Further Insights on the Carotenoid Profile of the Echinoderm Marthasterias glacialis L.
Abstract
:1. Introduction
2. Results and Discussion
| Peak a | Carotenoid | tR b(min) | λmax c (nm) | % III/II | [M + H]+ (m/z) | MS/MS fragment ions (positive mode) (m/z) | M−• or [M − H]− (m/z) | MS/MS fragment ions (negative mode) (m/z) |
|---|---|---|---|---|---|---|---|---|
| 1 | not identified 1 | 8.7–8.8 | 419, 445, 469 | nc d | 597 | 579 [M + H − 18]+, 561 [M + H − 18 − 18]+, 505 [M + H − 92]+ | 595 f | 577 [M − H − 18]−, 559 [M − H − 18 − 18]− |
| 2 | crustaxanthin | 9.1–9.2 | 422, 449, 470 | nc | 601 | 583 [M + H − 18]+ | 599 f | 581 [M − H − 18]−, 563 [M − H − 18 − 18]−, 507 [M − H − 92]− |
| 3 | 13-cis-astaxanthin | 10.4–10.5 | 371, 466 | 0 | 597 | 579 [M + H − 18]+, 561 [M + H − 18 − 18]+, 379, 285, 173 | 596 g | 581 [M − 15]−, 578 [M − 18]−, 504 [M − 92]−, 429 [M − 167]−, 389, 363 [M − 233]−, 337 [M − 167 − 92]−, 323, 297, 233 |
| 4 | all-trans-astaxanthin | 11.2–11.6 | 475 | 0 | 597 | 579 [M + H − 18]+, 561 [M + H − 18 − 18]+, 505 [M + H − 92]+, 379, 285 | 596 g595 f | 581 [M − 15]−, 577 [M − H − 18]−, 559 [M − H − 18 − 18]−, 541 [M − H − 18 − 18 − 18]−, 504 [M − 92]−, 429 [M − 167]−, 490 [M − 106]−, 389, 363 [M − 233]−, 337 [M − 167 − 92]−, 323, 297, 233, 203 |
| 5 | all-trans-lutein | 11.9–12.2 | 421, 445, 472 | 67–71 | nd e | 551 h [M + H − 18]+, 533 [M + H − 18 − 18]+, 495 [M + H − 18 − 56]+, 459 [M + H − 18 − 92]+, 430, 175 | 568 g567 f | 549 [M − H − 18]−, 531 [M − H − 18 − 18]−, 535 [M − 18 − 15]−, 429 |
| 6 | astaxanthin derivative 1 | 13.0–13.2 | 472 | 0 | 597 | 579 [M + H − 18]+, 561 [M + H − 18 − 18]+, 379, 285, 173 | 596 g | 581 [M − 15]−, 578 [M − 18]−, 542 [M − 18 − 18 − 18]−, 429 [M − 167]−, 490 [M − 106]−, 389, 363 [M − 233]−, 337 [M − 167 − 92]−, 323, 297, 233, 203 |
