Value of Serum Sirtuin-1 (SIRT1) Levels and SIRT1 Gene Variants in Periodontitis Patients
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Control and Patients with Periodontitis Group Formation
2.3. Odontological Examination
2.4. DNA Extraction, Genotyping, and Enzyme-Linked Immunosorbent Assay
2.5. Statistical Analysis
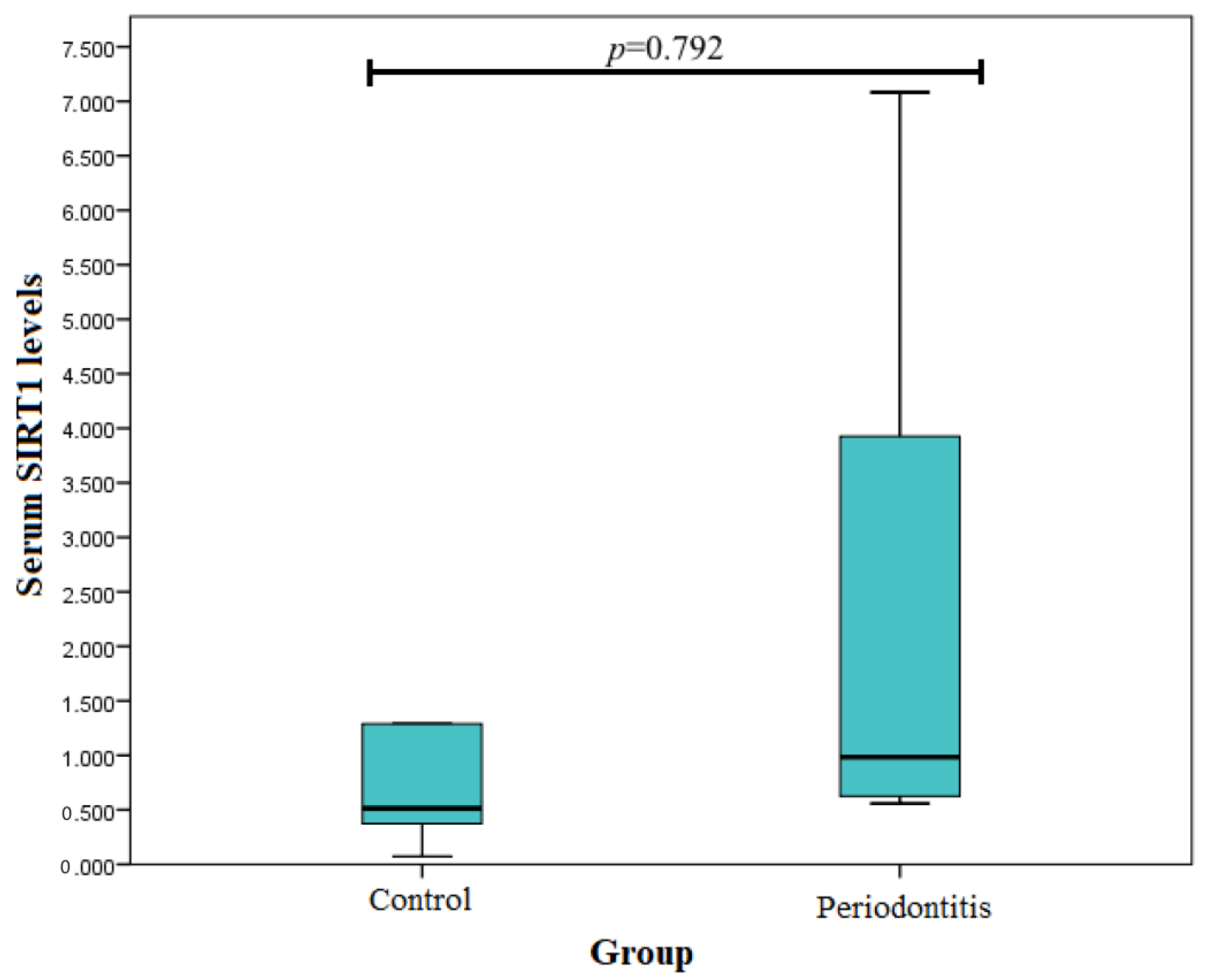
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Colombo, A.P.V.; Magalhães, C.B.; Hartenbach, F.A.R.R.; Souto, R.M.D.; da Silva-Boghossian, C.M. Periodontal-disease-associated biofilm: A reservoir for pathogens of medical importance. Microb. Pathog. 2016, 94, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Kassebaum, N.J.; Smith, A.G.C.; Bernabé, E.; Fleming, T.D.; Reynolds, A.E.; Vos, T.; Murray, C.J.L.; Marcenes, W. GBD 2015 Oral Health Collaborators. Global, Regional, and National Prevalence, Incidence, and Disability-Adjusted Life Years for Oral Conditions for 195 Countries, 1990–2015: A Systematic Analysis for the Global Burden of Diseases, Injuries, and Risk Factors. J. Dent. Res. 2017, 96, 380–387. [Google Scholar] [CrossRef] [PubMed]
- Gasparoni, L.; Alves, F.; Holzhausen, M.; Pannuti, C.; Serpa, M. Periodontitis as a risk factor for head and neck cancer. Med. Oral Patol. Oral Cir. Bucal. 2021, 26, e430–e436. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, A.S. Genetics of periodontitis: Discovery, biology, and clinical impact. Periodontology 2000 2018, 78, 162–173. [Google Scholar] [CrossRef] [PubMed]
- Islam, S.; Abiko, Y.; Uehara, O.; Chiba, I. Sirtuin 1 and oral cancer (Review). Oncol. Lett. 2019, 17, 729–738. [Google Scholar] [CrossRef] [Green Version]
- Carafa, V.; Rotili, D.; Forgione, M.; Cuomo, F.; Serretiello, E.; Hailu, G.S.; Jarho, E.; Lahtela-Kakkonen, M.; Mai, A.; Altucci, L. Sirtuin functions and modulation: From chemistry to the clinic. Clin. Epigenet. 2016, 8, 61. [Google Scholar] [CrossRef]
- Wątroba, M.; Dudek, I.; Skoda, M.; Stangret, A.; Rzodkiewicz, P.; Szukiewicz, D. Sirtuins, epigenetics and longevity. Ageing Res. Rev. 2017, 40, 11–19. [Google Scholar] [CrossRef]
- O’Callaghan, C.; Vassilopoulos, A. Sirtuins at the crossroads of stemness, aging, and cancer. Aging Cell 2017, 16, 1208–1218. [Google Scholar] [CrossRef]
