Association Between HLA Class II Gene Polymorphisms and Cytokine Levels in PLWH with HIV-Related Dermatoses in Latvia
Abstract
1. Introduction
1.1. Cytokines
1.2. IL-10
1.3. IFN-γ
1.4. IL-1β
1.5. HLA and Cytokines
2. Materials and Methods
2.1. Study Design
2.2. Study Population
2.3. Genetic Analysis
2.4. Cytokine Analysis
2.5. Statistical Data Analysis
3. Results
3.1. Comparison of Dermatological Groups and Immunological Parameters
3.2. Polymorphism of HLA Class II Alleles
3.3. Association of CD4+ Cell Count and Other Markers in the Research Group
3.4. Relationship Between Immunogenetic Biomarkers in the Research and Control Groups
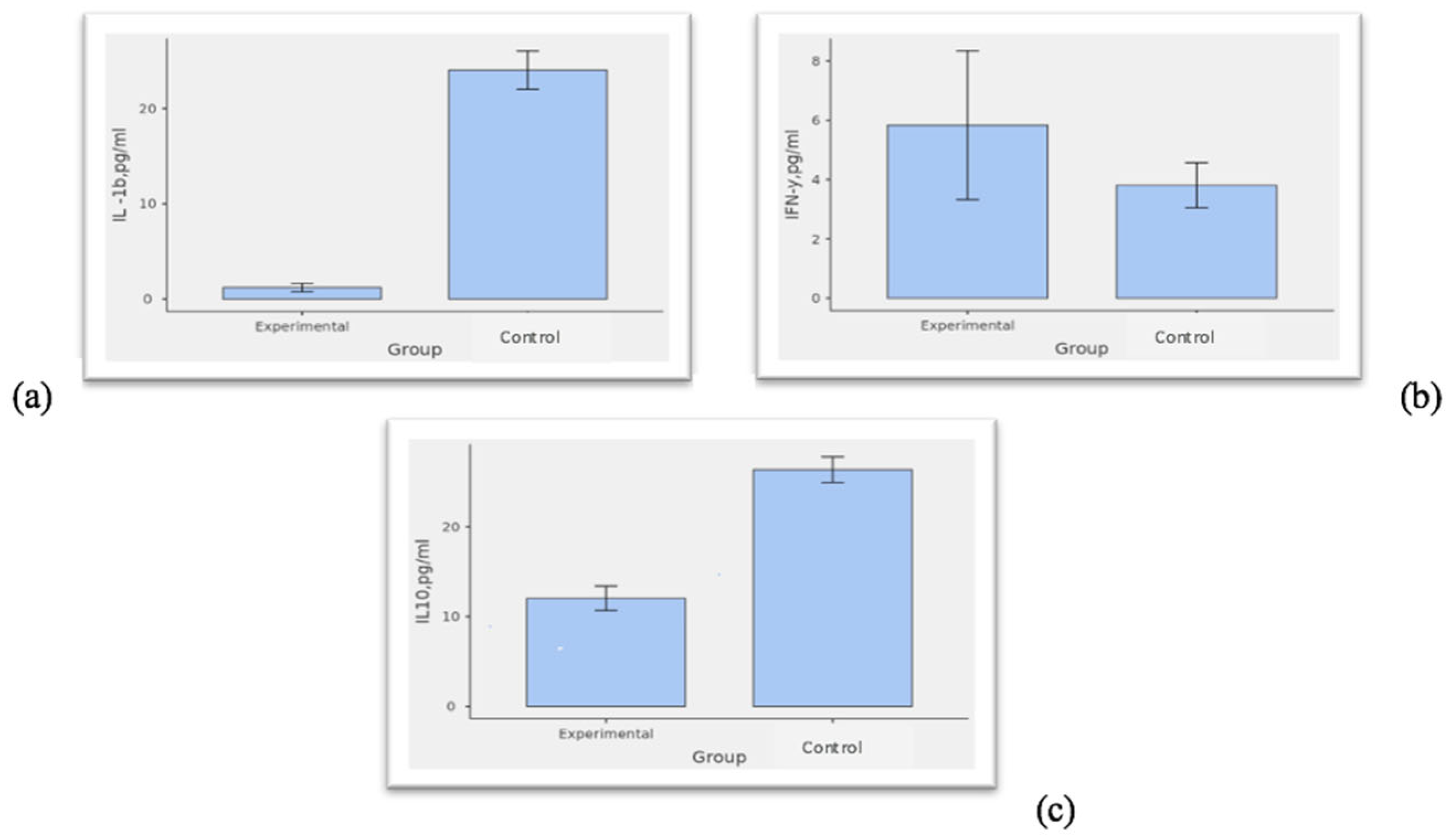
3.5. Expression Levels of Cytokines
3.6. Relationships Between Cytokines in the Research and Control Groups
3.7. Associations Between DRB1, DQA1, and DQB1 Alleles and Cytokine Responses
3.8. Predictive Role of Cytokines in HIV-Related Dermatoses
4. Discussion
4.1. Predictive Role of Cytokines
4.2. HLA
4.3. Clinical Implications
4.4. Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AMPs | antimicrobial peptides |
| ART | antiretroviral therapy |
| AD | atopic dermatitis |
| EBV | Epstein-Barr virus |
| IFN-γ-iMSC-EVs | IFN-y-induced mesenchymal stem cell extracellular vesicles |
| DCs | dendritic cells |
| HLA | human leucocyte antigen |
| RSY/JLCII | the Joint Laboratory of Clinical Immunology and Immunogenetics at Riga Stradiņš University |
| PPE | pruritic papular eruption |
| PLWH | patients living with HIV/AIDS |
| SD | seborrheic dermatitis |
| TB | tuberculosis |
References
- Navarrete-Dechent, C.; Ortega, R.; Fich, F.; Concha, M. Manifestaciones dermatológicas asociadas a la infección por VIH/SIDA [Dermatologic manifestations associated with HIV/AIDS]. Rev. Chilena Infectol. 2015, 32 (Suppl. S1), S57–S71. (In Spanish) [Google Scholar] [CrossRef] [PubMed][Green Version]
- Afshar, Z.M.; Goodarzi, A.; Emadi, S.N.; Miladi, R.; Shakoei, S.; Janbakhsh, A.; Aryanian, Z.; Hatami, P.; Khamesipour, F. A Comprehensive Review on HIV-Associated Dermatologic Manifestations: From Epidemiology to Clinical Management. Int. J. Microbiol. 2023, 2023, 6203193. [Google Scholar]
- Holman, D.M.; Kalaaji, A.N. Cytokines in dermatology. J. Drugs Dermatol. 2006, 5, 520–524. [Google Scholar]
- Mollazadeh, H.; Cicero, A.F.G.; Blesso, C.N.; Pirro, M.; Majeed, M.; Sahebkar, A. Immune modulation by curcumin: The role of interleukin-10. Crit. Rev. Food Sci. Nutr. 2019, 59, 89–101. [Google Scholar] [CrossRef]
