IgG Antibody Responses to Epstein-Barr Virus in Myalgic Encephalomyelitis/Chronic Fatigue Syndrome: Their Effective Potential for Disease Diagnosis and Pathological Antigenic Mimicry
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Participants
2.2. Basic Description of Serological Data
2.3. Statistical Analysis for Predicting the Disease Status
2.3.1. Dividing the Dataset into Train and Test Sets
2.3.2. Ranking Antibodies by Their Importance for Predicting the Disease Status
2.3.3. Individual Statistical and Machine Learning Methods for Predicting the Disease Status from the Anti-EBV Antibodies
2.3.4. Construction of Final Models for Predicting the Disease Status by Assembling Predictions from Individual Models
2.4. Bioinformatic Analysis to Test the Importance of Antigen Mimicry in Predicting the Disease Status
2.5. Statistical Software
3. Results
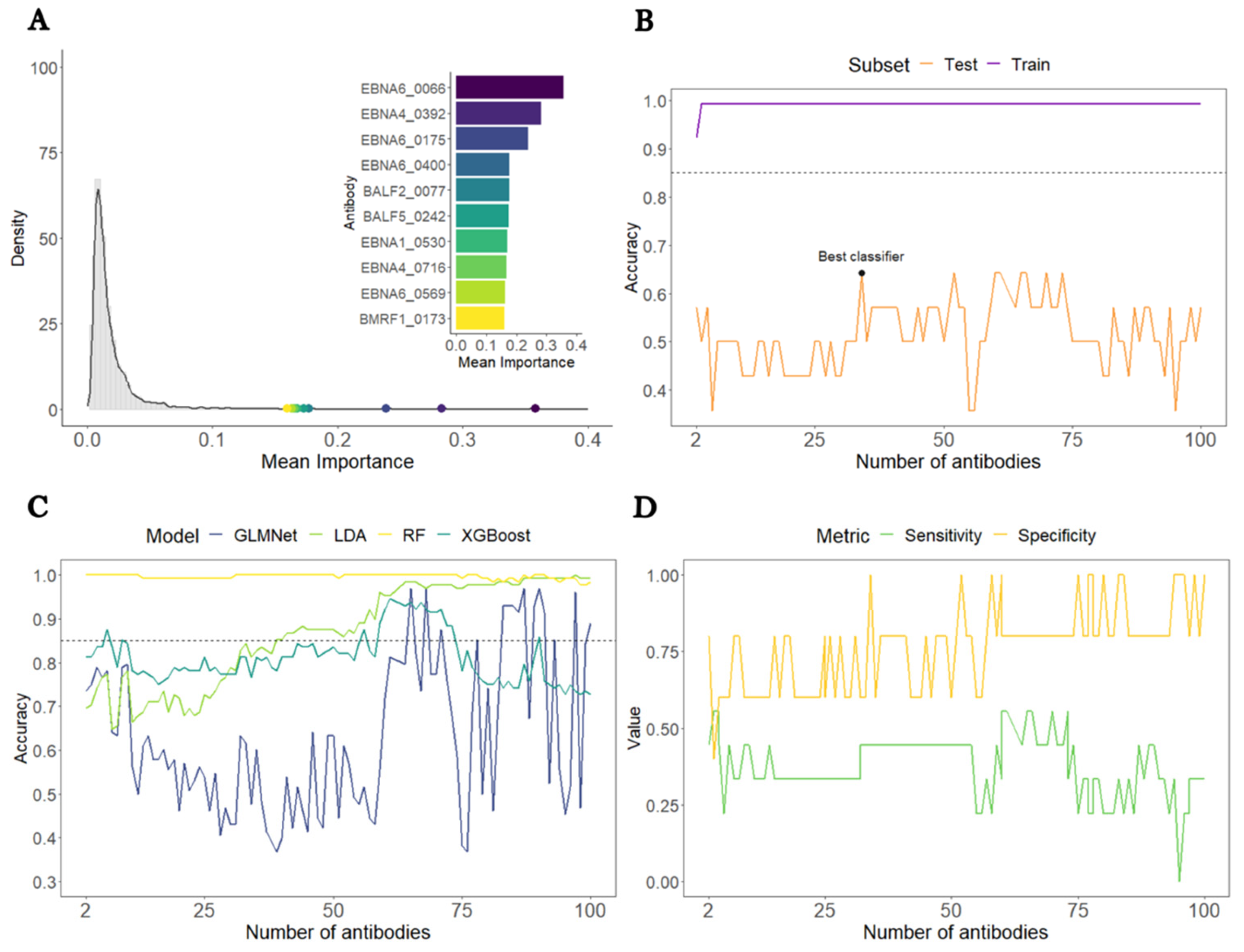
3.1. Construction of a Predictive Model to Distinguish All ME/CFS Patients from HCs
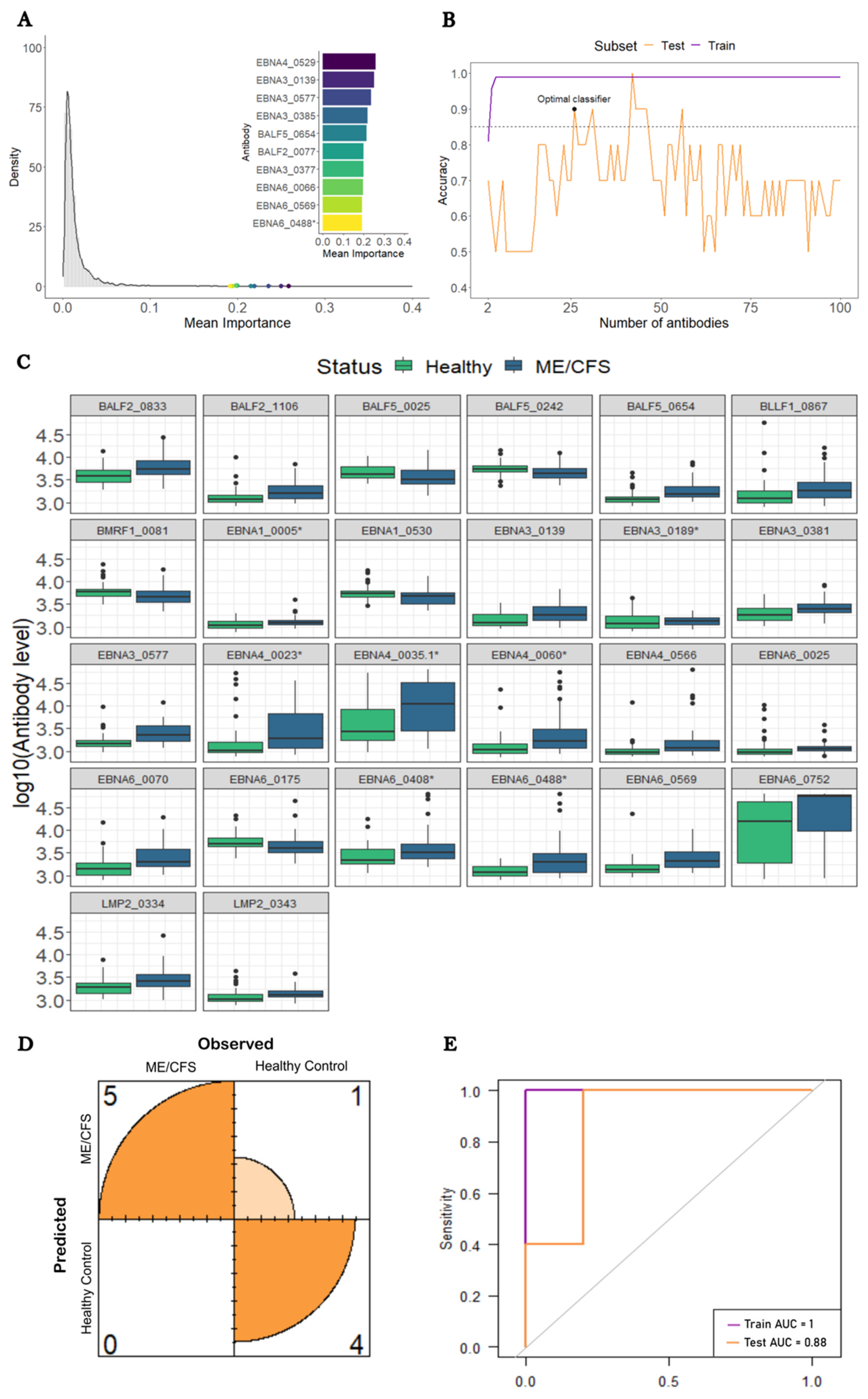
3.2. Construction of a Predictive Model to Distinguish ME/CFS Patients with Non-Infectious or Unknown Disease Triggers from HCs
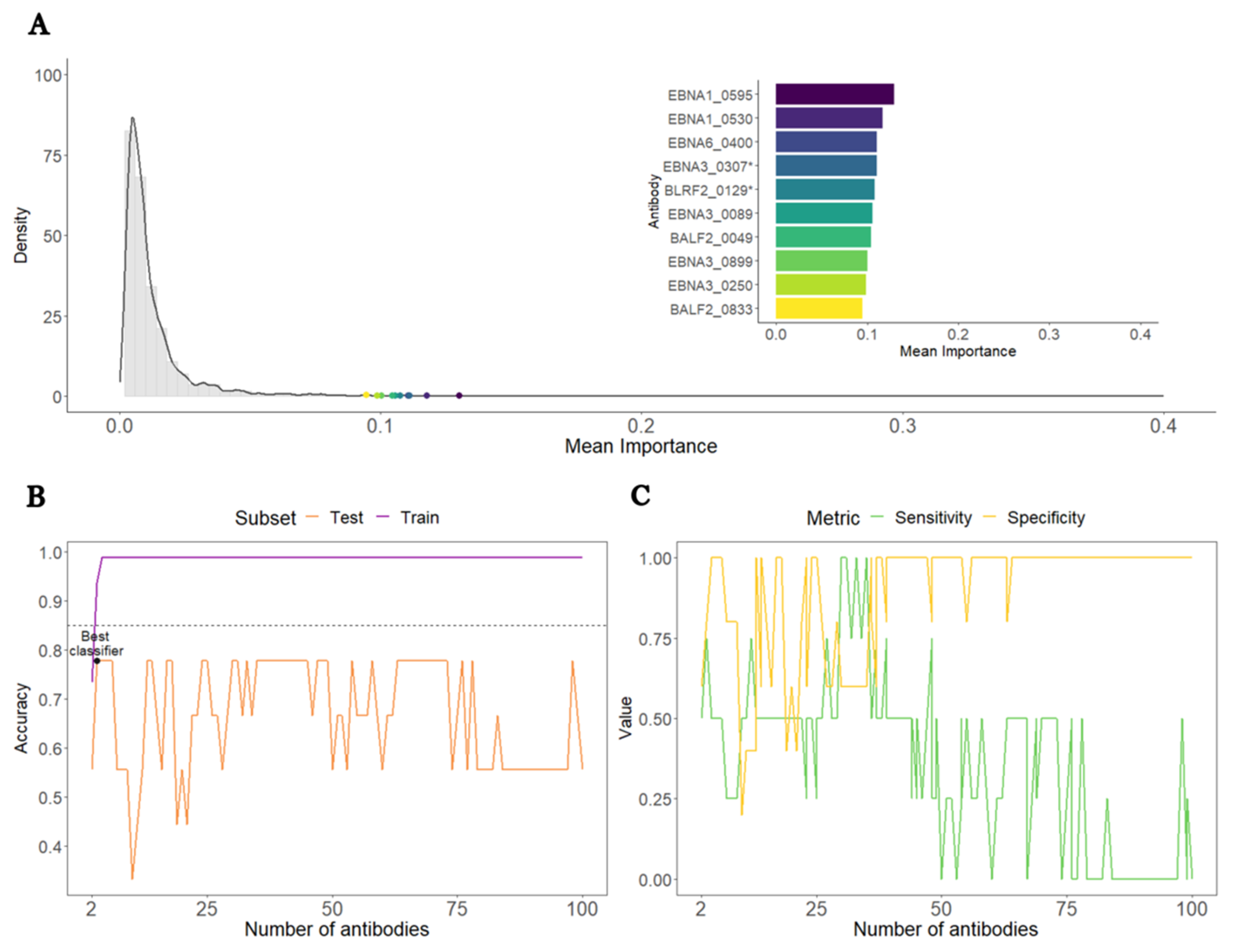
3.3. Construction of a Predictive Model to Distinguish ME/CFS Patients with a Putative Infectious Disease Trigger from HCs
3.4. Testing the Importance of Antigen Mimicry on Disease Prediction Using a Bioinformatic Approach
4. Discussion
4.1. General Comments
4.2. Clinical and Diagnostic Implications
4.3. EBV Antigenic Mimicry and Its Putative Role in ME/CFS Pathogenesis
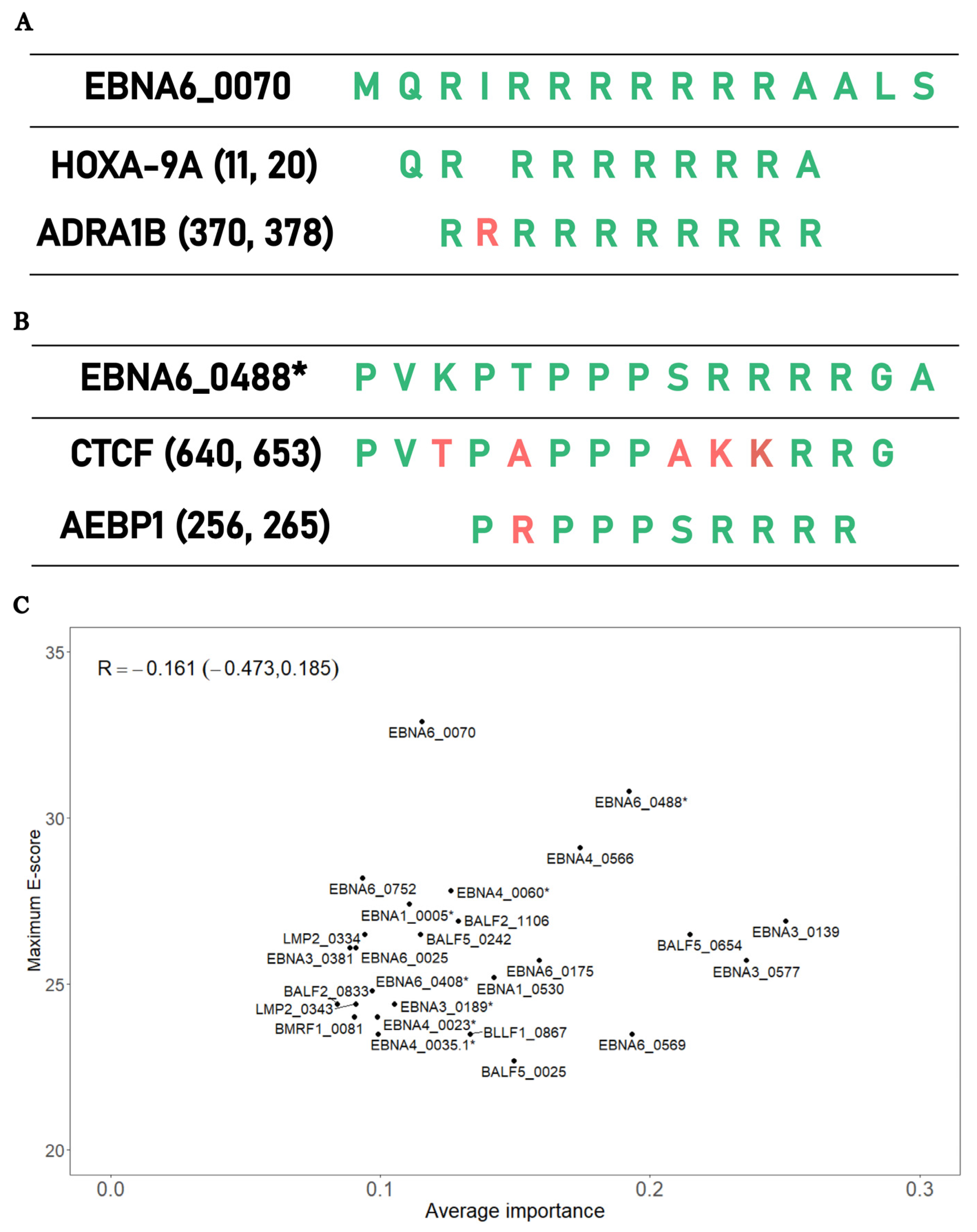
4.3.1. Replication of Previous Finding on EBNA6_0070 Peptide
4.3.2. EBNA6_0488 Peptide and the Antigenic Mimicry with CTCF and AEBP1
4.4. Interpretation of the Findings under the Lens of the Danger Theory
4.5. Potential Danger Signals in ME/CFS Pathogenesis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rivera, M.C.; Mastronardi, C.; Silva-Aldana, C.T.; Arcos-Burgos, M.; Lidbury, B.A. Myalgic encephalomyelitis/chronic fatigue syndrome: A comprehensive review. Diagnostics 2019, 9, 91. [Google Scholar] [CrossRef] [PubMed]
- Deumer, U.S.; Varesi, A.; Floris, V.; Savioli, G.; Mantovani, E.; Lopez-Carrasco, P.; Rosati, G.M.; Prasad, S.; Ricevuti, G. Myalgic Encephalomyelitis/Chronic Fatigue Syndrome (ME/CFS): An Overview. J. Clin. Med. 2021, 10, 4786. [Google Scholar] [CrossRef] [PubMed]
- Fluge, Ø.; Tronstad, K.J.; Mella, O. Pathomechanisms and possible interventions in myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS). J. Clin. Investig. 2021, 131, e150377. [Google Scholar] [CrossRef]
- Stanculescu, D.; Larsson, L.; Bergquist, J. Hypothesis: Mechanisms that Prevent Recovery in Prolonged ICU Patients Also Underlie Myalgic Encephalomyelitis/Chronic Fatigue Syndrome (ME/CFS). Front. Med. 2021, 8, 628029. [Google Scholar] [CrossRef]