- Ezhilarasan, D.; Lakshmi, T.; Subha, M.; Nallasamy, V.D.; Raghunandhakumar, S. The ambiguous role of sirtuins in head and neck squamous cell carcinoma. Oral Dis. 2022, 28, 559–567. [Google Scholar] [CrossRef]
- Chang, H.-C.; Guarente, L. SIRT1 and other sirtuins in metabolism. Trends Endocrinol. Metab. 2014, 25, 138–145. [Google Scholar] [CrossRef]
- Satoh, A.; Brace, C.S.; Rensing, N.; Cliften, P.; Wozniak, D.F.; Herzog, E.; Yamada, K.A.; Imai, S.-I. Sirt1 Extends Life Span and Delays Aging in Mice through the Regulation of Nk2 Homeobox 1 in the DMH and LH. Cell Metab. 2013, 18, 416–430. [Google Scholar] [CrossRef] [Green Version]
- Laddha, R.; Mahajania, M.; Khadse, A.; Bajaj, R.; Jawade, R.; Choube, S. Assessment of Sirtuin 3 and Sirtuin 4 Level in Pa-tients with Diabetes Mellitus and Periodontitis: A Clinical Study. J. Contemp. Dent. Pract. 2018, 19, 1199–1203. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Hou, K.; Kok, S.; Hong, C.; Lin, S.; Lai, E.H.; Wang, H.; Chang, J.Z.; Yang, H. Sirtuin 6 attenuates periapical lesion propagation by modulating hypoxia-induced chemokine (C-C motif) ligand 2 production in osteoblasts. Int. Endod. J. 2018, 51 (Suppl. 2), e74–e86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, C.; Lin, S.; Kok, S.; Wang, H.; Lee, Y.; Shun, C.; Chi, C.; Yang, H.; Hong, C. The possible role of sirtuin 5 in the pathogenesis of apical periodontitis. Oral Dis. 2021, 27, 1766–1774. [Google Scholar] [CrossRef] [PubMed]
- Mautone, N.; Zwergel, C.; Mai, A.; Rotili, D. Sirtuin modulators: Where are we now? A review of patents from 2015 to 2019. Expert Opin. Ther. Patents 2020, 30, 389–407. [Google Scholar] [CrossRef]
- Zu, Y.; Liu, L.; Lee, M.Y.; Xu, C.; Liang, Y.; Man, R.Y.; Vanhoutte, P.M.; Wang, Y. SIRT1 Promotes Proliferation and Prevents Senescence Through Targeting LKB1 in Primary Porcine Aortic Endothelial Cells. Circ. Res. 2010, 106, 1384–1393. [Google Scholar] [CrossRef] [Green Version]
- Tamaki, N.; Orihuela-Campos, R.C.; Inagaki, Y.; Fukui, M.; Nagata, T.; Ito, H.-O. Resveratrol improves oxidative stress and prevents the progression of periodontitis via the activation of the Sirt1/AMPK and the Nrf2/antioxidant defense pathways in a rat periodontitis model. Free Radic. Biol. Med. 2014, 75, 222–229. [Google Scholar] [CrossRef]