- Shin, H.D.; Winkler, C.; Stephens, J.C.; Bream, J.; Young, H.; Goedert, J.J.; O’BRien, T.R.; Vlahov, D.; Buchbinder, S.; Giorgi, J.; et al. Genetic restriction of HIV-1 pathogenesis to AIDS by promoter alleles of IL10. Proc. Natl. Acad. Sci. USA 2000, 97, 14467–14472. [Google Scholar] [CrossRef] [PubMed]
- Terzieva, V.I.; Popova, D.N.; Elenkov, I.I. IFN-γ Attenuates Spontaneous Lymphocyte Proliferation by Fuelling Regulatory T Cells in HIV-1-Infected Patients. Viral Immunol. 2017, 30, 157–166. [Google Scholar] [CrossRef]
- Hayashi, M.; Yanaba, K.; Umezawa, Y.; Yoshihara, Y.; Kikuchi, S.; Ishiuji, Y.; Saeki, H.; Nakagawa, H. IL-10-producing regulatory B cells are decreased in patients with psoriasis. J. Dermatol. Sci. 2016, 81, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Maspi, N.; Abdoli, A.; Ghaffarifar, F. Pro- and anti-inflammatory cytokines in cutaneous leishmaniasis: A review. Pathog. Glob. Health 2016, 110, 247–260. [Google Scholar] [CrossRef]
- Kim, J.; Lee, S.K.; Jung, M.; Jeong, S.-Y.; You, H.; Won, J.-Y.; Han, S.-D.; Cho, H.J.; Park, S.; Park, J.; et al. Extracellular vesicles from IFN-γ-primed mesenchymal stem cells repress atopic dermatitis in mice. J. Nanobiotechnol. 2022, 20, 526. [Google Scholar] [CrossRef]
- Okay, G.; Koc, M.M.; Guler, E.M.; Yabaci, A.; Kocyigit, A.; Akkoyunlu, Y. The Effect of Antiretroviral Therapy on IL-6, IL-1β, TNF-α, IFN-γ Levels and their Relationship with HIV-RNA and CD4+ T Cells in HIV Patients. Curr. HIV Res. 2020, 18, 354–361. [Google Scholar] [CrossRef]
- Lowes, M.A.; Suárez-Fariñas, M.; Krueger, J.G. Immunology of psoriasis. Annu. Rev. Immunol. 2014, 32, 227–255. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Choi, H.K.; N’deh, K.P.U.; Choi, Y.-J.; Fan, M.; Kim, E.-K.; Chung, K.-H.; An, J.H. Inhibitory Effect of Centella asiatica Extract on DNCB-Induced Atopic Dermatitis in HaCaT Cells and BALB/c Mice. Nutrients 2020, 12, 411. [Google Scholar] [CrossRef] [PubMed]
- Lima, T.S. Beyond an inflammatory mediator: Interleukin-1 in neurophysiology. Exp. Physiol. 2023, 108, 917–924. [Google Scholar] [CrossRef]
- Kaneko, N.; Kurata, M.; Yamamoto, T.; Morikawa, S.; Masumoto, J. The role of interleukin-1 in general pathology. Inflamm. Regen. 2019, 39, 12. [Google Scholar] [CrossRef]
- Boraschi, D. What Is IL-1 for? The Functions of Interleukin-1 Across Evolution. Front. Immunol. 2022, 13, 872155. [Google Scholar] [CrossRef]
- Galozzi, P.; Bindoli, S.; Doria, A.; Sfriso, P. The revisited role of interleukin-1 alpha and beta in autoimmune and inflammatory disorders and in comorbidities. Autoimmun. Rev. 2021, 20, 102785. [Google Scholar] [CrossRef] [PubMed]
- Trznadel-Grodzka, E.; Błaszkowski, M.; Rotsztejn, H. Investigations of seborrheic dermatitis. Part I. The role of selected cytokines in the pathogenesis of seborrheic dermatitis. Postepy Hig. Med. Dosw. 2012, 66, 843–847. [Google Scholar] [CrossRef]