- Stanculescu, D.; Sepúlveda, N.; Lim, C.L.; Bergquist, J. Lessons from Heat Stroke for Understanding Myalgic Encephalomyelitis/Chronic Fatigue Syndrome. Front. Neurol. 2021, 12, 789784. [Google Scholar] [CrossRef] [PubMed]
- Nacul, L.; O’Boyle, S.; Palla, L.; Nacul, F.E.; Mudie, K.; Kingdon, C.C.; Cliff, J.M.; Clark, T.G.; Dockrell, H.M.; Lacerda, E. How Myalgic Encephalomyelitis/Chronic Fatigue Syndrome (ME/CFS) Progresses: The Natural History of ME/CFS. Front. Neurol. 2020, 11, 826. [Google Scholar] [CrossRef]
- Blomberg, J.; Gottfries, C.G.; Elfaitouri, A.; Rizwan, M.; Rosén, A. Infection elicited autoimmunity and Myalgic encephalomyelitis/chronic fatigue syndrome: An explanatory model. Front. Immunol. 2018, 9, 229. [Google Scholar] [CrossRef]
- Sotzny, F.; Blanco, J.; Capelli, E.; Castro-Marrero, J.; Steiner, S.; Murovska, M.; Scheibenbogen, C. Myalgic Encephalomyelitis/Chronic Fatigue Syndrome–Evidence for an autoimmune disease. Autoimmun. Rev. 2018, 17, 601–609. [Google Scholar] [CrossRef]
- Morris, G.; Berk, M.; Galecki, P.; Maes, M. The emerging role of autoimmunity in myalgic encephalomyelitis/chronic fatigue syndrome (ME/cfs). Mol. Neurobiol. 2014, 49, 741–756. [Google Scholar] [CrossRef]
- Petracek, L.S.; Suskauer, S.J.; Vickers, R.F.; Patel, N.R.; Violand, R.L.; Swope, R.L.; Rowe, P.C. Adolescent and Young Adult ME/CFS After Confirmed or Probable COVID-19. Front. Med. 2021, 8, 668944. [Google Scholar] [CrossRef]
- Davis, H.E.; Assaf, G.S.; McCorkell, L.; Wei, H.; Low, R.J.; Re’em, Y.; Redfield, S.; Austin, J.P.; Akrami, A. Characterizing long COVID in an international cohort: 7 months of symptoms and their impact. EClinicalMedicine 2021, 38, 101019. [Google Scholar] [CrossRef]
- Jason, L.A.; Dorri, J.A. ME/CFS and Post-Exertional Malaise among Patients with Long COVID. Neurol. Int. 2022, 15, 1. [Google Scholar] [CrossRef] [PubMed]
- Vélez-Santamaría, R.; Fernández-Solana, J.; Méndez-López, F.; Domínguez-García, M.; González-Bernal, J.J.; Magallón-Botaya, R.; Oliván-Blázquez, B.; González-Santos, J.; Santamaría-Peláez, M. Functionality, physical activity, fatigue and quality of life in patients with acute COVID-19 and Long COVID infection. Sci. Rep. 2023, 13, 19907. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Pablos, M.; Paiva, B.; Montero-Mateo, R.; Garcia, N.; Zabaleta, A. Epstein-Barr Virus and the Origin of Myalgic Encephalomyelitis or Chronic Fatigue Syndrome. Front. Immunol. 2021, 12, 656797. [Google Scholar] [CrossRef] [PubMed]
- Ariza, M.E. Myalgic encephalomyelitis/chronic fatigue syndrome: The human herpesviruses are back! Biomolecules 2021, 11, 185. [Google Scholar] [CrossRef] [PubMed]