- Caribé, P.M.V.; Villar, C.; Romito, G.A.; Pacanaro, A.P.; Strunz, C.M.C.; Takada, J.Y.; Cesar, L.A.M.; Mansur, A.D.P. Influence of the treatment of periodontal disease in serum concentration of sirtuin 1 and mannose-binding lectin. J. Periodontol. 2020, 91, 900–905. [Google Scholar] [CrossRef]
- Caribé, P.M.V.; Villar, C.C.; Romito, G.A.; Takada, J.Y.; Pacanaro, A.P.; Strunz, C.M.C.; César, L.A.M.; Mansur, A.D.P. Prospective, case-controlled study evaluating serum concentration of sirtuin-1 and mannose-binding lectin in patients with and without periodontal and coronary artery disease. Ther. Adv. Chronic Dis. 2020, 11, 2040622320919621. [Google Scholar] [CrossRef]
- Papapanou, P.N.; Sanz, M.; Buduneli, N.; Dietrich, T.; Feres, M.; Fine, D.H.; Flemmig, T.F.; Garcia, R.; Giannobile, W.V.; Graziani, F.; et al. Periodontitis: Consensus report of workgroup 2 of the 2017 World Workshop on the Classification of Periodontal and Peri-Implant Diseases and Conditions. J. Clin. Periodontol. 2018, 45 (Suppl. 20), S162–S170. [Google Scholar] [CrossRef]
- Glebauskiene, B.; Vilkeviciute, A.; Liutkeviciene, R.; Jakstiene, S.; Kriauciuniene, L.; Zemaitiene, R.; Zaliuniene, D. Association of FGFR2 rs2981582, SIRT1 rs12778366, STAT3 rs744166 gene polymorphisms with pituitary adenoma. Oncol. Lett. 2017, 13, 3087–3099. [Google Scholar] [CrossRef] [PubMed]
- Kudo, H.; Takeichi, O.; Hatori, K.; Makino, K.; Himi, K.; Ogiso, B. A potential role for the silent information regulator 2 homologue 1 (SIRT1) in periapical periodontitis. Int. Endod. J. 2018, 51, 747–757. [Google Scholar] [CrossRef] [PubMed]
- Qu, L.; Yu, Y.; Qiu, L.; Yang, D.; Yan, L.; Guo, J.; Jahan, R. Sirtuin 1 regulates matrix metalloproteinase-13 expression induced by Porphyromonas endodontalis lipopolysaccharide via targeting nuclear factor–κB in osteoblasts. J. Oral Microbiol. 2017, 9, 1317578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.-M.; Shin, S.-I.; Shin, K.-S.; Park, B.-H.; Kim, E.-C. The role of sirtuin 1 in osteoblastic differentiation in human periodontal ligament cells. J. Periodontal Res. 2011, 46, 712–721. [Google Scholar] [CrossRef]
- Zhang, W.-G.; Bai, X.-J.; Chen, X.-M. SIRT1 variants are associated with aging in a healthy Han Chinese population. Clin. Chim. Acta 2010, 411, 1679–1683. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Xie, X.; Tu, Q.; Li, J.; Ding, J.; Shao, G.; Jiang, Q.; Yuan, L.; Lai, X. SIRT1 gene polymorphisms are associated with nondiabetic type 1 cardiorenal syndrome. Ann. Hum. Genet. 2019, 83, 445–453. [Google Scholar] [CrossRef] [PubMed]