| 7 | all-trans-zeaxanthin | 13.8–14.2 | 423, 451, 476 | 25–40 | 569 | 551 [M + H − 18]+, 533[M + H − 18 − 18]+, 459, 416, 175 | 567 f | 549 [M − H − 18]−, 534 [M − H − 18 − 15]−, 531 [M − H − 18 − 18]−, 465, 201, 187 |
| 8 | astaxanthin derivative 2 | 14.8–15.0 | 459 | nc | 597 | 579 [M + H − 18]+, 561 [M + H − 18 − 18]+, 285, 173 | 595 f596 g | 581 [M − 15]−, 577 [M−H − 18]−, 429 [M − 167]−, 389, 363 [M − 233]−, 337 [M − 167 − 92]−, 323, 297, 233, 203 |
| 9 | 9-cis-astaxanthin | 15.5 | 470 | 0 | 597 | 579 [M + H − 18]+, 561 [M + H − 18 − 18]+ | 596 g | 581 [M − 15]−, 578 [M − 18]−, 560 [M − 18 − 18]−, 504 [M − 92]−, 429 [M − 167]−, 490 [M − 106]−, 363 [M − 233]−, 337 [M − 167 − 92]−, 323, 297, 233, 203 |
| 10 | astaxanthin derivative 3 | 16.2–16.4 | 454, 475 | nc | 597 | 579 [M + H − 18]+, 379, 285 | 596 g | 581 [M − 15]−, 578 [M − 18]−, 504 [M − 92]−, 429 [M − 167]−, 490 [M − 106]−, 389, 337 [M − 167 − 92]−, 233 |
| 11 | not identified 2 | 16.8 | 422, 452, 472 | nc | 565 | nd | 564 g | nd |
| 12 | all-trans-canthaxanthin | 18.3 | 470 | 0 | 565 | nd | 564 g | nd |
| 13 | 5,6-epoxy-β-cryptoxanthin | 19.2 | 419, 447, 472 | 67 | 569 | 551 [M + H − 18]+, 221 | 567 f | 549 [M − H − 18]− |
| 14 | not identified 3 | 20.5–20.7 | 423, 452, 478 | 20–25 | 601 | 585, 548, 507, 441, 413 | 600 g | 581, 543, 416 |
| 15 | all-trans-β-cryptoxanthin | 22.6 | 422, 450, 476 | 25 | nd | nd | 552 g | 534 [M − 18]−, 519 [M − 18 − 15]−, 269, 243 |
| 16 | not identified 4 | 23.3 | 422, 450, 472 | 25 | nd | nd | nd | nd |
