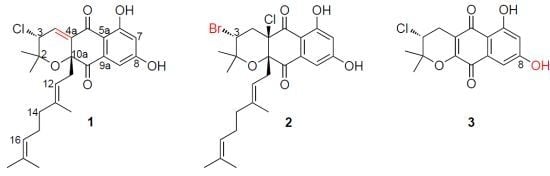
Antibacterial and Cytotoxic New Napyradiomycins from the Marine-Derived Streptomyces sp. SCSIO 10428
Abstract
:1. Introduction
2. Results and Discussion
2.1. Strain Identification and Compounds Isolation
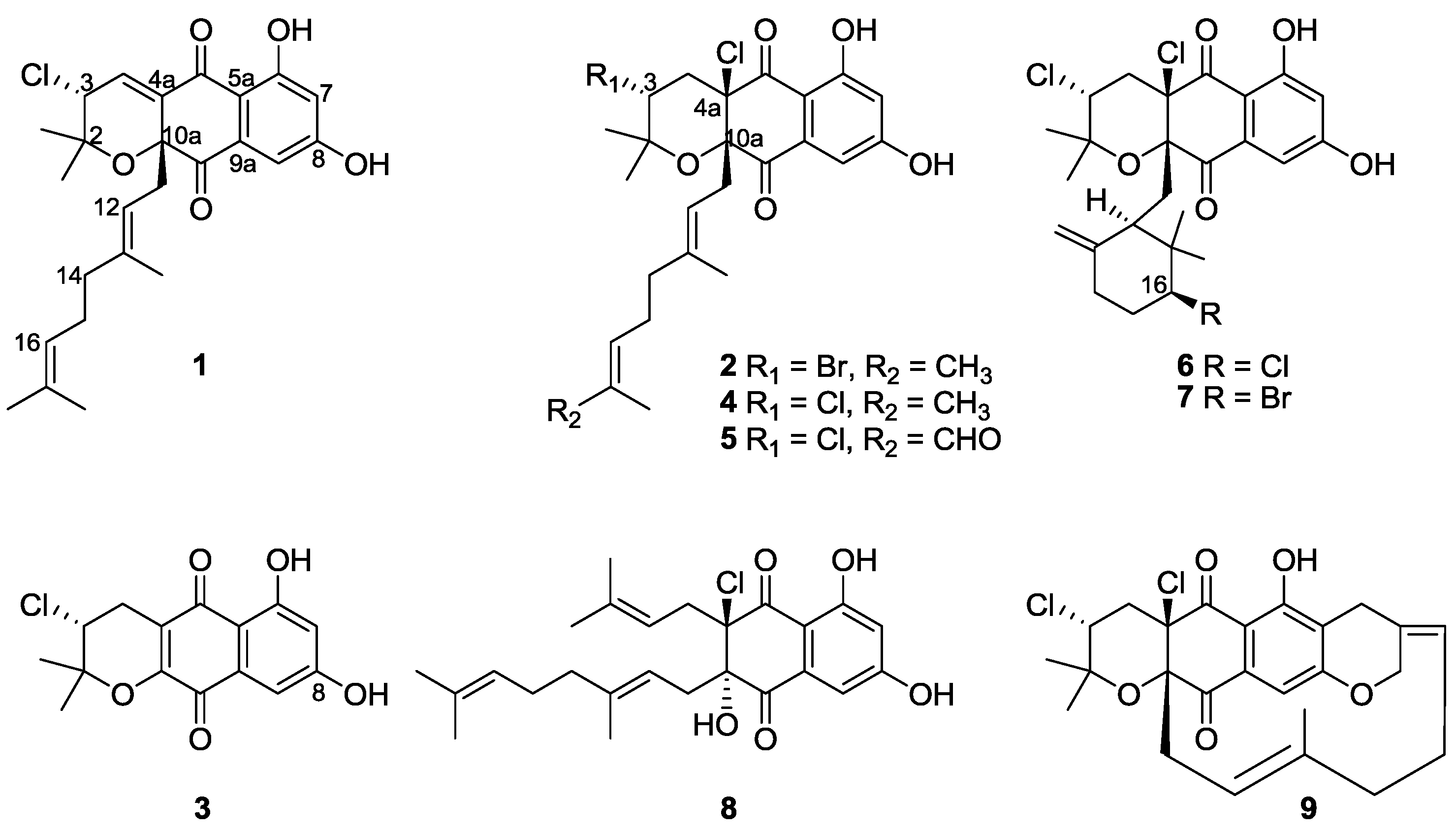
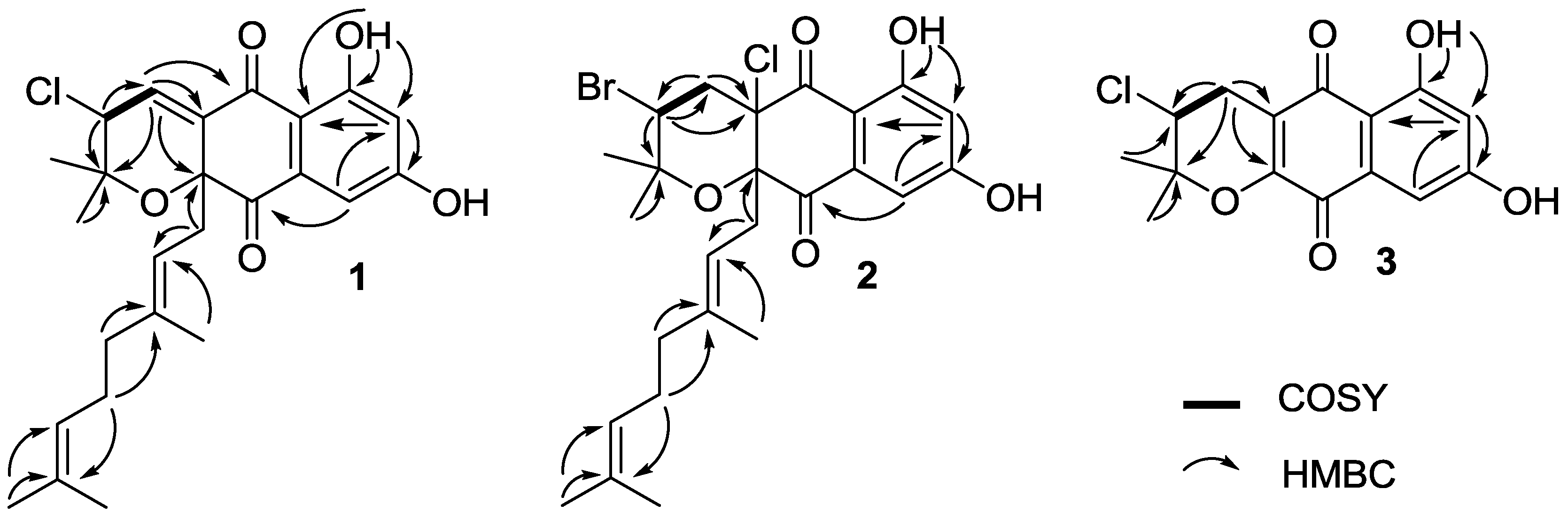
2.2. Structure Elucidation
| No. | 1 | 2 | 3 | |||
|---|---|---|---|---|---|---|
| δC | δH multi (J in Hz) | δC | δH multi (J in Hz) | δC | δH multi (J in Hz) | |
| 2 | 76.9 | 78.7 | 80.7 | |||
| 2-CH3 | 20.4 | 1.09 s | 23.6 | 1.26 s | 22.4 | 1.51 s |
| 2-CH3 | 27.4 | 1.53 s | 29.5 | 1.50 s | 25.8 | 1.53 s |
| 3 | 59.6 | 4.39 d (2.0) | 51.0 | 4.55 dd (8.0, 8.0) | 57.9 | 4.08 dd (5.5, 7.0) |
| 4 | 136.9 | 6.92 d (2.0) | 43.9 | 2.58 br d (8.0) | 27.2 | 2.87 dd (7.0, 19.0) |
| 3.10 dd (5.5, 19.0) | ||||||
| 4a | 137.1 | 79.7 | 117.8 | |||
| 5 | 188.6 | 193.9 | 188.0 | |||
| 5a | 112.0 | 110.5 | 108.8 | |||
| 6 | 165.5 | 165.0 | 162.5 | |||
| 6-OH | 12.57 s | 11.84 s | 12.30 s | |||
| 7 | 109.2 | 6.71 s | 109.7 | 6.73 d (2.0) | 108.9 | 6.62, d (2.0) |
| 8 | 164.0 | 163.7 | 163.9 | |||