- Sepúlveda, N.; Carneiro, J.; Lacerda, E.; Nacul, L. Myalgic Encephalomyelitis/Chronic Fatigue Syndrome as a Hyper-Regulated Immune System Driven by an Interplay Between Regulatory T Cells and Chronic Human Herpesvirus Infections. Front. Immunol. 2019, 10, 2684. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Pablos, M.; Paiva, B.; Zabaleta, A. Epstein–Barr virus-acquired immunodeficiency in myalgic encephalomyelitis—Is it present in long COVID? J. Transl. Med. 2023, 21, 633. [Google Scholar] [CrossRef] [PubMed]
- Cox, B.S.; Alharshawi, K.; Mena-Palomo, I.; Lafuse, W.P.; Ariza, M.E. EBV/HHV-6A dUTPases contribute to myalgic encephalomyelitis/chronic fatigue syndrome pathophysiology by enhancing TFH cell differentiation and extrafollicular activities. JCI Insight 2022, 7, e158193. [Google Scholar] [CrossRef] [PubMed]
- Capone, G.; Calabrò, M.; Lucchese, G.; Fasano, C.; Girardi, B.; Polimeno, L.; Kanduc, D. Peptide matching between Epstein-Barr virus and human proteins. Pathog. Dis. 2013, 69, 205–212. [Google Scholar] [CrossRef]
- Baboonian, C.; Venables, P.J.W.; Williams, D.G.; Williams, R.O.; Maini, R.N. Cross reaction of antibodies to a glycine/alanine repeat sequence of Epstein-Barr virus nuclear antigen-i with collagen, cytokeratin, and actin. Ann. Rheum. Dis. 1991, 50, 772–775. [Google Scholar] [CrossRef]
- Blomberg, J.; Rizwan, M.; Böhlin-Wiener, A.; Elfaitouri, A.; Julin, P.; Zachrisson, O.; Rosén, A.; Gottfries, C.G. Antibodies to human herpesviruses in myalgic encephalomyelitis/chronic fatigue syndrome patients. Front. Immunol. 2019, 10, 1946. [Google Scholar] [CrossRef]
- Lerner, A.M.; Ariza, M.E.; Williams, M.; Jason, L.; Beqaj, S.; Fitzgerald, J.T.; Lemeshow, S.; Glaser, R. Antibody to Epstein-Barr virus deoxyuridine triphosphate nucleotidohydrolase and deoxyribonucleotide polymerase in a chronic fatigue syndrome subset. PLoS ONE 2012, 7, e47891. [Google Scholar] [CrossRef]
- Kerr, J.R. Epstein-Barr Virus Induced Gene-2 Upregulation Identifies a Particular Subtype of Chronic Fatigue Syndrome/Myalgic Encephalomyelitis. Front. Pediatr. 2019, 7, 59. [Google Scholar] [CrossRef] [PubMed]
- Domingues, T.D.; Grabowska, A.D.; Lee, J.S.; Ameijeiras-Alonso, J.; Westermeier, F.; Scheibenbogen, C.; Cliff, J.M.; Nacul, L.; Lacerda, E.M.; Mouriño, H.; et al. Herpesviruses Serology Distinguishes Different Subgroups of Patients from the United Kingdom Myalgic Encephalomyelitis/Chronic Fatigue Syndrome Biobank. Front. Med. 2021, 8, 686736. [Google Scholar] [CrossRef] [PubMed]
- Malato, J.; Graça, L.; Sepúlveda, N. Impact of Misdiagnosis in Case-Control Studies of Myalgic Encephalomyelitis/Chronic Fatigue Syndrome. Diagnostics 2023, 13, 531. [Google Scholar] [CrossRef]
- Nacul, L.; Lacerda, E.M.; Kingdon, C.C.; Curran, H.; Bowman, E.W. How have selection bias and disease misclassification undermined the validity of myalgic encephalomyelitis/chronic fatigue syndrome studies? J. Health Psychol. 2019, 24, 1765–1769. [Google Scholar] [CrossRef]