- Vaiciulis, P.; Liutkeviciene, R.; Liutkevicius, V.; Vilkeviciute, A.; Gedvilaite, G.; Uloza, V. Association of SIRT1 single gene nucleotide polymorphisms and serum SIRT1 levels with laryngeal squamous cell carcinoma patient survival rate. Cancer Biomark. 2021, 1–14. [Google Scholar] [CrossRef]
- Kluknavska, J.; Krajcikova, K.; Bolerazska, B.; Maslankova, J.; Ohlasova, J.; Timkova, S.; Drotarova, Z.; Vaskova, J. Possible prognostic biomarkers of periodontitis in saliva. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 3154–3161. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, Y.; Gao, J.; Li, T.; Gan, X.; Yu, H. Sirtuin 3 deficiency exacerbates age-related periodontal disease. J. Periodontal Res. 2021, 56, 1163–1173. [Google Scholar] [CrossRef]
- Alhazzazi, T.Y.; Kamarajan, P.; Xu, Y.; Ai, T.; Chen, L.; Verdin, E.; Kapila, Y.L. A Novel Sirtuin-3 Inhibitor, LC-0296, Inhibits Cell Survival and Proliferation, and Promotes Apoptosis of Head and Neck Cancer Cells. Anticancer Res. 2016, 36, 49–60. [Google Scholar]
- Kok, S.-H.; Hou, K.-L.; Hong, C.-Y.; Chao, L.-H.; Lai, E.H.-H.; Wang, H.-W.; Yang, H.; Shun, C.-T.; Wang, J.-S.; Lin, S.-K. Sirtuin 6 Modulates Hypoxia-induced Apoptosis in Osteoblasts via Inhibition of Glycolysis: Implication for Pathogenesis of Periapical Lesions. J. Endod. 2015, 41, 1631–1637. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Sun, H.; Song, F.; Cao, Z.; Jiang, X.; Zhang, L.; Li, Z.; Huang, C. SIRT6 overexpression inhibits cementogenesis by suppressing glucose transporter 1. J. Cell. Physiol. 2019, 234, 4005–4014. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zhu, D.; Qin, S. SIRT7 suppresses the epithelial-to-mesenchymal transition in oral squamous cell carcinoma metastasis by promoting SMAD4 deacetylation. J. Exp. Clin. Cancer Res. 2018, 37, 148. [Google Scholar] [CrossRef] [PubMed]
| Characteristic | Group | p-Value | |
|---|---|---|---|
| Periodontitis n = 201 | Control Group n = 500 | ||
| Males, n (%) | 85 (42.73%) | 250 (50%) | 0.065 |
| Females, n (%) | 116 (57.7%) | 250 (50%) | |
| Age, median (IQR) | 70.0 (16) | 66.00 (15) | 0.082 |
| Genotype/Allele | Periodontitis n = 201 n (%) | Control Group n = 500 n (%) | HWE p-Value | p-Value |
|---|---|---|---|---|