| 9 | 108.4 | 7.19 s | 107.9 | 7.22 d (2.0) | 109.3 | 7.14 d (2.0) |
| 9a | 136.0 | 135.6 | 133.0 | |||
| 10 | 195.4 | 196.2 | 178.8 | |||
| 10a | 83.3 | 83.9 | 153.7 | |||
| 11 | 39.9 | 2.48 d (7.5) | 41.6 | 2.70 d (8.0) | ||
| 2.69 d (8.0) | ||||||
| 12 | 115.9 | 4.98 br t (7.5) | 115.0 | 4.70 br t (8.0) | ||
| 13 | 141.7 | 143.0 | ||||
| 13-CH3 | 16.7 | 1.44 s | 16.6 | 1.31 s | ||
| 14 | 39.8 | 1.94 m | 39.9 | 1.60 m | ||
| 15 | 26.6 | 2.00 m | 26.1 | 1.60 m | ||
| 16 | 124.0 | 5.02 m | 123.9 | 4.89 br s | ||
| 17 | 132.1 | 131.9 | ||||
| 17-CH3 | 18.0 | 1.58 s | 17.7 | 1.52 s | ||
| 17-CH3 | 25.9 | 1.68 s | 25.8 | 1.62 s | ||
 +23°, c 0.30 in MeOH); for 4,
+23°, c 0.30 in MeOH); for 4,  +51°, c 0.3 in MeOH).
+51°, c 0.3 in MeOH). −22°, c 0.14 in CHCl3) and (R)-3-chloro-6-hydroxy-8-methoxy-α-lapachone (
−22°, c 0.14 in CHCl3) and (R)-3-chloro-6-hydroxy-8-methoxy-α-lapachone (  −8°, c 0.12 in CHCl3).
−8°, c 0.12 in CHCl3).2.3. Biological Activities
| MIC (μg mL−1) | IC50 (μM) | |||||||
|---|---|---|---|---|---|---|---|---|
| Sa | Bs | Bt | Ec | SF-268 | MCF-7 | NCI-H460 | HepG-2 | |
| 1 | 4 | 4 | 8 | >128 | 22.8 ± 0.3 | 20.6 ± 0.1 | 22.4 ± 0.1 | 21.8 ± 0.5 |
