Accurate Prediction of Sudden Cardiac Death Based on Heart Rate Variability Analysis Using Convolutional Neural Network
Abstract
1. Introduction
- The development of a 1D-CNN model to predict the risk of SCD based on HRV Features.
- The expansion of the number of comparison subjects in predicting SCD, that is, CAD, VT, CHF, and NSR.
2. Materials and Methods
2.1. Database
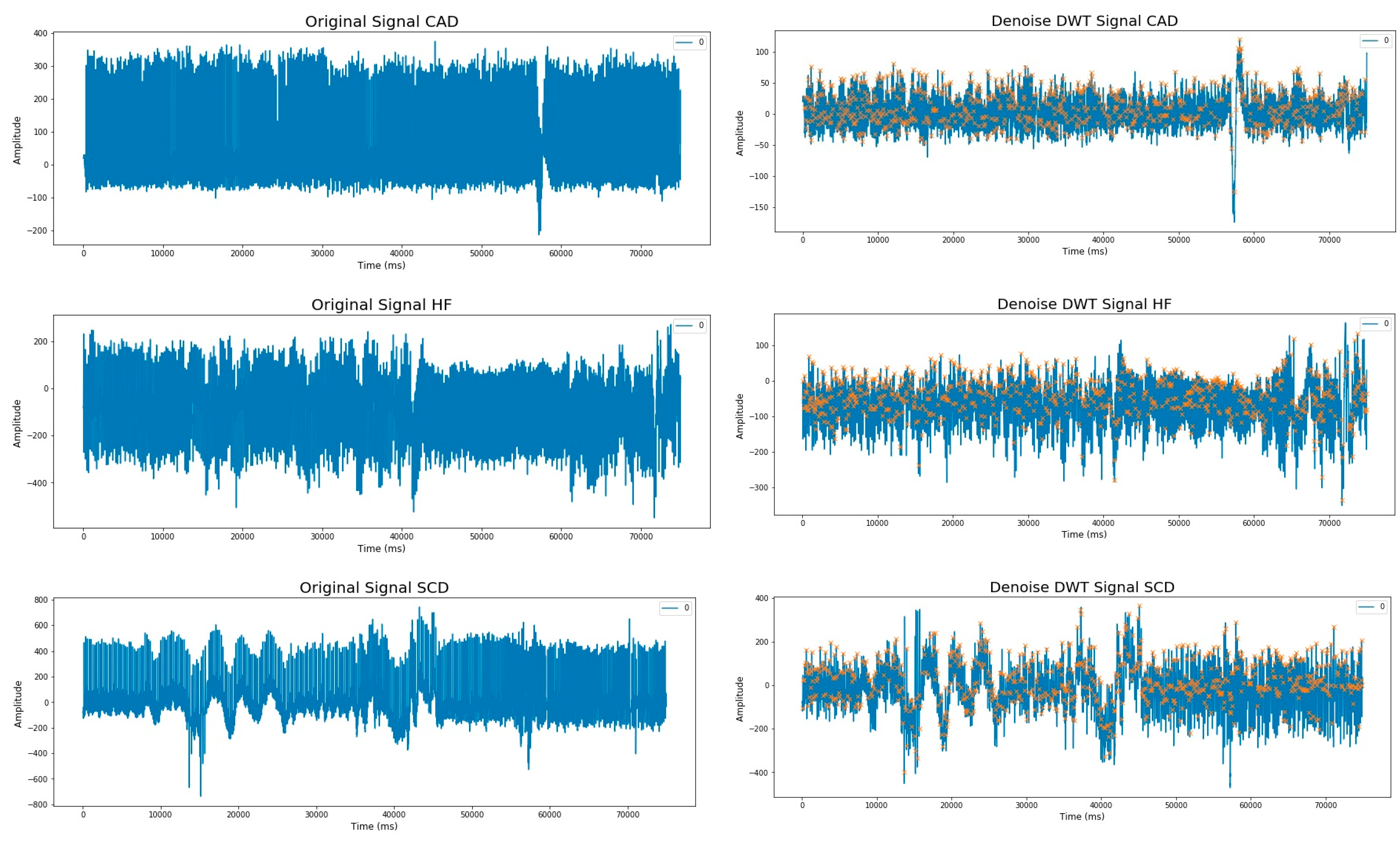
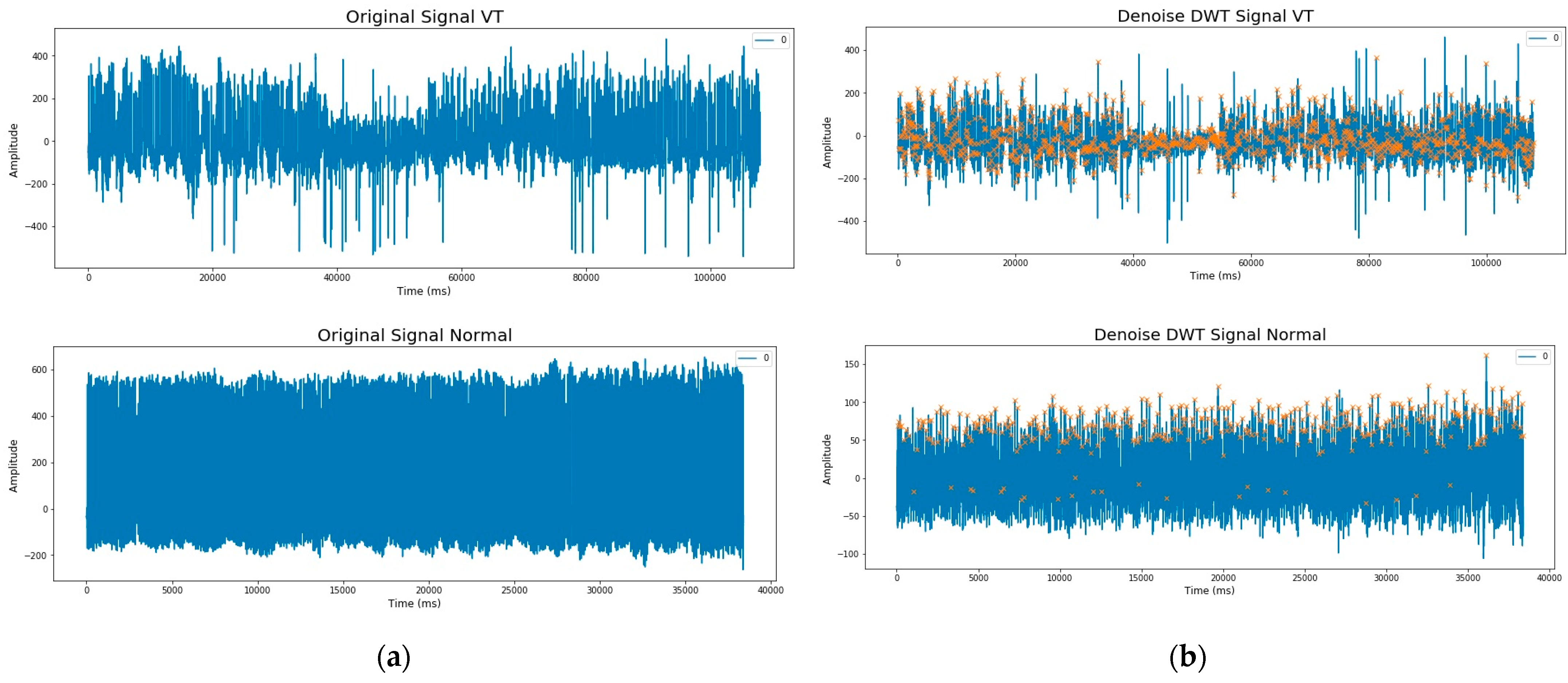
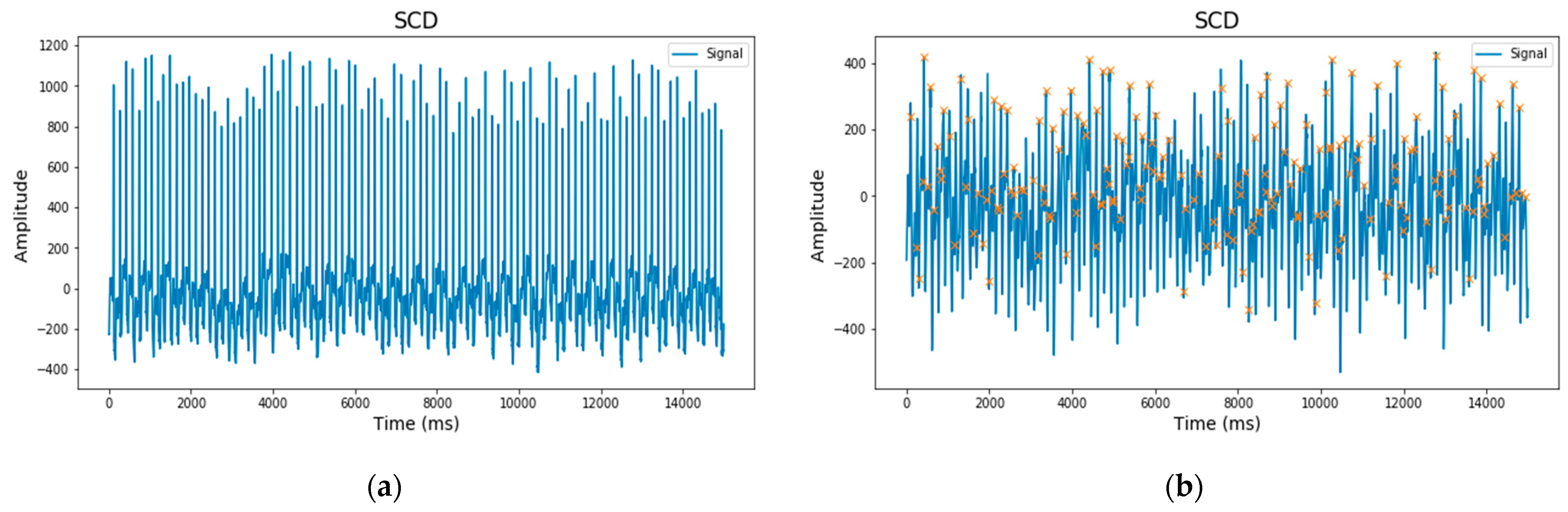
2.2. Preprocessing
2.3. Feature Extraction
2.4. Classification
- Input layer: The input to the network is a sequence of the length 9.
- Reshape layer: reshapes the input sequence to have a shape of (9, 1), converting it into a 1D Signal.
- Conv1D layers: There are several hidden layers in Conv1D layers, each with different dilation rates. The dilation rate determines the spacing between the values in the kernel. Higher dilation rates allow the network to capture larger patterns while retaining fewer parameters. The Conv1D layers have 64 filters each and a kernel size of 2. They use the “causal” padding, which ensures that the output at each time step only depends on the current and past inputs. The dilation rates for these layers are based on the values in the dilatation_rates list, which are powers of 2 ranging from 1 to 64. The activation function used in these Conv1D layers is ReLU.
- Multiplication and activation layers: Following each Conv1D layer, two activation functions are applied independently: tanh and sigmoid. The outputs of these activation functions are then element-wise multiplied together.