- Turchin, I.; Bourcier, M. The Role of Interleukins in the Pathogenesis of Dermatological Immune-Mediated Diseases. Adv. Ther. 2022, 39, 4474–4508. [Google Scholar] [CrossRef]
- Kuniholm, M.H.; Gao, X.; Xue, X.; Kovacs, A.; Anastos, K.; Marti, D.; Greenblatt, R.M.; Cohen, M.H.; Minkoff, H.; Gange, S.J.; et al. Human leukocyte antigen genotype and risk of HIV disease progression before and after initiation of antiretroviral therapy. J. Virol. 2011, 85, 10826–10833. [Google Scholar] [CrossRef]
- Xu, S. Modelling Role of Protective and Nonprotective HLA Allele Inducing Different HIV Infection Outcomes. Bull. Math. Biol. 2024, 86, 107. [Google Scholar] [CrossRef]
- Taneja, V. Cytokines pre-determined by genetic factors are involved in pathogenesis of Rheumatoid arthritis. Cytokine 2015, 75, 216–221. [Google Scholar] [CrossRef] [PubMed]
- Ovsyannikova, I.G.; Jacobson, R.M.; Ryan, J.E.; Dhiman, N.; Vierkant, R.A.; Poland, G.A. Relationship between HLA polymorphisms and gamma interferon and interleukin-10 cytokine production in healthy individuals after rubella vaccination. Clin. Vaccine Immunol. 2007, 14, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Pereira, L.M.S.; dos Santos França, E.; Costa, I.B.; Lima, I.T.; Jorge, E.V.O.; de Souza Mendonça Mattos, P.J.; Freire, A.B.C.; de Paula Ramos, F.L.; Monteiro, T.A.F.; Macedo, O.; et al. DRB1 locus alleles of HLA class II are associated with modulation of the immune response in different serological profiles of HIV-1/Epstein-Barr virus coinfection in the Brazilian Amazon region. Front. Med. 2024, 11, 1408290. [Google Scholar] [CrossRef]
- Bahls, L.; Yamakawa, R.; Zanão, K.; Alfieri, D.; Flauzino, T.; Delongui, F.; De Abreu, A.; Souza, R.; Gimenes, F.; Reiche, E.; et al. Human Leukocyte Antigen Class I and Class II Polymorphisms and Serum Cytokine Profiles in Cervical Cancer. Int. J. Mol. Sci. 2017, 18, 1478. [Google Scholar] [CrossRef]
- Soha, A.; Azina, I.; Miskina, D.A.; Murasko, V.; Zolovs, M.; Rubins, A. Association of HLA class II gene polymorphisms and expression levels of ORAI1/STIM1 genes in HIV-1-positive patients with HIV-related dermatoses in Latvia. Acta Dermatovenerol. Alp. Pannonica Adriat. 2024, 33, 89–94. [Google Scholar] [CrossRef]
- Weiss, E.; Mamelak, A.J.; La Morgia, S.; Wang, B.; Feliciani, C.; Tulli, A.; Sauder, D.N. The role of interleukin 10 in the pathogenesis and potential treatment of skin diseases. J. Am. Acad. Dermatol. 2004, 50, 657–675. [Google Scholar] [CrossRef]