| 2 | 0.5 | 1 | 1 | >128 | 11.5 ± 1.2 | 16.2 ± 0.7 | 18.1 ± 0.3 | 17.1 ± 1.0 |
| 3 | >128 | 8 | 16 | >128 | 23.8 ± 2.2 | 71.1 ± 0.4 | 127.1 ± 0.9 | 59.4 ± 0.7 |
| 4 | 1 | 2 | 2 | >128 | 18.5 ± 0.3 | 9.8 ± 0.2 | 19.0 ± 1.0 | 18.9 ± 0.3 |
| 5 | 32 | 8 | 16 | >128 | 132.7 ± 0.1 | 138.2 ± 0.8 | 137.1 ± 1.5 | 149.1 ± 1.4 |
| 6 | 1 | 2 | 0.5 | >128 | 11.1 ± 0.1 | 17.0 ± 0.2 | 18.6 ± 0.4 | 17.9 ± 0.7 |
| 7 | 0.5 | 0.25 | 0.5 | >128 | 15.3 ± 1.1 | 11.2 ± 0.5 | 17.2 ± 0.4 | 10.5 ± 1.6 |
| 8 | 1 | 2 | 2 | >128 | 29.6 ± 0.2 | 22.2 ± 0.7 | 26.9 ± 0.3 | 61.2 ± 0.1 |
| 9 | >128 | >128 | >128 | >128 | 98.1 ± 1.7 | 40.5 ± 1.4 | 163.7 ± 1.0 | 157.2 ± 4.5 |
| CK * | 2 | 1 | 2 | 2 | 7.3 ± 0.9 | 4.1 ± 0.3 | 4.4 ± 0.1 | 5.6 ± 0.3 |
2.4. Discussion
3. Experimental Section
3.1. General Experimental Procedures
3.2. Collection and Phylogenetic Analysis of Strain
3.3. Fermentation
3.4. Extraction and Isolation
3.5. Characterization Data
 −51° (c 0.30, MeOH); UV (MeOH) λmax (log ε): 202 (4.61), 257 (4.39), 312 (4.09), 362 (4.15) nm; IR νmax 3294, 2920, 1701, 1616, 1373, 1261, 1145, 802 cm−1; 1H and 13C NMR data see Table 1; HRESIMS m/z 467.1595 [M + Na]+ (calcd for C25H29ClO5Na, 467.1596).