| rs3818292 | ||||
| AA | 162 (80.6) 1 | 446 (89.2) 1 | 0.715 | 0.009 |
| AG | 37 (18.4) 2 | 52 (10.4) 2 | ||
| GG | 2 (1.0) | 2 (0.4) | ||
| Total | 201 (100.0) | 500 (100.0) | ||
| A | 361 (89.8) | 976 (94.6) | 0.001 | |
| G | 41 (10.2) | 56 (5.4) | ||
| rs3758391 | ||||
| CC | 103 (51.2) | 288 (57.6) | 0.179 | 0.272 |
| CT | 86 (42.8) | 190 (38.0) | ||
| TT | 12 (6.0) | 22 (4.4) | ||
| Total | 201 (100.0) | 500 (100.0) | ||
| C | 292 (72.6) | 766 (76.6) | 0.119 | |
| T | 110 (27.4) | 234 (23.4) | ||
| rs7895833 | ||||
| AA | 133 (66.2) 3 | 383 (76.6) 3 | 0.517 | 0.018 |
| AG | 65 (32.3) 4 | 111 (22.2) 4 | ||
| GG | 3 (1.5) | 6 (1.2) | ||
| Total | 201 (100.0) | 500 (100.0) | ||
| A | 331 (82.3) | 877 (87.7) | 0.009 | |
| G | 71 (17.7) | 123 (12.3) |
| Model | Genotype/Allele | OR (95% CI) | p-Value | AIC |
|---|---|---|---|---|
| Periodontitis | ||||
| SIRT1 rs3818292 | ||||
| Co-dominant | AG vs. AA GG vs. AA | 1.959 (1.239–3.098) 2.753 (0.385–19.706) | 0.004 0.313 | 835.292 |
| Dominant | AG + GG vs. AA | 1.988 (1.269–3.116) | 0.003 | 833.402 |
| Recessive | GG vs. AG + AA | 2.503 (0.350–17.888) | 0.361 | 841.271 |
| Overdominant | AG vs. AA + GG | 1.944 (1.230–3.073) | 0.004 | 834.269 |
| Additive | G | 1.905 (1.249–2906) | 0.003 | 833.384 |
| SIRT1 rs3758391 | ||||
| Co-dominant | CT vs. CC TT vs. CC | 1.266 (0.901–1.778) 1.525 (0.729–3.192) | 0.174 0.263 | 841.503 |
| Dominant | CT + TT vs. CC | 1.293 (0.931–1.795) | 0.126 | 839.739 |
| Recessive | TT vs. CT + CC | 1.380 (0.669–2.843) | 0.383 | 841.343 |
| Overdominant | CT vs. CC + TT | 1.220 (0.875–1.702) | 0.241 | 840.711 |
| Additive | T | 1.252 (0.952–1.646) | 0.108 | 839.515 |
| SIRT1 rs7895833 | ||||
| Co-dominant | AG vs. AA GG vs. AA | 1.686 (1.172–2.427) 1.440 (0.355–5.838) | 0.005 0.610 | 836.236 |
| Dominant | AG + GG vs. AA | 1.674 (1.170–2.394) | 0.005 | 834.284 |
| Recessive | GG vs. AG + AA | 1.247 (0.309–5.037) | 0.756 | 841.985 |
| Overdominant | AG vs. AA + GG | 1.675 (1.165–2.408) | 0.005 | 834.486 |
| Additive | G | 1.572 (1.129–2.188) | 0.007 | 835.030 |
| Genotype/Allele | Periodontitis n = 85 n (%) | Control Group n = 250 n (%) | p-Value |
|---|---|---|---|
| Males | |||
| rs3818292 | |||
| AA | 66 (77.6) 1 | 222 (88.8) 1 | 0.024 |
| AG | 19 (22.4) 2 | 27 (10.8) 2 | |
| GG | 0 (0) | 1 (0.4) | |
| Total | 85 (100.0) | 250 (100.0) | |