- Fife, D.J.; Waller, J.M.; Jeffes, E.W.; Koo, J.Y. Unraveling the paradoxes of HIV-associated psoriasis: A review of T-cell subsets and cytokine profiles. Dermatol. Online J. 2007, 13, 4. [Google Scholar] [CrossRef]
- Brockman, M.A.; Kwon, D.S.; Tighe, D.P.; Pavlik, D.F.; Rosato, P.C.; Sela, J.; Porichis, F.; Le Gall, S.; Waring, M.T.; Moss, K.; et al. IL-10 is up-regulated in multiple cell types during viremic HIV infection and reversibly inhibits virus-specific T cells. Blood 2009, 114, 346–356. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, L.; Coutsoudis, A.; Moodley, D.; Mngqundaniso, N.; Trabattoni, D.; Shearer, G.M.; Clerici, M.; Coovadia, H.M. Interferon-gamma and interleukin-10 production among HIV-1-infected and uninfected infants of HIV-1-infected mothers. Pediatr. Res. 2001, 50, 412–416. [Google Scholar] [CrossRef]
- Giunta, E.; Barra, G.; De Falco, V.; Vitale, P.; Zanaletti, N.; Terminiello, M.; Caputo, V.; Napolitano, S.; Vitiello, P.; Ciardiello, D.; et al. IFN-γ/IL-10 ratio as predictive biomarker for response to anti-PD-1 therapy in metastatic melanoma patients. Ann. Oncol. 2019, 30, v40. [Google Scholar] [CrossRef]
- Breuer-McHam, J.N.; Ledbetter, L.S.; Sarris, A.H.; Duvic, M. Cytokine expression patterns distinguish HIV associated skin diseases. Exp. Dermatol. 2000, 9, 341–350. [Google Scholar] [CrossRef] [PubMed]
- Breuer-McHAM, J.N.; Marshall, G.D.; Lewis, D.E.; Duvic, M. Distinct serum cytokines in AIDS-Related skin diseases. Viral Immunol. 1998, 11, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Shao, S.; Tsoi, L.C.; Sarkar, M.K.; Xing, X.; Xue, K.; Uppala, R.; Berthier, C.C.; Zeng, C.; Patrick, M.; Billi, A.C.; et al. IFN-γ enhances cell-mediated cytotoxicity against keratinocytes via JAK2/STAT1 in lichen planus. Sci. Transl. Med. 2019, 11, eaav7561. [Google Scholar] [CrossRef]
- Kedzierska, K.; Crowe, S.M. Cytokines and HIV-1: Interactions and Clinical Implications. Antivir. Chem. Chemother. 2001, 12, 133–150. [Google Scholar] [CrossRef]
- Kumar, H.; Desai, K.; Naveen, S.; Somanna, P. Vitiligo: Correlation with Cytokine Profiles and its Role in Novel Therapeutic Strategies: A Case–Control Study. Indian Dermatol. Online J. 2023, 14, 361–365. [Google Scholar] [CrossRef]
- Soha, A.; Azina, I.; Rozentale, B.; Kramicha, K.; Sture, G.; Savicka, O.; Titovica, G. HLA class II DRB1, DQA1, DQB1 loci in patients with HIV infection and tuberculosis in a Latvian cohort group. Cent. Eur. J. Immunol. 2024, 49, 37–44. [Google Scholar] [CrossRef]
- Dinarello, C.A. Interleukin-1 in the pathogenesis and treatment of inflammatory diseases. Blood 2011, 117, 3720–3732. [Google Scholar] [CrossRef]