−51° (c 0.30, MeOH); UV (MeOH) λmax (log ε): 202 (4.61), 257 (4.39), 312 (4.09), 362 (4.15) nm; IR νmax 3294, 2920, 1701, 1616, 1373, 1261, 1145, 802 cm−1; 1H and 13C NMR data see Table 1; HRESIMS m/z 467.1595 [M + Na]+ (calcd for C25H29ClO5Na, 467.1596). +23° (c 0.30, MeOH); UV (MeOH) λmax (log ε): 202 (4.62), 253 (4.35), 297 (4.19), 364 (4.06) nm; IR νmax 3348, 2939, 1701, 1612, 1450, 1369, 1253, 1072, 1018, 732 cm−1; 1H and 13C NMR data see Table 1; HRESIMS m/z 525.1047 [M + H]+ (calcd for C25H31ClBrO5, 525.1038).
+23° (c 0.30, MeOH); UV (MeOH) λmax (log ε): 202 (4.62), 253 (4.35), 297 (4.19), 364 (4.06) nm; IR νmax 3348, 2939, 1701, 1612, 1450, 1369, 1253, 1072, 1018, 732 cm−1; 1H and 13C NMR data see Table 1; HRESIMS m/z 525.1047 [M + H]+ (calcd for C25H31ClBrO5, 525.1038). −22° (c 0.14, CHCl3); UV (MeOH) λmax (log ε): 217 (4.42), 263 (4.19), 305 (3.95), 450 (3.42) nm; IR νmax 3348, 2939, 1674, 1608, 1280, 1095, 1022, 887, 779 cm−1; 1H and 13C NMR data see Table 1; HRESIMS m/z 309.0532 [M + H]+ (calcd for C15H14ClO5, 309.0524).