- Ariza, M.E. Commentary: Antibodies to Human Herpesviruses in Myalgic Encephalomyelitis/Chronic Fatigue Syndrome Patients. Front. Immunol. 2020, 11, 1945. [Google Scholar] [CrossRef]
- Loebel, M.; Eckey, M.; Sotzny, F.; Hahn, E.; Bauer, S.; Grabowski, P.; Zerweck, J.; Holenya, P.; Hanitsch, L.G.; Wittke, K.; et al. Serological profiling of the EBV immune response in Chronic Fatigue Syndrome using a peptide microarray. PLoS ONE 2017, 12, e0179124. [Google Scholar] [CrossRef]
- Sepúlveda, N.; Malato, J.; Sotzny, F.; Grabowska, A.D.; Fonseca, A.; Cordeiro, C.; Graça, L.; Biecek, P.; Behrends, U.; Mautner, J.; et al. Revisiting IgG Antibody Reactivity to Epstein-Barr Virus in Myalgic Encephalomyelitis/Chronic Fatigue Syndrome and Its Potential Application to Disease Diagnosis. Front. Med. 2022, 9, 921101. [Google Scholar] [CrossRef] [PubMed]
- Roshan, V.; Stewart, J.H.M.; Joseph, R.; Stewart, H.M. Optimal ratio for data splitting. Stat. Anal. Data Min. ASA Data Sci. J. 2022, 15, 531–538. [Google Scholar] [CrossRef]
- Van Der Laan, M.J.; Polley, E.C.; Hubbard, A.E. Super learner. Stat. Appl. Genet. Mol. Biol. 2007, 6, 25. [Google Scholar] [CrossRef]
- López-Ratón, M.; Rodríguez-Álvarez, M.X.; Cadarso-Suárez, C.; Gude-Sampedro, F. Optimalcutpoints: An R package for selecting optimal cutpoints in diagnostic tests. J. Stat. Softw. 2014, 61, 1–36. [Google Scholar] [CrossRef]
- Valletta, J.J.; Recker, M. Identification of immune signatures predictive of clinical protection from malaria. PLoS Comput. Biol. 2017, 13, e1005812. [Google Scholar] [CrossRef] [PubMed]
- Wilson, R.C.; Shenhav, A.; Straccia, M.; Cohen, J.D. The Eighty Five Percent Rule for optimal learning. Nat. Commun. 2019, 10, 4646. [Google Scholar] [CrossRef] [PubMed]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef] [PubMed]
- Karlin, S.; Altschul, S.F. Applications and statistics for multiple high-scoring segments in molecular sequences. Proc. Natl. Acad. Sci. USA 1993, 90, 5873–5877. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, M. Building Predictive Models in R Using the caret Package. J. Stat. Softw. 2008, 28, 1–26. [Google Scholar] [CrossRef]
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.C.; Müller, M. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011, 12, 77. [Google Scholar] [CrossRef]
- Wright, M.N.; Ziegler, A. ranger: A Fast Implementation of Random Forests for High Dimensional Data in C++ and R. J. Stat. Softw. 2017, 77, 1–17. [Google Scholar] [CrossRef]
- Polley, E.; LeDell, E.; Kennedy, C.; van der Laan, M. SuperLearner: Super. Learner Prediction 2021. R Package Version 2.0-28. Available online: https://cran.r-project.org/package=SuperLearner (accessed on 21 November 2023).