- TimeDistributed Dense layer: Each output from the multiplication and activation layers is passed through a TimeDistributed Dense layer with 64 units and ReLU activation. The TimeDistributed layer applies the same Dense layer to each time step of the sequence.
- Skip Connections: The outputs from the TimeDistributed Dense layers are appended to a list called skips. These skip connections allow information from earlier layers to be propagated and combined with later layers.
- Add and Activation layers: The elements in the skips list are summed using an Add layer. Then, the ReLU activation function is applied to the summed output. After the Conv1D layers, the code continues with other layers, such as Flatten, Dropout, Dense, and the final output layer. This model uses the 1D-CNN Wavenet architecture with a total of 11 layers (Table 2).
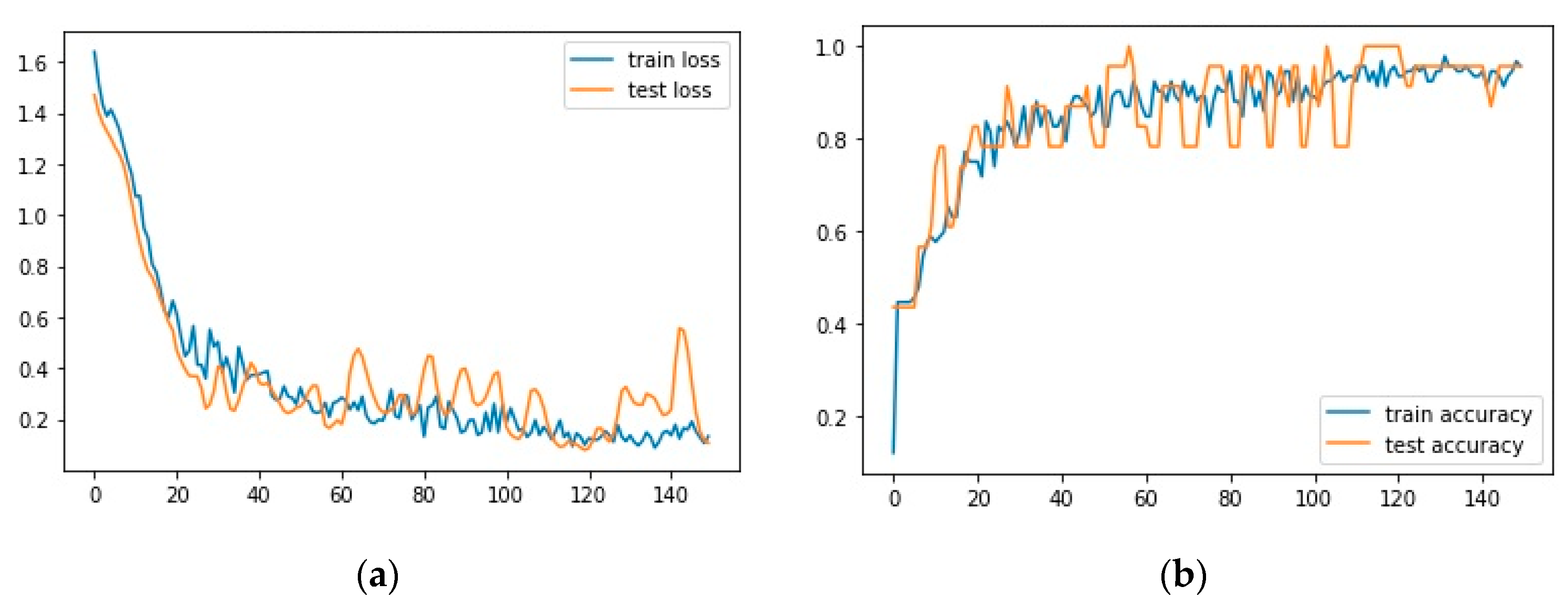
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Acharya, U.R.; Fujita, H.; Sudarshan, V.K.; Ghista, D.N.; Lim, W.J.E.; Koh, J.E. Automated Prediction of Sudden Cardiac Death Risk Using Kolmogorov Complexity and Recurrence Quantification Analysis Features Extracted from HRV Signals. In Proceedings of the 2015 IEEE International Conference on Systems, Man, and Cybernetics, Hong Kong, China, 9–12 October 2015; pp. 1110–1115. [Google Scholar] [CrossRef]
- Ebrahimzadeh, E.; Foroutan, A.; Shams, M.; Baradaran, R.; Rajabion, L.; Joulani, M.; Fayaz, F. An optimal strategy for prediction of sudden cardiac death through a pioneering feature-selection approach from HRV signal. Comput. Methods Programs Biomed. 2019, 169, 19–36. [Google Scholar] [CrossRef]
- Devi, R.; Kumar, H.; Kumar, D. ScienceDirect A novel multi-class approach for early-stage prediction of sudden cardiac death. Integr. Med. Res. 2019, 39, 586–598. [Google Scholar] [CrossRef]
- Wong, C.X.; Brown, A.; Lau, D.H.; Chugh, S.S.; Albert, C.M.; Kalman, J.M.; Sanders, P. Epidemiology of Sudden Cardiac Death: Global and Regional Perspectives. Heart Lung Circ. 2019, 28, 6–14. [Google Scholar] [CrossRef] [PubMed]
- Amezquita-Sanchez, J.P.; Valtierra-Rodriguez, M.; Adeli, H.; Perez-Ramirez, C.A. A Novel Wavelet Transform-Homogeneity Model for Sudden Cardiac Death Prediction Using ECG Signals. J. Med. Syst. 2018, 42, 176. [Google Scholar] [CrossRef] [PubMed]
- Kaplan Berkaya, S.; Uysal, A.K.; Sora Gunal, E.; Ergin, S.; Gunal, S.; Gulmezoglu, M.B. A survey on ECG analysis. Biomed. Signal Process. Control 2018, 43, 216–235. [Google Scholar] [CrossRef]
- Siontis, K.C.; Noseworthy, P.A.; Attia, Z.I.; Friedman, P.A. Artificial intelligence-enhanced electrocardiography in cardiovascular disease management. Nat. Rev. Cardiol. 2021, 18, 465–478. [Google Scholar] [CrossRef]
- Rai, H.M.; Trivedi, A.; Shukla, S. ECG signal processing for abnormalities detection using multi-resolution wavelet transform and Artificial Neural Network classifier. Meas. J. Int. Meas. Confed. 2013, 46, 3238–3246. [Google Scholar] [CrossRef]
- Sessa, F.; Anna, V.; Messina, G.; Cibelli, G.; Monda, V.; Marsala, G.; Ruberto, M.; Biondi, A.; Cascio, O.; Bertozzi, G.; et al. Heart rate variability as predictive factor for sudden cardiac death. Aging 2018, 10, 166–177. [Google Scholar] [CrossRef]
- Kiuchi, M.G.; Nolde, J.M.; Villacorta, H.; Carnagarin, R.; Chan, J.J.S.-Y.; Lugo-Gavidia, L.M.; Ho, J.K.; Matthews, V.B.; Dwivedi, G.; Schlaich, M.P. New approaches in the management of sudden cardiac death in patients with heart failure—Targeting the sympathetic nervous system. Int. J. Mol. Sci. 2019, 20, 2430. [Google Scholar] [CrossRef]