−22° (c 0.14, CHCl3); UV (MeOH) λmax (log ε): 217 (4.42), 263 (4.19), 305 (3.95), 450 (3.42) nm; IR νmax 3348, 2939, 1674, 1608, 1280, 1095, 1022, 887, 779 cm−1; 1H and 13C NMR data see Table 1; HRESIMS m/z 309.0532 [M + H]+ (calcd for C15H14ClO5, 309.0524).3.6. Antibacterial, Cytotoxic and Antioxidative Assays
4. Conclusions
Acknowledgments
Conflict of Interest
References
- Jensen, P.R.; Dwight, R.; Fenical, W. Distribution of actinomycetes in near-shore tropical marine-sediments. Appl. Environ. Microbiol. 1991, 57, 1102–1108. [Google Scholar]
- Fenical, W.; Jensen, P.R. Developing a new resource for drug discovery: Marine actinomycete bacteria. Nat. Chem. Biol. 2006, 2, 666–673. [Google Scholar] [CrossRef]
- Bhatnagar, I.; Kim, S.K. Marine antitumor drugs: Status, shortfalls and strategies. Mar. Drugs 2010, 8, 2702–2720. [Google Scholar] [CrossRef]
- Olano, C.; Mendez, C.; Salas, J.A. Antitumor compounds from marine actinomycetes. Mar. Drugs 2009, 7, 210–248. [Google Scholar] [CrossRef]
- Rahman, H.; Austin, B.; Mitchell, W.J.; Morris, P.C.; Jamieson, D.J.; Adams, D.R.; Spragg, A.M.; Schweizer, M. Novel anti-infective compounds from marine bacteria. Mar. Drugs 2010, 8, 498–518. [Google Scholar] [CrossRef]
- Fattorusso, E.; Taglialatela-Scafati, O. Marine antimalarials. Mar. Drugs 2009, 7, 130–152. [Google Scholar] [CrossRef]
- Huang, H.B.; Yao, Y.L.; He, Z.X.; Yang, T.T.; Ma, J.Y.; Tian, X.P.; Li, Y.Y.; Huang, C.G.; Chen, X.P.; Li, W.J.; et al. Antimalarial beta-carboline and indolactam alkaloids from Marinactinospora thermotolerans, a deep sea isolate. J. Nat. Prod. 2011, 74, 2122–2127. [Google Scholar] [CrossRef]
- Li, S.M.; Tian, X.P.; Niu, S.W.; Zhang, W.J.; Chen, Y.C.; Zhang, H.B.; Yang, X.W.; Zhang, W.M.; Li, W.J.; Zhang, S.; et al. Pseudonocardians A–C, new diazaanthraquinone derivatives from a deap-sea actinomycete Pseudonocardia sp. SCSIO 01299. Mar. Drugs 2011, 9, 1428–1439. [Google Scholar] [CrossRef]
- Niu, S.W.; Li, S.M.; Chen, Y.C.; Tian, X.P.; Zhang, H.B.; Zhang, G.T.; Zhang, W.M.; Yang, X.H.; Zhang, S.; Ju, J.H.; et al. Lobophorins E and F, new spirotetronate antibiotics from a South China Sea-derived Streptomyces sp. SCSIO 01127. J. Antibiot. 2011, 64, 711–716. [Google Scholar] [CrossRef]
- Wang, F.Z.; Tian, X.P.; Huang, C.G.; Li, Q.X.; Zhang, S. Marinactinones A–C, new gamma-pyrones from marine actinomycete Marinactinospora thermotolerans SCSIO 00606. J. Antibiot. 2011, 64, 189–192. [Google Scholar] [CrossRef]
- Huang, H.B.; Yang, T.T.; Ren, X.M.; Liu, J.; Song, Y.X.; Sun, A.J.; Ma, J.Y.; Wang, B.; Zhang, Y.; Huang, C.G.; et al. Cytotoxic angucycline alass glycosides from the deep sea actinomycete Streptomyces lusitanus SCSIO LR32. J. Nat. Prod. 2012, 75, 202–208. [Google Scholar] [CrossRef]
- Li, H.X.; Zhang, Q.B.; Li, S.M.; Zhu, Y.G.; Zhang, G.T.; Zhang, H.B.; Tian, X.P.; Zhang, S.; Ju, J.H.; Zhang, C.S. Identification and characterization of xiamycin A and oxiamycin gene cluster reveals an oxidative cyclization strategy tailoring indolosesquiterpene biosynthesis. J. Am. Chem. Soc. 2012, 134, 8996–9005. [Google Scholar] [CrossRef]