| 17 | 15-cis-β-carotene | 25.4 | 420, 448, 471 | nc | 537 | 413 [M + H − 124]+ | 536 g | 295, 269, 189 |
| 18 | 13-cis-β-carotene | 26.5 | 418, 447, 471 | nc | 537 | 269 | 536 g | 444 [M − 92]−, 295, 269 |
| 19 | all-trans-β-carotene | 33.1–34.7 | 422, 451, 477 | 20 | 537 | 444 [M + H − 92]+, 413 [M + H − 124]+, 400 [M + H − 137]+, 269, 177 | 536 g | 444 [M − 92]−, 295, 269 |
| 20 | 9-cis-β-carotene | 35.1–36.8 | 419, 448, 472 | nc | 537 | 444 [M + H − 92]+, 269 | 535 f | 295, 269 |
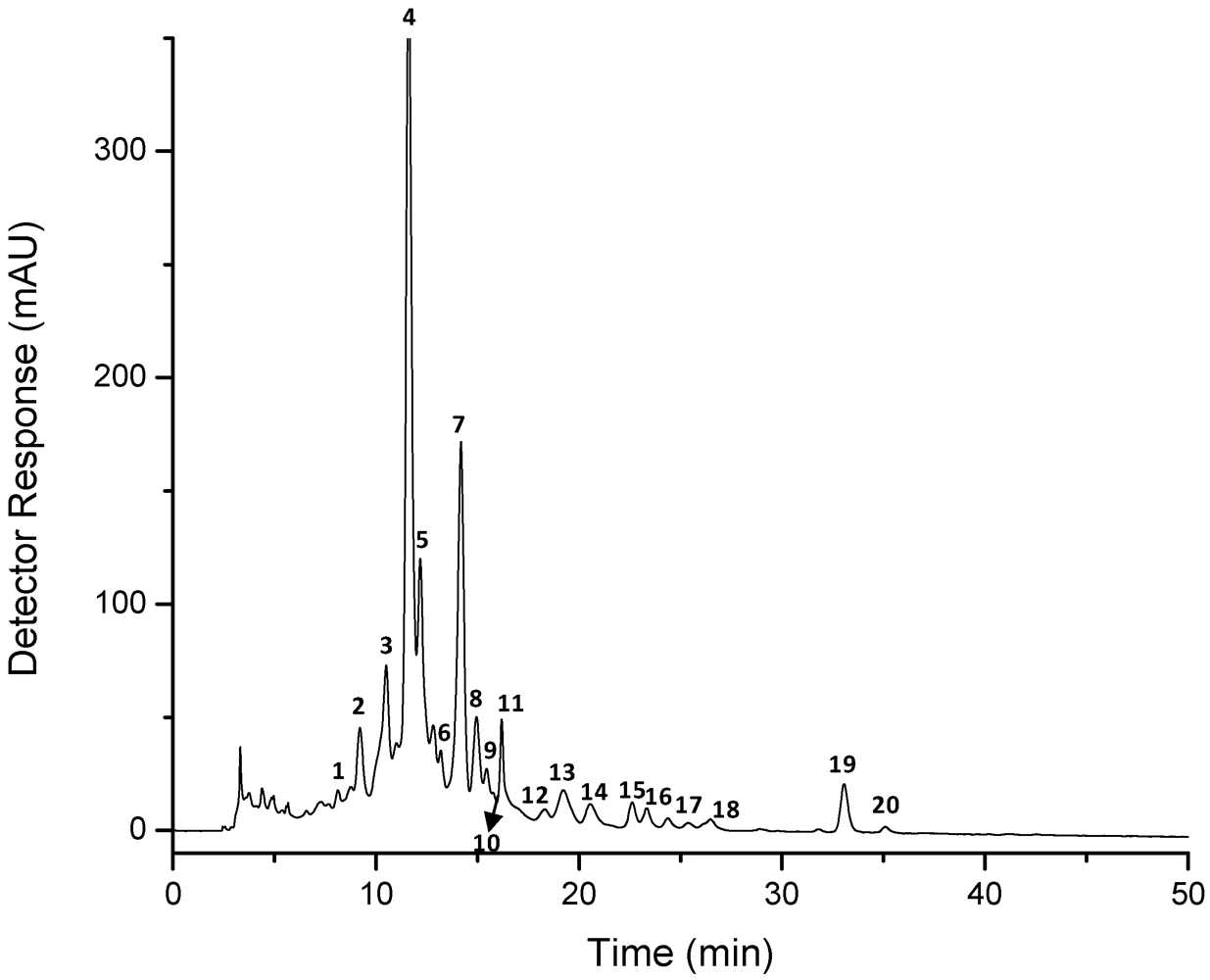
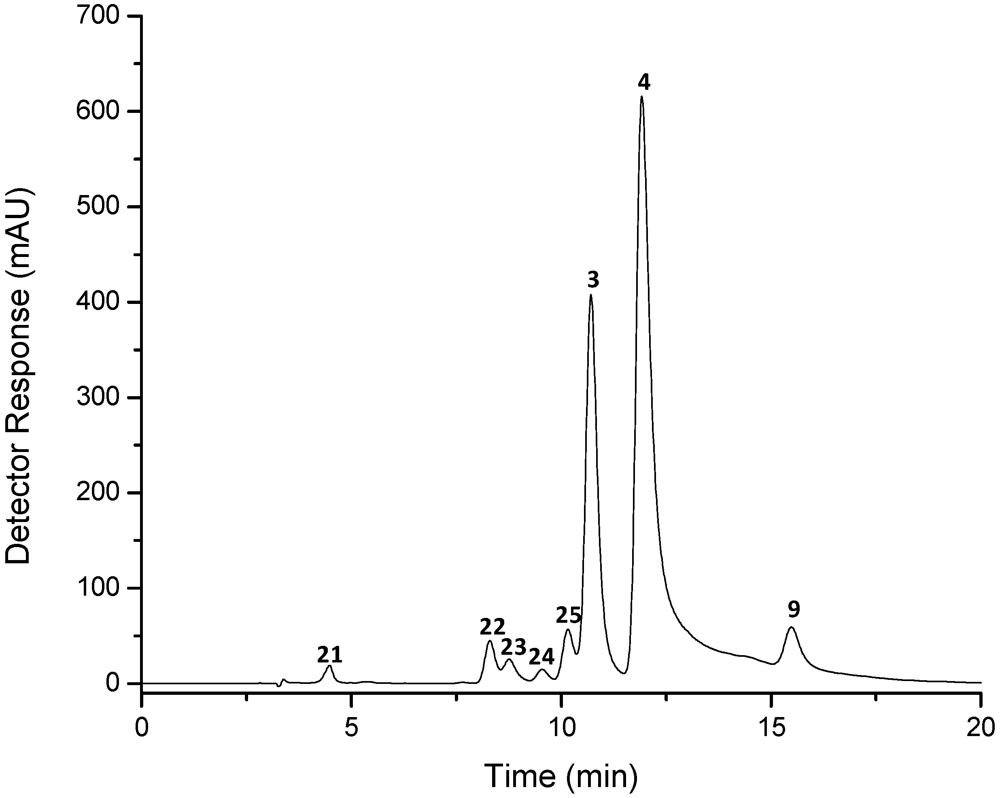
| Peak a | Carotenoid | tR b (min) | λmax c (nm) | Δλ | % III/II | % AB/AII | [M + H]+ (m/z) | MS/MS fragment ions (positive mode) (m/z) | M−• (m/z) | MS/MS fragment ions (negative mode)(m/z) |
|---|---|---|---|---|---|---|---|---|---|---|
| 21 | apo-10′-astaxanthinal | 4.4–4.5 | 428 | 48 | 0 | 407 | 389 [M + H − 18]+ | 406 | 388 [M − 18]− | |
| 22 | di-cis-astaxanthin 1 | 8.2–8.3 | 457 | 19 | 0 | nd d | nd | 596 | 578 [M − 18]−, 564 [M − 18 − 18]−, 504 [M − 92]− | |
| 23 | di-cis-astaxanthin 2 | 8.6–8.8 | 455–457 | 21 | 0 | nd | nd | nd | nd | |
| 24 | di-cis-astaxanthin 3 | 9.5 | 459–457 | 17 | 0 | nd | nd | 596 | nd | |
| 25 | cis-astaxanthin | 10.2–10.3 | 368, 469 | 7 | 0 | 58 | 597 | 285 | 596 | 581 [M − 15]−, 578 [M − 18]−, 504 [M − 92]−, 429 [M − 167]−, 389, 363 [M − 233]−, 337 [M − 167 − 92]−, 233 |
| 3 | 13-cis-astaxanthin | 10.7–10.8 | 370, 468 | 8 | 0 | 51 | 597 | 579 [M + H − 18]+, 561 [M + H − 18 − 18]+, 379, 285, 173 | 596 | 581 [M − 15]−, 578 [M − 18]−, 504 [M − 92]−, 429 [M − 167]−, 389, 363 [M − 233]−, 337 [M − 167 − 92]−, 233 |