- Task Force of the European Society of Cardiology the North American Society of Pacing Electrophysiology. Heart rate variability: Standards of measurement, physiological interpretation, and clinical use. Circulation 1996, 93, 1043–1065. [Google Scholar] [CrossRef]
- Sassi, R.; Cerutti, S.; Lombardi, F.; Malik, M.; Huikuri, H.V.; Peng, C.-K.; Schmidt, G.; Yamamoto, Y.; Reviewers:, D.; Gorenek, B. Advances in heart rate variability signal analysis: Joint position statement by the e-Cardiology ESC Working Group and the European Heart Rhythm Association co-endorsed by the Asia Pacific Heart Rhythm Society. Ep Eur. 2015, 17, 1341–1353. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimzadeh, E.; Manuchehri, M.S.; Amoozegar, S.; Araabi, B.N.; Soltanian-zadeh, H. A time local subset feature selection for prediction of sudden cardiac death from ECG signal A time local subset feature selection for prediction of sudden cardiac death from ECG signal. Med. Biol. Eng. Comput. 2018, 56, 1253–1270. [Google Scholar] [CrossRef] [PubMed]
- Parsi, A.; Byrne, D.; Glavin, M.; Jones, E. Heart rate variability feature selection method for automated prediction of sudden cardiac death. Biomed. Signal Process. Control 2021, 65, 102310. [Google Scholar] [CrossRef]
- Rohila, A.; Sharma, A. Detection of sudden cardiac death by a comparative study of heart rate variability in normal and abnormal heart conditions. Biocybern. Biomed. Eng. 2020, 40, 1140–1154. [Google Scholar] [CrossRef]
- Mjahad, A.; Frances-Villora, J.V.; Bataller-Mompean, M.; Rosado-Muñoz, A. Ventricular Fibrillation and Tachycardia Detection Using Features Derived from Topological Data Analysis. Appl. Sci. 2022, 12, 7248. [Google Scholar] [CrossRef]
- Kaspal, R.; Alsadoon, A.; Prasad, P.W.C.; Al-Saiyd, N.A.; Nguyen, T.Q.V.; Pham, D.T.H. A novel approach for early prediction of sudden cardiac death (SCD) using hybrid deep learning. Multimed. Tools Appl. 2021, 80, 8063–8090. [Google Scholar] [CrossRef]
- Nurmaini, S.; Tondas, A.E.; Darmawahyuni, A.; Rachmatullah, M.N.; Partan, R.U.; Firdaus, F.; Tutuko, B.; Pratiwi, F.; Juliano, A.H.; Khoirani, R. Robust detection of atrial fibrillation from short-term electrocardiogram using convolutional neural networks. Futur. Gener. Comput. Syst. 2020, 113, 304–317. [Google Scholar] [CrossRef]
- Wang, J. A deep learning approach for atrial fibrillation signals classification based on convolutional and modified Elman neural network. Futur. Gener. Comput. Syst. 2020, 102, 670–679. [Google Scholar] [CrossRef]
- Acharya, U.R.; Fujita, H.; Oh, S.L.; Hagiwara, Y.; Tan, J.H.; Adam, M. Application of deep convolutional neural network for automated detection of myocardial infarction using ECG signals. Inf. Sci. 2017, 415, 190–198. [Google Scholar] [CrossRef]
- Faust, O.; Hagiwara, Y.; Hong, T.J.; Lih, O.S.; Acharya, U.R. Deep learning for healthcare applications based on physiological signals: A review. Comput. Methods Programs Biomed. 2018, 161, 1–13. [Google Scholar] [CrossRef]
- PhysioNet. Available online: https://physionet.org/ (accessed on 15 June 2023).
- Hammad, M.; Maher, A.; Wang, K.; Jiang, F.; Amrani, M. Detection of abnormal heart conditions based on characteristics of ECG signals. Meas. J. Int. Meas. Confed. 2018, 125, 634–644. [Google Scholar] [CrossRef]
- MIT-BIH Arrhythmia Database v1.0.0. Available online: https://physionet.org/content/mitdb/1.0.0/ (accessed on 27 June 2022).