- Zhang, Q.B.; Mandi, A.; Li, S.M.; Chen, Y.C.; Zhang, W.J.; Tian, X.P.; Zhang, H.B.; Li, H.X.; Zhang, W.M.; Zhang, S.; et al. N–N-coupled indolo-sesquiterpene atropo-diastereomers from a marine-derived actinomycete. Eur. J. Org. Chem. 2012, 2012, 5256–5262. [Google Scholar] [CrossRef]
- Zhang, W.J.; Liu, Z.; Li, S.M.; Yang, T.T.; Zhang, Q.B.; Ma, L.; Tian, X.P.; Zhang, H.B.; Huang, C.G.; Zhang, S.; et al. Spiroindimicins A–D: New bisindole alkaloids from a deep-sea derived actinomycete. Org. Lett. 2012, 14, 3364–3367. [Google Scholar] [CrossRef]
- Zhou, X.; Huang, H.B.; Chen, Y.C.; Tan, J.H.; Song, Y.X.; Zou, J.H.; Tian, X.P.; Hua, Y.; Ju, J.H. Marthiapeptide A, an anti-infective and cytotoxic polythiazole cyclopeptide from a 60 L scale fermentation of the deep sea derived Marinactinospora thermotolerans SCSIO 00652. J. Nat. Prod. 2012, 75, 2251–2255. [Google Scholar] [CrossRef]
- Zhang, Q.B.; Li, S.M.; Chen, Y.C.; Tian, X.P.; Zhang, H.B.; Zhang, G.T.; Zhu, Y.G.; Zhang, S.; Zhang, W.M.; Zhang, C.S. New diketopiperazine derivatives from a deep-sea-derived Nocardiopsis alba SCSIO 03039. J. Antibiot. 2013, 66, 31–36. [Google Scholar] [CrossRef]
- Wu, Z.C.; Li, S.M.; Nam, S.J.; Liu, Z.; Zhang, C.S. Nocardiamides A and B, two cyclohexapeptides from the marine-derived actinomycete Nocardiopsis sp. CNX037. J. Nat. Prod. 2013, 76, 694–701. [Google Scholar] [CrossRef]
- Shiomi, K.; Nakamura, H.; Iinuma, H.; Naganawa, H.; Isshiki, K.; Takeuchi, T.; Umezawa, H. Structures of new antibiotics napyradiomycins. J. Antibiot. 1986, 39, 494–501. [Google Scholar] [CrossRef]
- Motohashi, K.; Sue, M.; Furihata, K.; Ito, S.; Seto, H. Terpenoids produced by actinomycetes: Napyradiomycins from Streptomyces antimycoticus NT17. J. Nat. Prod. 2008, 71, 595–601. [Google Scholar] [CrossRef]
- Henkel, T.; Zeeck, A. Secondary metabolites by chemical screening. 15. Structure and absolute configuration of naphthomevalin, a new dihydro-naphthoquinone antibiotic from Streptomyces sp. J. Antibiot. 1991, 44, 665–669. [Google Scholar] [CrossRef]
- Shiomi, K.; Nakamura, H.; Iinuma, H.; Naganawa, H.; Takeuchi, T.; Umezawa, H.; Iitaka, Y. New antibiotic napyradiomycins A2 and B4 and stereochemistry of napyradiomycins. J. Antibiot. 1987, 40, 1213–1219. [Google Scholar] [CrossRef]
- Soria-Mercado, I.E.; Jensen, P.R.; Fenical, W.; Kassel, S.; Golen, J. 3,4a-Dichloro-10a-(3-chloro-6-hydroxy-2,2,6-trimethylcyclohexylmethyl)-6,8-dihydroxy-2,2,7-trimethyl-3,4,4a,10a-tetrahydro-2H-benzo[g]chromene-5,10-dione. Acta Crystallogr. Sect. E 2004, 60, o1627–o1629. [Google Scholar] [CrossRef]
- Soria-Mercado, I.E.; Prieto-Davo, A.; Jensen, P.R.; Fenical, W. Antibiotic terpenoid chloro-dihydroquinones from a new marine actinomycete. J. Nat. Prod. 2005, 68, 904–910. [Google Scholar] [CrossRef]