- Tengvall, K.; Huang, J.; Hellström, C.; Kammer, P.; Biström, M.; Ayoglu, B.; Lima Bomfim, I.; Stridh, P.; Butt, J.; Brenner, N.; et al. Molecular mimicry between Anoctamin 2 and Epstein-Barr virus nuclear antigen 1 associates with multiple sclerosis risk. Proc. Natl. Acad. Sci. USA 2019, 116, 16955–16960. [Google Scholar] [CrossRef]
- Sepúlveda, N. Impact of genetic variation on the molecular mimicry between Anoctamin-2 and Epstein-Barr virus nuclear antigen 1 in Multiple Sclerosis. Immunol. Lett. 2021, 238, 29–31. [Google Scholar] [CrossRef]
- O’Neal, A.J.; Glass, K.A.; Emig, C.J.; Vitug, A.A.; Henry, S.J.; Shungu, D.C.; Mao, X.; Levine, S.M.; Hanson, M.R. Survey of Anti-Pathogen Antibody Levels in Myalgic Encephalomyelitis/Chronic Fatigue Syndrome. Proteomes 2022, 10, 21. [Google Scholar] [CrossRef]
- Jason, L.A.; Corradi, K.; Torres-Harding, S.; Taylor, R.R.; King, C. Chronic fatigue syndrome: The need for subtypes. Neuropsychol. Rev. 2005, 15, 29–58. [Google Scholar] [CrossRef]
- Ruprecht, K.; Wunderlich, B.; Gieß, R.; Meyer, P.; Loebel, M.; Lenz, K.; Hofmann, J.; Rosche, B.; Wengert, O.; Paul, F.; et al. Multiple sclerosis: The elevated antibody response to Epstein-Barr virus primarily targets, but is not confined to, the glycine-alanine repeat of Epstein-Barr nuclear antigen-1. J. Neuroimmunol. 2014, 272, 56–61. [Google Scholar] [CrossRef]
- Bjornevik, K.; Cortese, M.; Healy, B.C.; Kuhle, J.; Mina, M.J.; Leng, Y.; Elledge, S.J.; Niebuhr, D.W.; Scher, A.I.; Munger, K.L.; et al. Longitudinal analysis reveals high prevalence of Epstein-Barr virus associated with multiple sclerosis. Science 2022, 375, 296–301. [Google Scholar] [CrossRef] [PubMed]
- Jason, L.; Ohanian, D.; Brown, A.; Sunnquist, M.; McManimen, S.; Klebek, L.; Fox, P.; Sorenson, M. Differentiating Multiple Sclerosis from Myalgic Encephalomyelitis and Chronic Fatigue Syndrome. Insights Biomed. 2017, 2, 11. [Google Scholar] [CrossRef]
- Ohanian, D.; Brown, A.; Sunnquist, M.; Furst, J.; Nicholson, L.; Klebek, L.; Jason, L.A. Identifying Key Symptoms Differentiating Myalgic Encephalomyelitis and Chronic Fatigue Syndrome from Multiple Sclerosis. Neurology 2016, 4, 41. [Google Scholar] [PubMed]
- Domingues, T.D.; Malato, J.; Grabowska, A.D.; Lee, J.S.; Ameijeiras-Alonso, J.; Biecek, P.; Graça, L.; Mouriño, H.; Scheibenbogen, C.; Westermeier, F.; et al. Association analysis between symptomology and herpesvirus IgG antibody concentrations in myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS) and multiple sclerosis. Heliyon 2023, 9, E18250. [Google Scholar] [CrossRef] [PubMed]
- Steiner, S.; Fehrer, A.; Hoheisel, F.; Schoening, S.; Aschenbrenner, A.; Babel, N.; Bellmann-Strobl, J.; Finke, C.; Fluge, Ø.; Froehlich, L.; et al. Understanding, diagnosing, and treating Myalgic encephalomyelitis/chronic fatigue syndrome-State of the art: Report of the 2nd international meeting at the Charité Fatigue Center. Autoimmun. Rev. 2023, 22, 103452. [Google Scholar] [CrossRef] [PubMed]
- Scheibenbogen, C.; Loebel, M.; Freitag, H.; Krueger, A.; Bauer, S.; Antelmann, M.; Doehner, W.; Scherbakov, N.; Heidecke, H.; Reinke, P.; et al. Immunoadsorption to remove ß2 adrenergic receptor antibodies in Chronic Fatigue Syndrome CFS/ME. PLoS ONE 2018, 13, e0193672. [Google Scholar] [CrossRef]
- Tölle, M.; Freitag, H.; Antelmann, M.; Hartwig, J.; Schuchardt, M.; van der Giet, M.; Eckardt, K.U.; Grabowski, P.; Scheibenbogen, C. Myalgic Encephalomyelitis/Chronic Fatigue Syndrome: Efficacy of Repeat Immunoadsorption. J. Clin. Med. 2020, 9, 2443. [Google Scholar] [CrossRef]
- Stein, E.; Heindrich, C.; Wittke, K.; Kedor, C.; Kim, L.; Freitag, H.; Krüger, A.; Tölle, M.; Scheibenbogen, C. Observational Study of Repeat Immunoadsorption (RIA) in Post-COVID ME/CFS Patients with Elevated ß2-Adrenergic Receptor Autoantibodies—An Interim Report. J. Clin. Med. 2023, 12, 6428. [Google Scholar] [CrossRef]
- Fluge, Ø.; Risa, K.; Lunde, S.; Alme, K.; Rekeland, I.G.; Sapkota, D.; Kristoffersen, E.K.; Sørland, K.; Bruland, O.; Dahl, O.; et al. B-Lymphocyte Depletion in Myalgic Encephalopathy/Chronic Fatigue Syndrome. An Open-Label Phase II Study with Rituximab Maintenance Treatment. PLoS ONE 2015, 10, e0129898. [Google Scholar] [CrossRef] [PubMed]
- Fluge, Ø.; Bruland, O.; Risa, K.; Storstein, A.; Kristoffersen, E.K.; Sapkota, D.; Næss, H.; Dahl, O.; Nyland, H.; Mella, O. Benefit from B-Lymphocyte Depletion Using the Anti-CD20 Antibody Rituximab in Chronic Fatigue Syndrome. A Double-Blind and Placebo-Controlled Study. PLoS ONE 2011, 6, e26358. [Google Scholar] [CrossRef] [PubMed]
- Fluge, Ø.; Rekeland, I.G.; Lien, K.; Thürmer, H.; Borchgrevink, P.C.; Schäfer, C.; Sørland, K.; Aßmus, J.; Ktoridou-Valen, I.; Herder, I.; et al. B-Lymphocyte Depletion in Patients with Myalgic Encephalomyelitis/Chronic Fatigue Syndrome: A Randomized, Double-Blind, Placebo-Controlled Trial. Ann. Intern. Med. 2019, 170, 585–593. [Google Scholar] [CrossRef] [PubMed]
- Rekeland, I.G.; Fosså, A.; Lande, A.; Ktoridou-Valen, I.; Sørland, K.; Holsen, M.; Tronstad, K.J.; Risa, K.; Alme, K.; Viken, M.K.; et al. Intravenous Cyclophosphamide in Myalgic Encephalomyelitis/Chronic Fatigue Syndrome. An Open-Label Phase II Study. Front. Med. 2020, 7, 162. [Google Scholar] [CrossRef] [PubMed]
- Patti, F.; Lo Fermo, S. Lights and Shadows of Cyclophosphamide in the Treatment of Multiple Sclerosis. Autoimmune Dis. 2011, 2011, 14. [Google Scholar] [CrossRef]