- Zhu, Y.; Qin, H.; Ye, K.; Sun, C.; Qin, Y.; Li, G.; Wang, H.; Wang, H. Dual role of IL-37 in the progression of tumors. Cytokine 2022, 150, 155760. [Google Scholar] [CrossRef] [PubMed]
- Sundaramurthi, J.C.; Ashokkumar, M.; Swaminathan, S.; Hanna, L.E. HLA based selection of epitopes offers a potential window of opportunity for vaccine design against HIV. Vaccine 2017, 35, 5568–5575. [Google Scholar] [CrossRef]
- Shaker, O.; Bassiony, H.; El Raziky, M.; El-Kamary, S.S.; Esmat, G.; El-Ghor, A.M.; Mohamed, M.M. Human leukocyte antigen class II alleles (DQB1 and DRB1) as predictors for response to interferon therapy in HCV genotype 4. Mediat. Inflamm. 2013, 2013, 392746. [Google Scholar] [CrossRef]
- Soha, A.; Azina, I.; Zolovs, M.; Miskina, D.A.; Murasko, V.; Rozentale, B.; Hartmane, I.; Rubins, A. HLA Class II, Cytokines, HIV; Rīga Stradiņš University Institutional Repository Dataverse: Riga, Latvia, 2025. [Google Scholar] [CrossRef]
| Parameter | Skin Group I (n = 77) Median (IQR) | Skin Group II (n = 38) Median (IQR) | p-Value |
|---|---|---|---|
| HIV RNS III, copy/mL | 81.00 (20.00–131,000.00) | 55.00 (20.00–126,250.00) | 0.949 |
| CD4+ III, cells/mL | 222.00 (80.00–364.00) | 196.00 (86.25–373.00) | 0.704 |
| IL-1b, pg/mL | 0.00 (0.00–0.00) | 0.00 (0.00–0.00) | 0.543 |
| IFN-y, pg/mL | 0.00 (0.00–3.50) | 0.00 (0.00–1.00) | 0.831 |
| IL-10, pg/mL | 8.00 (6.00–12.90) | 9.10 (4.00–14.90) | 0.813 |
| Alleles | Experimental Relative Frequency | Control Relative Frequency | OR (95% CI) | p |
|---|---|---|---|---|
| DRB1 07-13 | 0.02 | 0.09 | 0.19 (0.04–0.91) | 0.022 |
| DRB1 01-13 | 0.01 | 0.09 | 0.09 (0.01–0.76) | 0.006 |
| DRB1 04-11 | 0 | 0.05 | 0.07 (0.001–1.39) | 0.015 |
| DQA1 0101-0501 | 0.22 | 0.06 | 4.2 (1.5–11.4) | 0.003 |
| DQA1 0501-0501 | 0.03 | 0.10 | 0.24 (0.06–0.94) | 0.028 |
| DQB1 0201-0301 | 0.10 | 0 | 19.4 (1.1–333) | 0.003 |
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Age | — | |||||||||||
| 2 | Gender | 0.105 | — | ||||||||||
| 3 | Post TB | −0.307 *** | −0.013 | — | |||||||||
| 4 | IL-1b | 0.295 *** | −0.096 | −0.697 *** | — | ||||||||
| 5 | IFN-y | 0.089 | 0.116 | −0.182 * | 0.169 * | — | |||||||
| 6 | IL10 | 0.219 ** | −0.077 | −0.566 *** | 0.591 *** | 0.225 ** | — | ||||||
| 7 | DQA1*0101:0501 | −0.106 | 0.064 | 0.258 *** | −0.260 ** | −0.077 | −0.247 ** | — | |||||
| 8 | DQA1*0501:0501 | 0.076 | 0.039 | −0.137 | 0.192 * | −0.009 | 0.153 | −0.102 | — | ||||
| 9 | DRB1*07:13 | 0.003 | 0.007 | −0.114 | 0.103 | 0.015 | 0.120 | −0.092 | 0.158 * | — | |||
| 10 | DRB1*01:13 | 0.037 | 0.043 | −0.101 | 0.084 | −0.110 | 0.110 | −0.014 | −0.051 | −0.045 | — | ||
| 11 | DRB1*04:11 | −0.051 | 0.030 | −0.108 | 0.054 | 0.164 | 0.033 | −0.060 | −0.035 | −0.032 | −0.030 | — | |