- Farnaes, L.L.L. Novel Analogs and a Protein Target for the Napyradiomycins. Ph.D. Thesis, University of California, San Diego, CA, USA, 2009. [Google Scholar]
- Winter, J.M.; Moffitt, M.C.; Zazopoulos, E.; McAlpine, J.B.; Dorrestein, P.C.; Moore, B.S. Molecular basis for chloronium-mediated meroterpene cyclization. Cloning, sequencing, and heterologous expression of the napyradiomycin biosynthetic gene cluster. J. Biol. Chem. 2007, 282, 16362–16368. [Google Scholar]
- Haste, N.M.; Farnaes, L.L.L.; Perera, V.R.; Fenical, W.; Nizet, V.; Hensler, M.E. Bactericidal kinetics of marine-derived napyradiomycins against contemporary methicillin-resistant Staphylococcus aureus. Mar. Drugs 2011, 9, 680–689. [Google Scholar] [CrossRef]
- Mensor, L.L.; Menezes, F.S.; Leitao, G.G.; Reis, A.S.; dos Santos, T.C.; Coube, C.S.; Leitao, S.G. Screening of Brazilian plant extracts for antioxidant activity by the use of DPPH free radical method. Phytother. Res. 2001, 15, 127–130. [Google Scholar]
- Shiomi, K.; Iinuma, H.; Hamada, M.; Naganawa, H.; Manabe, M.; Matsuki, C.; Takeuchi, T.; Umezawa, H. Novel antibiotics napyradiomycins. Production, isolation, physicochemical properties and biological activity. J. Antibiot. 1986, 39, 487–493. [Google Scholar] [CrossRef]
- Gomi, S.; Ohuchi, S.; Sasaki, T.; Itoh, J.; Sezaki, M. Studies on new antibiotics SF2415. 2. The structural elucidation. J. Antibiot. 1987, 40, 740–749. [Google Scholar] [CrossRef]
- Fukuda, D.S.; Mynderse, J.S.; Baker, P.J.; Berry, D.M.; Boeck, L.D.; Yao, R.C.; Mertz, F.P.; Nakatsukasa, W.M.; Mabe, J.; Ott, J.; et al. A80915, a new antibiotic complex produced by Streptomyces aculeolatus. Discovery, taxonomy, fermentation, isolation, characterization, and antibacterial evaluation. J. Antibiot. 1990, 43, 623–633. [Google Scholar] [CrossRef]
- Umezawa, K.; Masuoka, S.; Ohse, T.; Naganawa, H.; Kondo, S.; Ikeda, Y.; Kinoshita, N.; Hamada, M.; Sawa, T.; Takeuchi, T. Isolation from Streptomyces of a novel naphthoquinone compound, naphthablin, that inhibits Abl oncogene functions. J. Antibiot. 1995, 48, 604–607. [Google Scholar] [CrossRef]
- Cho, J.Y.; Kwon, H.C.; Williams, P.G.; Jensen, P.R.; Fenical, W. Azamerone, a terpenoid phthalazinone from a marine-derived bacterium related to the genus Streptomyces (actinomycetales). Org. Lett. 2006, 8, 2471–2474. [Google Scholar] [CrossRef]
- Motohashi, K.; Irie, K.; Toda, T.; Matsuo, Y.; Kasai, H.; Sue, M.; Furihata, K.; Seto, H. Studies on terpenoids produced by actinomycetes. 5-Dimethylallylindole-3-carboxylic acid and A80915G-8″-acid produced by marine-derived Streptomyces sp. MS239. J. Antibiot. 2008, 61, 75–80. [Google Scholar] [CrossRef]
- Shiomi, K.; Iinuma, H.; Naganawa, H.; Isshiki, K.; Takeuchi, T.; Umezawa, H. Biosynthesis of napyradiomycins. J. Antibiot. 1987, 40, 1740–1745. [Google Scholar] [CrossRef]