| 4 | all-trans-astaxanthin | 11.8–11.9 | 476 | 0 | 597 | 579 [M + H − 18]+, 505 [M + H − 92]+, 379, 285, 173 | 596 | 581 [M − 15]−, 578 [M − 18]−, 504 [M − 92]−, 429 [M − 167]−, 389, 363 [M − 233]−, 337 [M − 167 − 92]−, 297, 233, 167 | ||
| 9 | 9-cis-astaxanthin | 15.5–15.7 | 471 | 5 | 0 | 597 | 579 [M + H − 18]+, 561 [M + H − 18 − 18]+, 379, 285 | 596 | 581 [M − 15]−, 578 [M − 18]−, 504 [M − 92]−, 429 [M − 167]−, 389, 363 [M − 233]−, 337 [M − 167 − 92]−, 233 |
| Peak a | Carotenoid | tR b (min) | λmax c (nm) | % III/II | % AB/AII | [M + H]+ (m/z) | MS/MS fragment ions (positive mode) (m/z) | [M − H]− (m/z) | MS/MS fragment ions (negative mode)(m/z) |
|---|---|---|---|---|---|---|---|---|---|
| 26 | crustaxanthin | 7.9 | 422, 449, 476 | 40 | 0 | 601 | 583 [M + H − 18]+, 565 d [M + H − 18 − 18]+, 547 [M + H − 18-18-18]+, 509 [M + H − 92]+ | 599 | 581 [M − 18]−, 563 [M − 18 − 18]−, 545 [M − 18 − 18 − 18]−, 507 [M − 92]−, 493 [M − 106]− |
| 27 | crustaxanthin | 8.5 | 422, 449, 476 | 35 | 0 | 601 | 583 [M + H − 18]+, 565 [M + H − 18 − 18]+, 547 d [M + H − 18 − 18 − 18]+, 509 [M + H − 92]+ | 599 | 581 [M − 18]−, 563 [M − 18 − 18]−, 545 [M − 18 − 18 − 18]−, 507 [M − 92]− |
| 28 | cis-crustaxanthin | 9.0 | 337, 422, 449, 476 | 40 | 6 | 601 | 583 [M + H − 18]+, 565 [M + H − 18 − 18]+, 547 [M + H − 18 − 18 − 18]+, 509 [M + H − 92]+ | 599 | 581 [M − 18]−, 563 [M − 18 − 18]−, 545 [M − 18 − 18 − 18]−, 507 [M − 92]− |
| 29 | cis-crustaxanthin | 9.3 | 337, 420, 444, 470 | 20 | 39 | 601 | 583 [M + H − 18]+, 565 [M + H − 18 − 18]+, 547 [M + H − 18 − 18 − 18]+, 509 [M + H − 92]+ | 599 | 581 [M − 18]− |

3. Experimental Section
3.1. Reagents and Standards
3.2. Sample
3.3. Carotenoid Extraction
3.4. Preparation of Reference Compounds
 = 2100; 470 nm) [19].