- Willers, J.M.; Sluis, E. The influence of cyclophosphamide on antibody formation in the mouse. Ann. Immunol. 1975, 126, 267–279. [Google Scholar]
- Ahlmann, M.; Hempel, G. The effect of cyclophosphamide on the immune system: Implications for clinical cancer therapy. Cancer Chemother. Pharmacol. 2016, 78, 661–671. [Google Scholar] [CrossRef]
- Hurd, E.R.; Giuliano, V.J. The effect of cyclophosphamide on B and T lymphocytes in patients with connective tissue diseases. Arthritis Rheum. 1975, 18, 67–75. [Google Scholar] [CrossRef]
- Nacul, L.C.; Lacerda, E.M.; Pheby, D.; Campion, P.; Molokhia, M.; Fayyaz, S.; Leite, J.C.; Poland, F.; Howe, A.; Drachler, M.L. Prevalence of myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS) in three regions of England: A repeated cross-sectional study in primary care. BMC Med. 2011, 9, 91. [Google Scholar] [CrossRef] [PubMed]
- Chu, L.; Valencia, I.J.; Garvert, D.W.; Montoya, J.G. Onset patterns and course of myalgic encephalomyelitis/chronic fatigue syndrome. Front. Pediatr. 2019, 7, 12. [Google Scholar] [CrossRef] [PubMed]
- Longley, R.J.; White, M.T.; Takashima, E.; Brewster, J.; Morita, M.; Harbers, M.; Obadia, T.; Robinson, L.J.; Matsuura, F.; Liu, Z.S.J.; et al. Development and validation of serological markers for detecting recent Plasmodium vivax infection. Nat. Med. 2020, 26, 741–749. [Google Scholar] [CrossRef]
- Helb, D.A.; Tetteh, K.K.A.; Felgner, P.L.; Skinner, J.; Hubbard, A.; Arinaitwe, E.; Mayanja-Kizza, H.; Ssewanyana, I.; Kamya, M.R.; Beeson, J.G.; et al. Novel serologic biomarkers provide accurate estimates of recent Plasmodium falciparum exposure for individuals and communities. Proc. Natl. Acad. Sci. USA 2015, 112, E4438–E4447. [Google Scholar] [CrossRef]
- Brooks, S.P.; Dunnett, S.B. Tests to assess motor phenotype in mice: A user’s guide. Nat. Rev. Neurosci. 2009, 10, 519–529. [Google Scholar] [CrossRef] [PubMed]
- Goebel, A.; Krock, E.; Gentry, C.; Israel, M.R.; Jurczak, A.; Urbina, C.M.; Sandor, K.; Vastani, N.; Maurer, M.; Cuhadar, U.; et al. Passive transfer of fibromyalgia symptoms from patients to mice. J. Clin. Investig. 2021, 131, e144201. [Google Scholar] [CrossRef]
- Janowski, A.; Lesnak, J.; Plumb, A.; Rasmussen, L.; Sluka, K. Development of a Mouse Model for Chronic Fatigue Syndrome. J. Pain. 2022, 23, 12. [Google Scholar] [CrossRef]
- Tamura, Y.; Yamato, M.; Kataoka, Y. Animal Models for Neuroinflammation and Potential Treatment Methods. Front. Neurol. 2022, 13, 890217. [Google Scholar] [CrossRef]
- Bastiaan Holwerda, S.J.; de Laat, W. CTCF: The protein, the binding partners, the binding sites and their chromatin loops. Philos. Trans. R. Soc. B Biol. Sci. 2013, 368, 20120369. [Google Scholar] [CrossRef]
- Dehingia, B.; Milewska, M.; Janowski, M.; Pe ZKowska, A. CTCF shapes chromatin structure and gene expression in health and disease. EMBO Rep. 2022, 23, e55146. [Google Scholar] [CrossRef]
- DiSpirito, J.R.; Zemmour, D.; Ramanan, D.; Cho, J.; Zilionis, R.; Klein, A.M.; Benoist, C.; Mathis, D. Molecular diversification of regulatory T cells in nonlymphoid tissues. Sci. Immunol. 2018, 3, 5861. [Google Scholar] [CrossRef]
- Chandrasekaran, V.; Oparina, N.; Garcia-Bonete, M.J.; Wasén, C.; Erlandsson, M.C.; Malmhäll-Bah, E.; Andersson, K.M.E.; Jensen, M.; Silfverswärd, S.T.; Katona, G.; et al. Cohesin-Mediated Chromatin Interactions and Autoimmunity. Front. Immunol. 2022, 13, 840002. [Google Scholar] [CrossRef]
- Kerr, J.R. Gene profiling of patients with chronic fatigue syndrome/myalgic encephalomyelitis. Curr. Rheumatol. Rep. 2008, 10, 482–491. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, N.; Fear, D.; Richards, S.C.M.; McDermott, C.R.; Nuwaysir, E.F.; Kellam, P.; Harrison, T.J.; Wilkinson, R.J.; Tyrrell, D.A.; Holgate, S.T.; et al. Gene expression in peripheral blood mononuclear cells from patients with chronic fatigue syndrome. J. Clin. Pathol. 2005, 58, 826–832. [Google Scholar] [CrossRef]
- Sweetman, E.; Ryan, M.; Edgar, C.; Mackay, A.; Vallings, R.; Tate, W. Changes in the transcriptome of circulating immune cells of a New Zealand cohort with myalgic encephalomyelitis/chronic fatigue syndrome. Int. J. Immunopathol. Pharmacol. 2019, 33, 2058738418820402. [Google Scholar] [CrossRef] [PubMed]
- Almenar-Pérez, E.; Ovejero, T.; Sánchez-Fito, T.; Espejo, J.A.; Nathanson, L.; Oltra, E. Epigenetic Components of Myalgic Encephalomyelitis/Chronic Fatigue Syndrome Uncover Potential Transposable Element Activation. Clin. Ther. 2019, 41, 675–698. [Google Scholar] [CrossRef] [PubMed]
- De Vega, W.C.; Herrera, S.; Vernon, S.D.; McGowan, P.O. Epigenetic modifications and glucocorticoid sensitivity in Myalgic Encephalomyelitis/Chronic Fatigue Syndrome (ME/CFS). BMC Med. Genom. 2017, 10, 11. [Google Scholar] [CrossRef] [PubMed]
- Lorusso, L.; Mikhaylova, S.V.; Capelli, E.; Ferrari, D.; Ngonga, G.K.; Ricevuti, G. Immunological aspects of chronic fatigue syndrome. Autoimmun. Rev. 2009, 8, 287–291. [Google Scholar] [CrossRef] [PubMed]
- Majdalawieh, A.F.; Massri, M.; Ro, H.S. AEBP1 is a Novel Oncogene: Mechanisms of Action and Signaling Pathways. J. Oncol. 2020, 2020, 8097872. [Google Scholar] [CrossRef]
- Angwin, C.; Ghali, N.; van Dijk, F.S. Case report: Two individuals with AEBP1-related classical-like EDS: Further clinical characterisation and description of novel AEBP1 variants. Front. Genet. 2023, 14, 1148224. [Google Scholar] [CrossRef]