- Darmawahyuni, A.; Nurmaini, S.; Yuwandini, M.; Rachmatullah, M.N.; Firdaus, F.; Tutuko, B. Congestive heart failure waveform classification based on short time-step analysis with recurrent network. Inform. Med. Unlocked 2020, 21, 100441. [Google Scholar] [CrossRef]
- Mahdavi, S.; Panamtash, H.; Norouzi, Y.; Norouzi, S. Atrial fibrillation detection method based on converting ECG to signal using both symptoms of AF. Comput. Res. Prog. Appl. Sci. Eng. 2020, 6, 90–94. [Google Scholar]
- Mayapur, P.; Student, B.E. A Review on Detection and Performance Analysis on R-R Interval Methods for ECG. J. Innov. Res. Sci. 2018, 7, 11019–11025. [Google Scholar] [CrossRef]
- Elola, A.; Aramendi, E.; Rueda, E.; Irusta, U.; Wang, H.; Idris, A. Towards the prediction of rearrest during out-of-hospital cardiac arrest. Entropy 2020, 22, 758. [Google Scholar] [CrossRef] [PubMed]
- Isasi, I.; Alonso, E.; Irusta, U.; Aramendi, E.; Zabihi, M.; Rad, A.B.; Eftestøl, T.; Kramer-Johansen, J.; Wik, L. A Machine Learning-Based Pulse Detection Algorithm for Use During Cardiopulmonary Resuscitation. In Proceedings of the 2021 Computing in Cardiology (CinC), Brno, Czech Republic, 13–15 September 2021; Volume 48, pp. 1–4. [Google Scholar]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; MIT Press: Cambridge, MA, USA, 2016; ISBN 0262337371. [Google Scholar]
- Banerjee, I.; Ling, Y.; Chen, M.C.; Hasan, S.A.; Langlotz, C.P.; Moradzadeh, N.; Chapman, B.; Amrhein, T.; Mong, D.; Rubin, D.L. Comparative effectiveness of convolutional neural network (CNN) and recurrent neural network (RNN) architectures for radiology text report classification. Artif. Intell. Med. 2019, 97, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Kilicarslan, S.; Celik, M.; Sahin, Ş. Hybrid models based on genetic algorithm and deep learning algorithms for nutritional Anemia disease classification. Biomed. Signal Process. Control 2021, 63, 102231. [Google Scholar] [CrossRef]
- Meng, Y.; Lin, L.; Qin, Z.; Qu, Y.; Qin, Y.; Li, Y. Biosignal Classification Based on Multi-Feature Multi-Dimensional WaveNet-LSTM Models. J. Commun. 2022, 17, 399–404. [Google Scholar] [CrossRef]