= 2100; 470 nm) [19]. 3.5. HPLC-DAD-MS/MS Analysis
4. Conclusions
Acknowledgments
References
- Britton, G.; Liaaen-Jensen, S.; Pfander, H. Carotenoids Handbook; Birkhauser Publishing: Basel, Switzerland, 2004. [Google Scholar]
- Moran, N.A.; Jarvik, T. Lateral transfer of genes from fungi underlies carotenoid production in aphids. Science 2010, 328, 624–627. [Google Scholar] [CrossRef]
- Umeno, D.; Tobias, A.V.; Arnold, F.H. Diversifying carotenoid biosynthetic pathways by directed evolution. Microbiol. Mol. Biol. Rev. 2005, 69, 51–78. [Google Scholar] [CrossRef]
- Muller, L.; Frohlich, K.; Bohm, V. Comparative antioxidant activities of carotenoids measured by ferric reducing antioxidant power (FRAP), ABTS bleaching assay (alpha TEAC), DPPH assay and peroxyl radical scavenging assay. Food Chem. 2011, 129, 139–148. [Google Scholar] [CrossRef]
- Harrison, E.H. Mechanisms of digestion and absorption of dietary vitamin A. Annu. Rev. Nutr. 2005, 25, 87–103. [Google Scholar] [CrossRef]
- Ma, L.; Dou, H.L.; Wu, Y.Q.; Huang, Y.M.; Huang, Y.B.; Xu, X.R.; Zou, Z.Y.; Lin, X.M. Lutein and zeaxanthin intake and the risk of age-related macular degeneration: A systematic review and meta-analysis. Br. J. Nutr. 2012, 107, 350–359. [Google Scholar] [CrossRef]
- Czeczuga, B. Investigations of carotenoids in some animals of the Adriatic Sea V. Echinodermata. Hydrobiologia 1977, 54, 177–180. [Google Scholar] [CrossRef]
- Ferreres, F.; Pereira, D.M.; Gil-Izquierdo, A.; Valentão, P.; Botelho, J.; Mouga, T.; Andrade, P.B. HPLC-PAD-atmospheric pressure chemical ionization-MS metabolite profiling of cytotoxic carotenoids from the echinoderm Marthasterias glacialis (spiny sea-star). J. Sep. Sci. 2010, 33, 2250–2257. [Google Scholar]
- De Rosso, V.V.; Mercadante, A.Z. Identification and quantification of carotenoids, by HPLC-PDA-MS/MS, from Amazonian fruits. J. Agric. Food Chem. 2007, 55, 5062–5072. [Google Scholar] [CrossRef]
- Van Breemen, R.B.; Huang, C.R.; Tan, Y.; Sander, L.C.; Schilling, A.B. Liquid chromatography/mass spectrometry of carotenoids using atmospheric pressure chemical ionization. J. Mass Spectrom. 1996, 31, 975–981. [Google Scholar] [CrossRef]
- Breithaupt, D.E. Identification and quantification of astaxanthin esters in shrimp (Pandalus borealis) and in a microalga (Haematococcus pluvialis) by liquid chromatography-mass spectrometry using negative ion atmospheric pressure chemical ionization. J. Agri. Food Chem. 2004, 52, 3870–3875. [Google Scholar] [CrossRef]
- Etoh, H.; Suhara, M.; Tokuyama, S.; Kato, H.; Nakahigashi, R.; Maejima, Y.; Ishikura, M.; Terada, Y.; Maoka, T. Auto-oxidation products of astaxanthin. J. Oleo Sci. 2012, 61, 17–21. [Google Scholar] [CrossRef]
- Hayakawa, T.; Kulkarni, A.; Terada, Y.; Maoka, T.; Etoh, H. Reaction of astaxanthin with peroxynitrite. Biosci. Biotechnol. Biochem. 2008, 72, 2716–2722. [Google Scholar] [CrossRef]