- Ritelli, M.; Cinquina, V.; Venturini, M.; Pezzaioli, L.; Formenti, A.M.; Chiarelli, N.; Colombi, M. Expanding the Clinical and Mutational Spectrum of Recessive AEBP1-Related Classical-Like Ehlers-Danlos Syndrome. Genes 2019, 10, 135. [Google Scholar] [CrossRef]
- Castori, M.; Celletti, C.; Camerota, F.; Grammatico, P. Chronic fatigue syndrome is commonly diagnosed in patients with Ehlers-Danlos syndrome hypermobility type/joint hypermobility syndrome. Clin. Exp. Rheumatol. 2011, 29, 597–598. [Google Scholar] [PubMed]
- Hakim, A.; De Wandele, I.; O’Callaghan, C.; Pocinki, A.; Rowe, P. Chronic fatigue in Ehlers-Danlos syndrome-Hypermobile type. Am. J. Med. Genet. C Semin. Med. Genet. 2017, 175, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Nacul, L.; Authier, F.J.; Scheibenbogen, C.; Lorusso, L.; Helland, I.B.; Martin, J.A.; Sirbu, C.A.; Mengshoel, A.M.; Polo, O.; Behrends, U.; et al. European Network on Myalgic Encephalomyelitis/Chronic Fatigue Syndrome (EUROMENE): Expert Consensus on the Diagnosis, Service Provision, and Care of People with ME/CFS in Europe. Medicina 2021, 57, 510. [Google Scholar] [CrossRef]
- Schlauch, K.A.; Khaiboullina, S.F.; De Meirleir, K.L.; Rawat, S.; Petereit, J.; Rizvanov, A.A.; Blatt, N.; Mijatovic, T.; Kulick, D.; Palotás, A.; et al. Genome-wide association analysis identifies genetic variations in subjects with myalgic encephalomyelitis/chronic fatigue syndrome. Transl. Psychiatry 2016, 6, e730. [Google Scholar] [CrossRef]
- Dibble, J.J.; McGrath, S.J.; Ponting, C.P. Genetic risk factors of ME/CFS: A critical review. Hum. Mol. Genet. 2020, 29, R118–R125. [Google Scholar] [CrossRef]
- Herrera, S.; de Vega, W.C.; Ashbrook, D.; Vernon, S.D.; McGowan, P.O. Genome-epigenome interactions associated with Myalgic Encephalomyelitis/Chronic Fatigue Syndrome. Epigenetics 2018, 13, 1174–1190. [Google Scholar] [CrossRef] [PubMed]
- Hajdarevic, R.; Lande, A.; Mehlsen, J.; Rydland, A.; Sosa, D.D.; Strand, E.B.; Mella, O.; Pociot, F.; Fluge, Ø.; Lie, B.A.; et al. Genetic association study in myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS) identifies several potential risk loci. Brain Behav. Immun. 2022, 102, 362–369. [Google Scholar] [CrossRef]
- Das, S.; Taylor, K.; Kozubek, J.; Sardell, J.; Gardner, S. Genetic risk factors for ME/CFS identified using combinatorial analysis. J. Transl. Med. 2022, 20, 598. [Google Scholar] [CrossRef]
- Haffke, M.; Freitag, H.; Rudolf, G.; Seifert, M.; Doehner, W.; Scherbakov, N.; Hanitsch, L.; Wittke, K.; Bauer, S.; Konietschke, F.; et al. Endothelial dysfunction and altered endothelial biomarkers in patients with post-COVID-19 syndrome and chronic fatigue syndrome (ME/CFS). J. Transl. Med. 2022, 20, 138. [Google Scholar] [CrossRef]
- Scherbakov, N.; Szklarski, M.; Hartwig, J.; Sotzny, F.; Lorenz, S.; Meyer, A.; Grabowski, P.; Doehner, W.; Scheibenbogen, C. Peripheral endothelial dysfunction in myalgic encephalomyelitis/chronic fatigue syndrome. ESC Hear. Fail. 2020, 7, 1064–1071. [Google Scholar] [CrossRef]
- Blauensteiner, J.; Bertinat, R.; León, L.E.; Riederer, M.; Sepúlveda, N.; Westermeier, F. Altered endothelial dysfunction-related miRs in plasma from ME/CFS patients. Sci. Rep. 2021, 11, 10604. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Jelcic, I.; Mühlenbruch, L.; Haunerdinger, V.; Toussaint, N.C.; Zhao, Y.; Cruciani, C.; Faigle, W.; Naghavian, R.; Foege, M.; et al. HLA-DR15 Molecules Jointly Shape an Autoreactive T Cell Repertoire in Multiple Sclerosis. Cell 2020, 183, 1264–1281.e20. [Google Scholar] [CrossRef] [PubMed]
- Pradeu, T.; Cooper, E.L. The danger theory: 20 years later. Front. Immunol. 2012, 3, 287. [Google Scholar] [CrossRef] [PubMed]
- Gallucci, S.; Matzinger, P. Danger signals: SOS to the immune system. Curr. Opin. Immunol. 2001, 13, 114–119. [Google Scholar] [CrossRef] [PubMed]
- Cohen, I.R.; Young, D.B. Autoimmunity, microbial immunity and the immunological homunculus. Immunol. Today 1991, 12, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Quintana, F.J.; Mimran, A.; Carmi, P.; Mor, F.; Cohen, I.R. HSP60 as a target of anti-ergotypic regulatory T cells. PLoS ONE 2008, 3, e4026. [Google Scholar] [CrossRef] [PubMed]
- Jammes, Y.; Steinberg, J.G.; Delliaux, S. Chronic fatigue syndrome: Acute infection and history of physical activity affect resting levels and response to exercise of plasma oxidant/antioxidant status and heat shock proteins. J. Intern. Med. 2012, 272, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Thambirajah, A.A.; Sleigh, K.; Stiver, H.G.; Chow, A.W. Differential heat shock protein responses to strenuous standardized exercise in chronic fatigue syndrome patients and matched healthy controls. Clin. Investig. Med. 2008, 31, 319–327. [Google Scholar] [CrossRef]
- Clayton, E.W. Beyond Myalgic Encephalomyelitis/Chronic Fatigue Syndrome: An IOM Report on Redefining an Illness. JAMA 2015, 313, 1101–1102. [Google Scholar] [CrossRef]
- Elfaitouri, A.; Herrmann, B.; Bölin-Wiener, A.; Wang, Y.; Gottfries, C.G.; Zachrisson, O.; Pipkorn, R.; Rönnblom, L.; Blomberg, J. Epitopes of microbial and human heat shock protein 60 and their recognition in myalgic encephalomyelitis. PLoS ONE 2013, 8, e81155. [Google Scholar] [CrossRef]
- Delneste, Y.; Herbault, N.; Galea, B.; Magistrelli, G.; Bazin, I.; Bonnefoy, J.-Y.; Jeannin, P. Vasoactive Intestinal Peptide Synergizes with TNF-α in Inducing Human Dendritic Cell Maturation. J. Immunol. 1999, 163, 3071–3075. [Google Scholar] [CrossRef] [PubMed]