- Van den Oord, A.; Dieleman, S.; Zen, H.; Simonyan, K.; Vinyals, O.; Graves, A.; Kalchbrenner, N.; Senior, A.; Kavukcuoglu, K. Wavenet: A generative model for raw audio. arXiv 2016, arXiv:1609.03499. [Google Scholar]
- Kalchbrenner, N.; Espeholt, L.; Vinyals, O.; Graves, A. Conditional image generation with PixelCNN decoders. arXiv 2016, arXiv:1606.05328. [Google Scholar]
- Abdul-Kadir, N.A.; Safri, N.M.; Othman, M.A. Effect of ECG episodes on parameters extraction for paroxysmal atrial fibrillation classification. In Proceedings of the 2014 IEEE Conference on Biomedical Engineering and Sciences (IECBES), Kuala Lumpur, Malaysia, 8–10 December 2014; pp. 874–877. [Google Scholar] [CrossRef]
- Powers, D.M.W. Evaluation: From precision, recall and F-measure to ROC, informedness, markedness and correlation. arXiv 2020, arXiv:2010.16061. [Google Scholar]

| Class | Patient | Time-Domain Features | |||||||
|---|---|---|---|---|---|---|---|---|---|
| MRR | SDRR | RMSSD | PNN50 | CVRR | NN50 | Min_RR | Max_RR | ||
| CAD | 20031 | 0.977 | 0.722 | 0.897 | 0.009 | 0.739 | 0.951 | 0.007 | 4.687 |
| HF | Chf01 | 0.787 | 0.425 | 0.581 | 0.009 | 0.540 | 0.928 | 0.007 | 2.617 |
| NSR | nsr001 | 0.769 | 0.315 | 0.442 | 0.005 | 0.409 | 0.586 | 0.412 | 2.078 |
| SCD | 30 | 0.736 | 0.482 | 0.632 | 0.009 | 0.655 | 0.919 | 0.007 | 3.578 |
| VT | 106 | 0.906 | 0.684 | 1.035 | 0.009 | 0.754 | 0.955 | 0.007 | 3.960 |
| No. | Layer | Number of Layers |
|---|---|---|
| 1 | Input | 1 |
| 2 | Reshape | 1 |
| 3 | TimeDistribute Dense (Loop) | hidden layers |
| 4 | Conv1D (Loop) | hidden layers |
| 5 | Multiply | 1 |
| 6 | TimeDistribute Dense | 1 |
| 7 | Add (Loop) | hidden layers |
| 8 | Flatten | 1 |
| 9 | Dropout | 2 |
| 10 | Dense | 2 |
| 11 | Output | 1 |
| Hyperparameters | List of Values |
|---|---|
| Number of hidden layers | 3, 5, 7, 9 |
| Learning rate | 0.1, 0.01, 0.001, 0.0001 |
| Batch Size | 32, 64, 128, 256 |
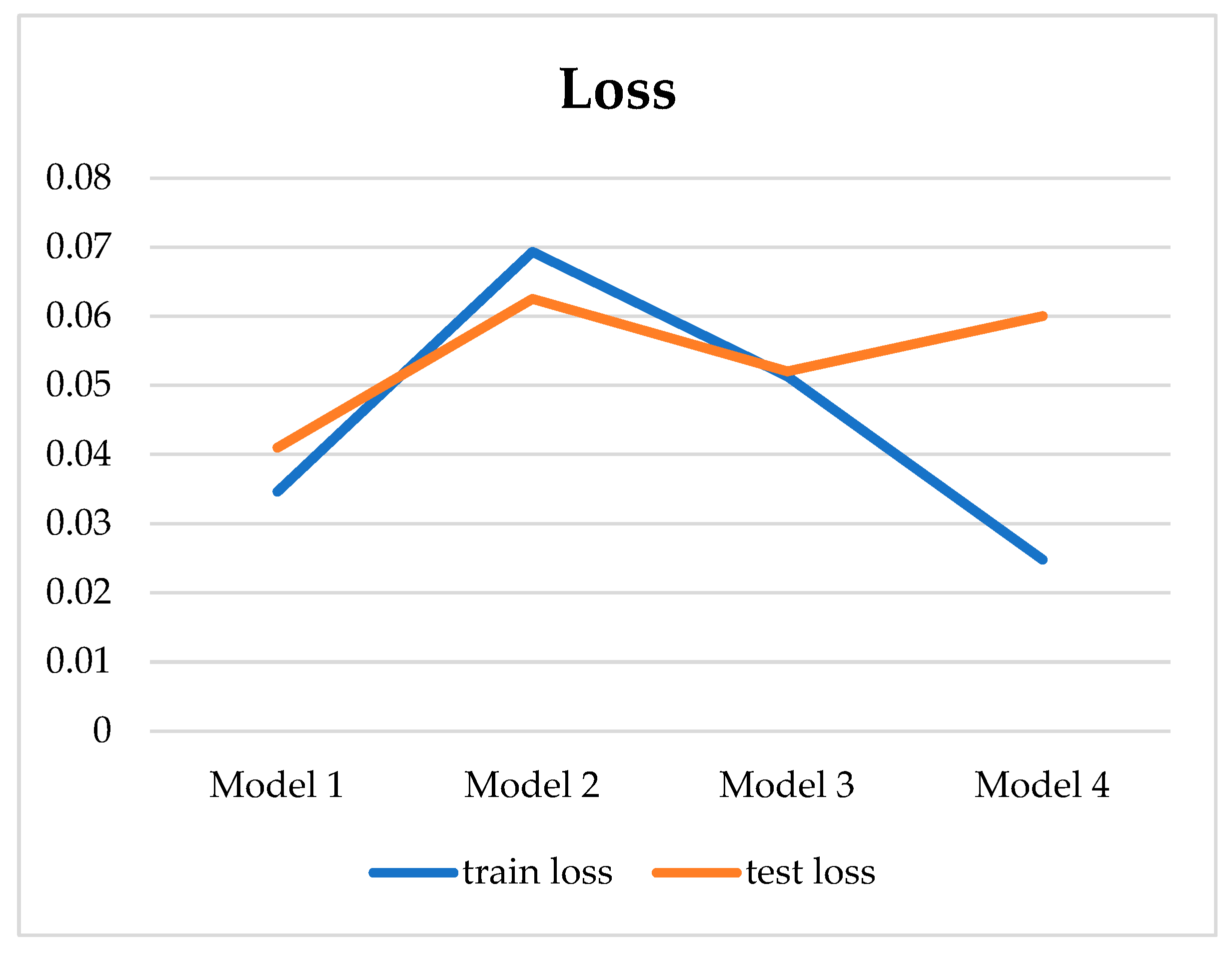
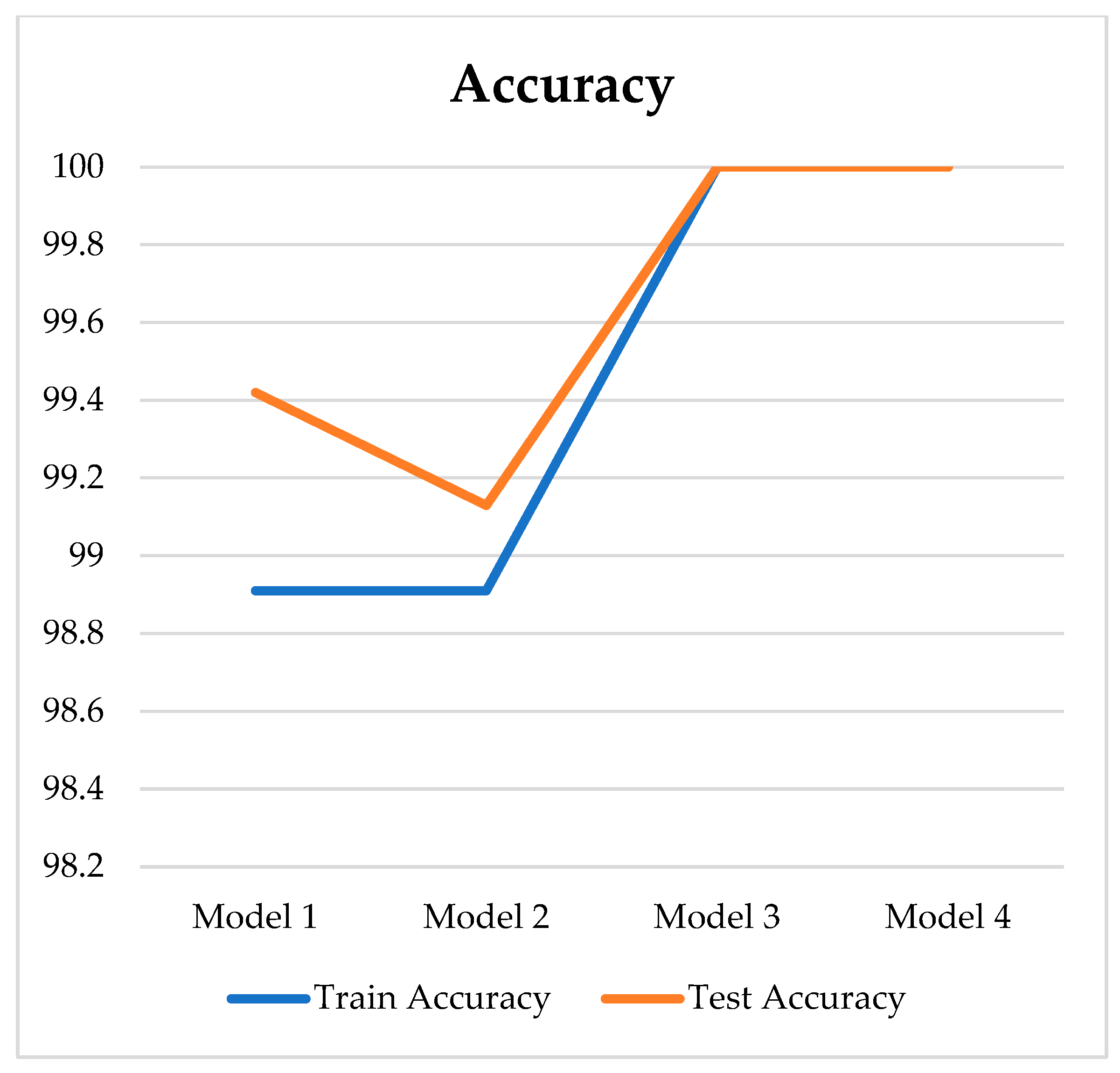