| 12 | DQB1*0201:0301 | −0.130 | 0.008 | 0.209** | −0.237 ** | −0.019 | −0.056 | −0.107 | −0.063 | −0.056 | −0.053 | −0.037 | — |
| Research (n = 115) | Control (n = 80) | |||||
|---|---|---|---|---|---|---|
| IL-1b, pg/mL | IFN-y, pg/mL | IL10, pg/mL | IL-1b, pg/mL | IFN-y, pg/mL | IL10, pg/mL | |
| IL-1b, pg/mL | - | - | ||||
| IFN-y, pg/mL | 0.059 | - | 0.101 | - | ||
| IL10, pg/mL | 0.189 | 0.518 *** | - | 0.402 *** | −0.097 | - |
| Parameter | DQA1 0101-0501 | DQA1 0501-0501 | p-Value | Effect Size |
|---|---|---|---|---|
| Median (Q1–Q3) | Median (Q1–Q3) | |||
| IL-1b, pg/mL | 0.00 (0.00–0.00) | 28.75 (19.10–32.60) | < 0.001 | 0.79 |
| IFN-y, pg/mL | 0.00 (0.00–3.50) | 0.00 (0.00–3.85) | 0.677 | NA |
| IL-10, pg/mL | 9.10 (4.80–16.00) | 31.30 (14.43–41.75) | 0.010 | 0.63 |
| Exp(B) 95% Confidence Intervals | |||||||
|---|---|---|---|---|---|---|---|
| Name | Estimate | SE | Exp(B) | Lower | Upper | z | p |
| (Intercept) | −2.4040 | 0.6611 | 0.0904 | 0.00484 | 0.237 | −3.64 | <0.001 |
| IL-1b, pg/mL | −0.2683 | 0.0542 | 0.7647 | 0.62555 | 0.824 | −4.95 | <0.001 |
| IL-10 | −0.0702 | 0.0294 | 0.9322 | 0.81632 | 0.991 | −2.38 | 0.017 |
| IFN-y | 0.0634 | 0.0313 | 1.0654 | 0.98488 | 1.176 | 2.02 | 0.043 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Published by MDPI on behalf of the Lithuanian University of Health Sciences. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soha, A.; Azina, I.; Zolovs, M.; Miskina, D.A.; Murasko, V.; Rozentale, B.; Hartmane, I.; Rubins, A. Association Between HLA Class II Gene Polymorphisms and Cytokine Levels in PLWH with HIV-Related Dermatoses in Latvia. Medicina 2025, 61, 1091. https://doi.org/10.3390/medicina61061091
Soha A, Azina I, Zolovs M, Miskina DA, Murasko V, Rozentale B, Hartmane I, Rubins A. Association Between HLA Class II Gene Polymorphisms and Cytokine Levels in PLWH with HIV-Related Dermatoses in Latvia. Medicina. 2025; 61(6):1091. https://doi.org/10.3390/medicina61061091
Chicago/Turabian StyleSoha, Alena, Inga Azina, Maksims Zolovs, Darja Arina Miskina, Viktorija Murasko, Baiba Rozentale, Ilona Hartmane, and Andris Rubins. 2025. "Association Between HLA Class II Gene Polymorphisms and Cytokine Levels in PLWH with HIV-Related Dermatoses in Latvia" Medicina 61, no. 6: 1091. https://doi.org/10.3390/medicina61061091
APA StyleSoha, A., Azina, I., Zolovs, M., Miskina, D. A., Murasko, V., Rozentale, B., Hartmane, I., & Rubins, A. (2025). Association Between HLA Class II Gene Polymorphisms and Cytokine Levels in PLWH with HIV-Related Dermatoses in Latvia. Medicina, 61(6), 1091. https://doi.org/10.3390/medicina61061091