- Winter, J.M.; Jansma, A.L.; Handel, T.M.; Moore, B.S. Formation of the pyridazine natural product azamerone by biosynthetic rearrangement of an aryl diazoketone. Angew. Chem. Int. Ed. 2009, 48, 767–770. [Google Scholar] [CrossRef]
- Bernhardt, P.; Okino, T.; Winter, J.M.; Miyanaga, A.; Moore, B.S. A stereoselective vanadium-dependent chloroperoxidase in bacterial antibiotic biosynthesis. J. Am. Chem. Soc. 2011, 133, 4268–4270. [Google Scholar]
- Dantzig, A.H.; Minor, P.L.; Garrigus, J.L.; Fukuda, D.S.; Mynderse, J.S. Studies on the mechanism of action of A80915A, a semi-naphthoquinone natural product, as an inhibitor of gastric (H+-K+)-ATPase. Biochem. Pharmacol. 1991, 42, 2019–2026. [Google Scholar] [CrossRef]
- Hori, Y.; Abe, Y.; Shigematsu, N.; Goto, T.; Okuhara, M.; Kohsaka, M. Napyradiomycin A and B1: Non-steroidal estrogen-receptor antagonists produced by a Streptomyces. J. Antibiot. 1993, 46, 1890–1893. [Google Scholar] [CrossRef]
- Sun, P.; Maloney, K.N.; Nam, S.J.; Haste, N.M.; Raju, R.; Aalbersberg, W.; Jensen, P.R.; Nizet, V.; Hensler, M.E.; Fenical, W. Fijimycins A–C, three antibacterial etamycin-class depsipeptides from a marine-derived Streptomyces sp. Bioorg. Med. Chem. 2011, 19, 6557–6562. [Google Scholar] [CrossRef]
- Xiao, Y.; Li, S.; Niu, S.; Ma, L.; Zhang, G.; Zhang, H.; Zhang, G.; Ju, J.; Zhang, C. Characterization of tiacumicin B biosynthetic gene cluster affording diversified tiacumicin analogs and revealing a tailoring Di-halogenase. J. Am. Chem. Soc. 2011, 133, 1092–1105. [Google Scholar] [CrossRef]
- Skehan, P.; Storeng, R.; Scudiero, D.; Monks, A.; McMahon, J.; Vistica, D.; Warren, J.T.; Bokesch, H.; Kenney, S.; Boyd, M.R. New colorimetric cytotoxicity assay for anticancer drug screening. J. Natl. Cancer Inst. 1990, 82, 1107–1112. [Google Scholar] [CrossRef]
Supplementary Files
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Wu, Z.; Li, S.; Li, J.; Chen, Y.; Saurav, K.; Zhang, Q.; Zhang, H.; Zhang, W.; Zhang, W.; Zhang, S.; et al. Antibacterial and Cytotoxic New Napyradiomycins from the Marine-Derived Streptomyces sp. SCSIO 10428. Mar. Drugs 2013, 11, 2113-2125. https://doi.org/10.3390/md11062113
Wu Z, Li S, Li J, Chen Y, Saurav K, Zhang Q, Zhang H, Zhang W, Zhang W, Zhang S, et al. Antibacterial and Cytotoxic New Napyradiomycins from the Marine-Derived Streptomyces sp. SCSIO 10428. Marine Drugs. 2013; 11(6):2113-2125. https://doi.org/10.3390/md11062113
Chicago/Turabian StyleWu, Zhengchao, Sumei Li, Jie Li, Yuchan Chen, Kumar Saurav, Qingbo Zhang, Haibo Zhang, Wenjun Zhang, Weimin Zhang, Si Zhang, and et al. 2013. "Antibacterial and Cytotoxic New Napyradiomycins from the Marine-Derived Streptomyces sp. SCSIO 10428" Marine Drugs 11, no. 6: 2113-2125. https://doi.org/10.3390/md11062113
APA StyleWu, Z., Li, S., Li, J., Chen, Y., Saurav, K., Zhang, Q., Zhang, H., Zhang, W., Zhang, W., Zhang, S., & Zhang, C. (2013). Antibacterial and Cytotoxic New Napyradiomycins from the Marine-Derived Streptomyces sp. SCSIO 10428. Marine Drugs, 11(6), 2113-2125. https://doi.org/10.3390/md11062113