| Model | Number of Hidden Layers | Learning Rate | Batch Size | Epoch | Loss | Acc. (%) | Sens. (%) | Spec. (%) | Pres. (%) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 3 | 0.001 | 32 | 140 | 0.041 | 99.42 | 96.67 | 100 | 98.46 |
| 2 | 5 | 0.0001 | 32 | 140 | 0.060 | 99.13 | 95.83 | 99.49 | 96.58 |
| 3 | 7 | 0.001 | 128 | 140 | 0.052 | 100 | 100 | 100 | 100 |
| 4 | 9 | 0.0001 | 32 | 140 | 0.060 | 100 | 100 | 100 | 100 |
| No. | Layer | Layer Parameter | Outpshape |
|---|---|---|---|
| 1 | Input | shape = (9,) | (None, 9) |
| 2 | Reshape | target_shape = (9, 1) | (None, 9, 1) |
| 3 | TimeDistributed (Dense) | units = 64, activation = ‘relu’ | (None, 9, 64) |
| 4 | Conv1D | filters = 64, kernel_size = 2 | (None, 9, 64) |
| 5 | Conv1D | filters = 64, kernel_size = 2 | (None, 9, 64) |
| 6 | Activation | activation = ‘tanh’ | (None, 9, 64) |
| 7 | Activation | activation = ‘sigmoid’ | (None, 9, 64) |
| 8 | TimeDistributed (Dense) | units = 64, activation = ‘relu’ | (None, 9, 64) |
| 9 | … (repeated dilations) | - | - |
| 10 | Add | - | (None, 9, 64) |
| 11 | Activation | activation = ‘relu’ | (None, 9, 64) |
| 12 | Flatten | - | (None, 576) |
| 13 | Dropout | rate = 0.5 | (None, 576) |
| 14 | Dense | units = 512, activation = ‘relu’ | (None, 512) |
| 15 | Dropout | rate = 0.5 | (None, 512) |
| 16 | Dense | units = 512, activation = ‘relu’ | (None, 512) |
| 17 | Dense | units = 512, activation = ‘relu’ | (None, 5) |
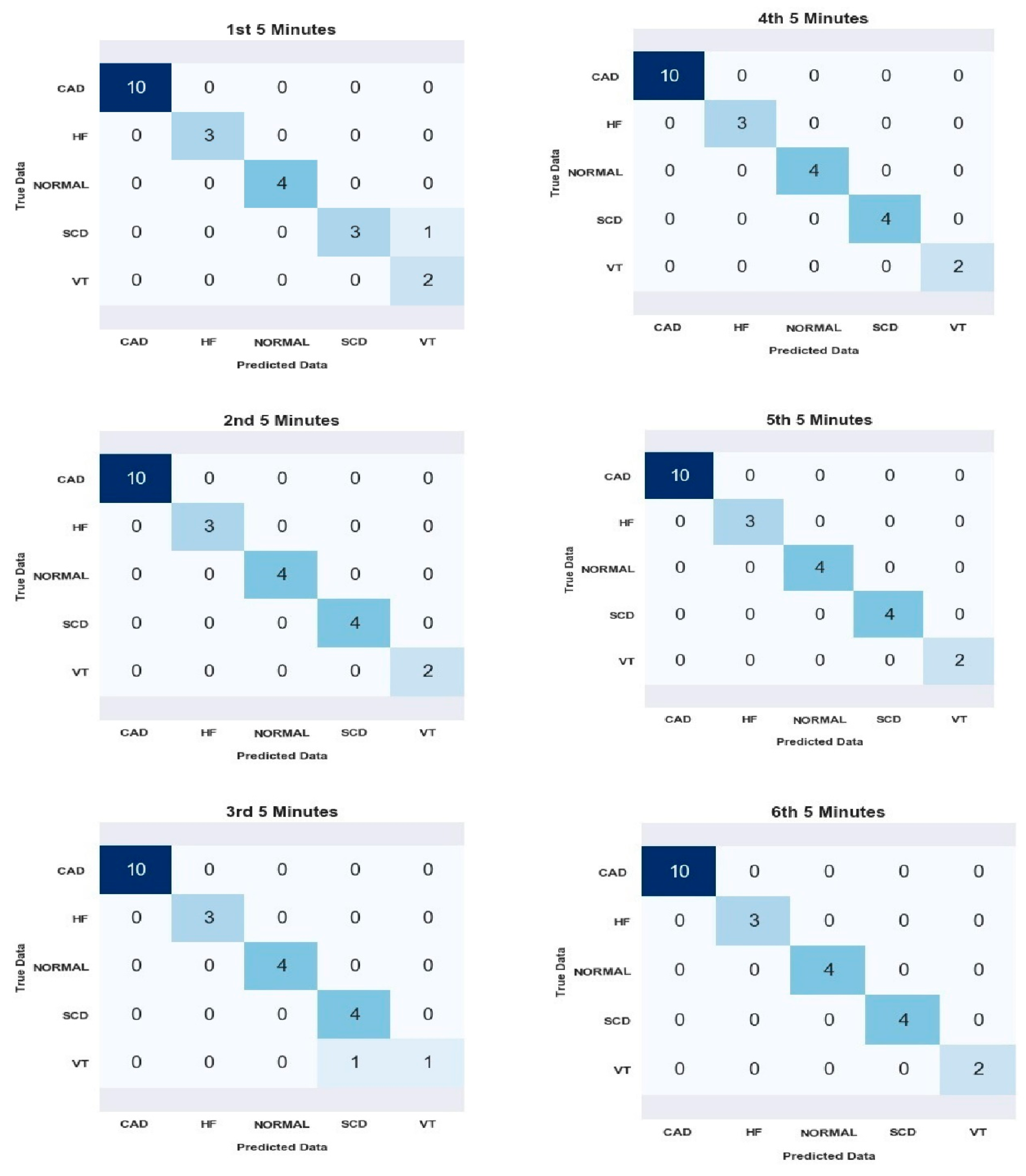
| Segments | Average for All Subject | |||
|---|---|---|---|---|
| Acc. (%) | Sens. (%) | Spec. (%) | Pres. (%) | |
| 1st | 98.26 | 95 | 99.05 | 93.33 |
| 2nd | 100 | 100 | 100 | 100 |
| 3rd | 98.26 | 90.00 | 98.95 | 96.00 |
| 4th | 100 | 100 | 100 | 100 |
| 5th | 100 | 100 | 100 | 100 |
| 6th | 100 | 100 | 100 | 100 |
| Subject | Acc. (%) | Sens. (%) | Spec. (%) | Pres. (%) |
|---|---|---|---|---|
| CAD | 100 | 100 | 100 | 100 |
| HF | 100 | 100 | 100 | 100 |
| SNR | 100 | 100 | 100 | 100 |
| SCD | 99.28 | 95 | 99.05 | 99.33 |
| VT | 99.28 | 90 | 98.95 | 96.00 |
| average | 99.30 | 97 | 99.60 | 97.87 |
| Authors (Year) | Signal Length Prediction Period | No. of Subject | Features | Method | Performance Result (%) | |||