- Gomariz, R.P.; Juarranz, Y.; Abad, C.; Arranz, A.; Leceta, J.; Martinez, C. VIP-PACAP system in immunity: New insights for multitarget therapy. Ann. N. Y. Acad. Sci. 2006, 1070, 51–74. [Google Scholar] [CrossRef]
- Staines, D.R. Is chronic fatigue syndrome an autoimmune disorder of endogenous neuropeptides, exogenous infection and molecular mimicry? Med. Hypotheses 2004, 62, 646–652. [Google Scholar] [CrossRef] [PubMed]
- Brenu, E.W.; van Driel, M.L.; Staines, D.R.; Ashton, K.J.; Ramos, S.B.; Keane, J.; Klimas, N.G.; Marshall-Gradisnik, S.M. Immunological abnormalities as potential biomarkers in Chronic Fatigue Syndrome/Myalgic Encephalomyelitis. J. Transl. Med. 2011, 9, 81. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Rey, E.; Delgado, M. Vasoactive intestinal peptide and regulatory T-cell induction: A new mechanism and therapeutic potential for immune homeostasis. Trends Mol. Med. 2007, 13, 241–251. [Google Scholar] [CrossRef]
- Blundell, S.; Ray, K.K.; Buckland, M.; White, P.D. Chronic fatigue syndrome and circulating cytokines: A systematic review. Brain Behav. Immun. 2015, 50, 186–195. [Google Scholar] [CrossRef]
- Carlo-Stella, N.; Badulli, C.; De Silvestri, A.; Bazzichi, L.; Martinetti, M.; Lorusso, L.; Bombardieri, S.; Salvaneschi, L.; Cuccia, M. A first study of cytokine genomic polymorphisms in CFS: Positive association of TNF-857 and IFNγ874 rare alleles. Clin. Exp. Rheumatol. 2006, 24, 179–182. [Google Scholar]
- Steiner, S.; Becker, S.C.; Hartwig, J.; Sotzny, F.; Lorenz, S.; Bauer, S.; Löbel, M.; Stittrich, A.B.; Grabowski, P.; Scheibenbogen, C. Autoimmunity-Related Risk Variants in PTPN22 and CTLA4 Are Associated with ME/CFS with Infectious Onset. Front. Immunol. 2020, 11, 578. [Google Scholar] [CrossRef]
- Sarode, A.Y.; Jha, M.K.; Zutshi, S.; Ghosh, S.K.; Mahor, H.; Sarma, U.; Saha, B. Residue-Specific Message Encoding in CD40-Ligand. IScience 2020, 23, 101441. [Google Scholar] [CrossRef]
- Burger, D.; Molnarfi, N.; Gruaz, L.; Dayer, J.-M. Differential induction of IL-1beta and TNF by CD40 ligand or cellular contact with stimulated T cells depends on the maturation stage of human monocytes. J. Immunol. 2004, 173, 1292–1297. [Google Scholar] [CrossRef]
- Andrade, R.M.; Wessendarp, M.; Subauste, C.S. CD154 activates macrophage antimicrobial activity in the absence of IFN-gamma through a TNF-alpha-dependent mechanism. J. Immunol. 2003, 171, 6750–6756. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.F.; Cook, W.J.; Lin, L.L.; Noelle, R.J. CD40 signaling through a newly identified tumor necrosis factor receptor-associated factor 2 (TRAF2) binding site. J. Biol. Chem. 2003, 278, 45414–45418. [Google Scholar] [CrossRef] [PubMed]
- Danese, S.; Sans, M.; Scaldaferri, F.; Sgambato, A.; Rutella, S.; Cittadini, A.; Piqué, J.M.; Panes, J.; Katz, J.A.; Gasbarrini, A.; et al. TNF-α Blockade Down-Regulates the CD40/CD40L Pathway in the Mucosal Microcirculation: A Novel Anti-Inflammatory Mechanism of Infliximab in Crohn’s Disease. J. Immunol. 2006, 176, 2617–2624. [Google Scholar] [CrossRef] [PubMed]
- White, A.T.; Light, A.R.; Hughen, R.W.; Bateman, L.; Martins, T.B.; Hill, H.R.; Light, K.C. Severity of symptom flare after moderate exercise is linked to cytokine activity in chronic fatigue syndrome. Psychophysiology 2010, 47, 615–624. [Google Scholar] [CrossRef] [PubMed]
- Hornig, M.; Montoya, J.G.; Klimas, N.G.; Levine, S.; Felsenstein, D.; Bateman, L.; Peterson, D.L.; Gottschalk, C.G.; Schultz, A.F.; Che, X.; et al. Distinct plasma immune signatures in ME/CFS are present early in the course of illness. Sci. Adv. 2015, 1, e1400121. [Google Scholar] [CrossRef] [PubMed]
- Luft, T.; Jefford, M.; Luetjens, P.; Hochrein, H.; Masterman, K.-A.; Maliszewski, C.; Shortman, K.; Cebon, J.; Maraskovsky, E. IL-1 beta enhances CD40 ligand-mediated cytokine secretion by human dendritic cells (DC): A mechanism for T cell-independent DC activation. J. Immunol. 2002, 168, 713–722. [Google Scholar] [CrossRef]
- Wesa, A.; Galy, A. Increased production of pro-inflammatory cytokines and enhanced T cell responses after activation of human dendritic cells with IL-1 and CD40 ligand. BMC Immunol. 2002, 3, 14. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fonseca, A.; Szysz, M.; Ly, H.T.; Cordeiro, C.; Sepúlveda, N. IgG Antibody Responses to Epstein-Barr Virus in Myalgic Encephalomyelitis/Chronic Fatigue Syndrome: Their Effective Potential for Disease Diagnosis and Pathological Antigenic Mimicry. Medicina 2024, 60, 161. https://doi.org/10.3390/medicina60010161
Fonseca A, Szysz M, Ly HT, Cordeiro C, Sepúlveda N. IgG Antibody Responses to Epstein-Barr Virus in Myalgic Encephalomyelitis/Chronic Fatigue Syndrome: Their Effective Potential for Disease Diagnosis and Pathological Antigenic Mimicry. Medicina. 2024; 60(1):161. https://doi.org/10.3390/medicina60010161
Chicago/Turabian StyleFonseca, André, Mateusz Szysz, Hoang Thien Ly, Clara Cordeiro, and Nuno Sepúlveda. 2024. "IgG Antibody Responses to Epstein-Barr Virus in Myalgic Encephalomyelitis/Chronic Fatigue Syndrome: Their Effective Potential for Disease Diagnosis and Pathological Antigenic Mimicry" Medicina 60, no. 1: 161. https://doi.org/10.3390/medicina60010161
APA StyleFonseca, A., Szysz, M., Ly, H. T., Cordeiro, C., & Sepúlveda, N. (2024). IgG Antibody Responses to Epstein-Barr Virus in Myalgic Encephalomyelitis/Chronic Fatigue Syndrome: Their Effective Potential for Disease Diagnosis and Pathological Antigenic Mimicry. Medicina, 60(1), 161. https://doi.org/10.3390/medicina60010161