- Holtin, K.; Kuehnle, M.; Rehbein, J.; Schuler, P.; Nicholson, G.; Albert, K. Determination of astaxanthin and astaxanthin esters in the microalgae Haematococcus pluvialis by LC-(APCI)MS and characterization of predominant carotenoid isomers by NMR spectroscopy. Anal. Bioanal. Chem. 2009, 395, 1613–1622. [Google Scholar] [CrossRef]
- Van Breemen, R.B.; Dong, L.L.; Pajkovic, N.D. Atmospheric pressure chemical ionization tandem mass spectrometry of carotenoids. Int. J. Mass. Spectrom. 2012, 312, 163–172. [Google Scholar] [CrossRef]
- Zechmeistee, L. Cis-trans isomerization and stereochemistry of carotenoids and diphenylpolyenes. Chem. Rev. 1944, 34, 267–344. [Google Scholar] [CrossRef]
- Faria, A.F.; de Rosso, V.V.; Mercadante, A.Z. Carotenoid composition of jackfruit (Artocarpus heterophyllus), determined by HPLC-PDA-MS/MS. Plant Food Hum. Nutr. 2009, 64, 108–115. [Google Scholar] [CrossRef]
- Faria, A.F.; Hasegawa, P.N.; Chagas, E.A.; Pio, R.; Purgatto, E.; Mercadante, A.Z. Cultivar influence on carotenoid composition of loquats from Brazil. J. Food Compos. Anal. 2009, 22, 196–203. [Google Scholar] [CrossRef]
- Schuep, W.; Schierle, J. Astaxanthin: Determination of Stabilized, Added Astaxanthin in Fish Feeds and Pre-mixes. In Carotenoids: Isolation and Analysis; Britton, G., Liaanen-Jensen, S., Pfander, H., Eds.; Birkauser Verlag: Basel, Switzerland, 1995; Volume 1A, pp. 273–276. [Google Scholar]
- Eugster, C.H. Chemichal derivatization: Microscale Tests for the Presence of Common Functional Groups in Carotenoids. In Carotenoids: Isolation and Analysis; Britton, G., Liaanen-Jensen, S., Pfander, H., Eds.; Birkauser Verlag: Basel, Switzerland, 1995; Volume 1A, pp. 71–80. [Google Scholar]
- Samples Availability: Available from the authors.
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Mariutti, L.R.B.; Pereira, D.M.; Mercadante, A.Z.; Valentão, P.; Teixeira, N.; Andrade, P.B. Further Insights on the Carotenoid Profile of the Echinoderm Marthasterias glacialis L. Mar. Drugs 2012, 10, 1498-1510. https://doi.org/10.3390/md10071498
Mariutti LRB, Pereira DM, Mercadante AZ, Valentão P, Teixeira N, Andrade PB. Further Insights on the Carotenoid Profile of the Echinoderm Marthasterias glacialis L. Marine Drugs. 2012; 10(7):1498-1510. https://doi.org/10.3390/md10071498
Chicago/Turabian StyleMariutti, Lilian R. B., David M. Pereira, Adriana Zerlotti Mercadante, Patrícia Valentão, Natércia Teixeira, and Paula B. Andrade. 2012. "Further Insights on the Carotenoid Profile of the Echinoderm Marthasterias glacialis L." Marine Drugs 10, no. 7: 1498-1510. https://doi.org/10.3390/md10071498
APA StyleMariutti, L. R. B., Pereira, D. M., Mercadante, A. Z., Valentão, P., Teixeira, N., & Andrade, P. B. (2012). Further Insights on the Carotenoid Profile of the Echinoderm Marthasterias glacialis L. Marine Drugs, 10(7), 1498-1510. https://doi.org/10.3390/md10071498