|---|---|---|---|---|---|---|---|---|
| Acc. | Sens. | Spec. | Pres. | |||||
| Ebrahimzadeh et al. (2018) [13] | 12 min before 1 min interval | 35 NSR 35 SCD | HRV Time-Domain, Frequency-Domain, Time-Frequency, Non-Linear | MLP | 83.87 | 82.66 | 85.09 | 84.72 |
| Ebrahimzadeh et al. (2019) [2] | 13 min before 1 min interval | 30 NSR 35 SCD | HRV Time-Domain, Frequency-Domain, Time-Frequency, Non-Linear | MLP | 84.28 | 85.71 | 82.85 | 83.33 |
| Devi et al. (2019) [3] | 10 min before 5 min interval | 18 NSR 15 HF 18 SCD | HRV Classical, non-linear, and CWT features | KNN | 83.33 | 75 | 87.5 | 75 |
| Ashish Rohila et al. (2020) [15] | 1 hour before 5 min interval | 18 NSR 23 CAD 15 HF 20 SCD | HRV Entropy, Poincare plot, S-transform | SVM and DT | 91.67 | 83.33 | 94.64 | 84.75 |
| A. Parsi et al. (2020) [14] | 5 min before 5 min interval | 106 VT 29 VF | HRV Time-Domain | SVM KNN RF | 90.2 | 88.8 | 94.2 | - |
| Our Work | 30 min before 5 min interval | 18 NSR 51 CAD 15 HF 11 VT 20 SCD | HRV Time-Domain | 1D-CNN Wavenet model | 99.30 | 97.00 | 99.60 | 97.87 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Panjaitan, F.; Nurmaini, S.; Partan, R.U. Accurate Prediction of Sudden Cardiac Death Based on Heart Rate Variability Analysis Using Convolutional Neural Network. Medicina 2023, 59, 1394. https://doi.org/10.3390/medicina59081394
Panjaitan F, Nurmaini S, Partan RU. Accurate Prediction of Sudden Cardiac Death Based on Heart Rate Variability Analysis Using Convolutional Neural Network. Medicina. 2023; 59(8):1394. https://doi.org/10.3390/medicina59081394
Chicago/Turabian StylePanjaitan, Febriyanti, Siti Nurmaini, and Radiyati Umi Partan. 2023. "Accurate Prediction of Sudden Cardiac Death Based on Heart Rate Variability Analysis Using Convolutional Neural Network" Medicina 59, no. 8: 1394. https://doi.org/10.3390/medicina59081394
APA StylePanjaitan, F., Nurmaini, S., & Partan, R. U. (2023). Accurate Prediction of Sudden Cardiac Death Based on Heart Rate Variability Analysis Using Convolutional Neural Network. Medicina, 59(8), 1394. https://doi.org/10.3390/medicina59081394