| A | 151 (88.8) | 471 (94.2) | 0.019 |
| G | 19 (11.2) | 29 (5.8) | |
| rs3758391 | |||
| CC | 43 (50.6) | 144 (57.6) | 0.631 |
| CT | 36 (42.4) | 96 (38.4) | |
| TT | 6 (7.1) | 10 (4.0) | |
| Total | 85 (100.0) | 250 (100.0) | |
| C | 122 (71.8) | 384 (76.8) | 0.187 |
| T | 48 (28.2) | 116 (23.2) | |
| rs7895833 | |||
| AA | 51 (60.0) 3 | 188 (75.2) 3 | 0.027 |
| AG | 32 (37.6) 4 | 59 (23.6) 4 | |
| GG | 2 (2.4) | 3 (1.2) | |
| Total | 85 (100.0) | 250 (100.0) | |
| A | 134 (78.8) | 435 (87.0) | 0.010 |
| G | 36 (21.2) | 65 (13.0) |
| Genotype/Allele | Periodontitis n = 116 n (%) | Control Group n = 250 n (%) | p-Value |
|---|---|---|---|
| Females | |||
| rs3818292 | |||
| AA | 96 (82.8) | 224 (89.6) | 0.124 |
| AG | 18 (15.5) | 25 (10.0) | |
| GG | 2 (1.7) | 1 (0.4) | |
| Total | 116 (100.0) | 250 (100.0) | |
| A | 210 (90.5) | 473 (94.6) | 0.039 |
| G | 22 (9.5) | 27 (5.4) | |
| rs3758391 | |||
| CC | 60 (51.7) | 144 (57.6) | 0.570 |
| CT | 50 (43.1) | 94 (37.6) | |
| TT | 6 (5.2) | 12 (4.8) | |
| Total | 116 (100.0) | 250 (100.0) | |
| C | 170 (73.3) | 382 (76.4) | 0.361 |
| T | 62 (26.7) | 118 (23.6) | |
| rs7895833 | |||
| AA | 82 (70.7) | 195 (78.0) | 0.267 |
| AG | 33 (28.4) | 52 (20.8) | |
| GG | 1 (0.9) | 3 (1.2) | |
| Total | 116 (100.0) | 250 (100.0) | |
| A | 197 (84.9) | 442 (88.4) | 0.188 |
| G | 35 (15.1) | 58 (11.6) |
| Model | Genotype/Allele | OR (95% CI) | p-Value | AIC |
|---|---|---|---|---|
| Males | ||||
| SIRT1 rs3818292 | ||||
| Co-dominant | AG vs. AA GG vs. AA | 2.367 (1.238–4.525) - | 0.009 - | 376.41 |
| Dominant | AG + GG vs. AA | 2.282 (1.199–4.347) | 0.012 | 375.46 |
| Recessive | GG vs. AG + AA | - | - | - |
| Overdominant | AG vs. AA + GG | 2.378 (1.244–4.545) | 0.009 | 374.93 |
| Additive | G | 2.110 (1.129–3.945) | 0.019 | 376.24 |
| SIRT1 rs3758391 | ||||
| Co-dominant | CT vs. CC TT vs. CC | 1.256 (0.752–2.097) 2.009 (0.691–5.846) | 0.384 0.200 | 381.53 |
| Dominant | CT + TT vs. CC | 1.327 (0.810–2.174) | 0.261 | 380.23 |
| Recessive | TT vs. CT + CC | 1.823 (0.642–5.175) | 0.259 | 380.28 |
| Overdominant | CT vs. CC + TT | 1.179 (0.715–1.943) | 0.520 | 381.07 |
| Additive | T | 1.329 (0.882–2.002) | 0.174 | 379.66 |
| SIRT1 rs7895833 | ||||
| Co-dominant | AG vs. AA GG vs. AA | 1.999 (1.177–3.397) 2.458 (0.400–15.103) | 0.010 0.332 | 376.55 |
| Dominant | AG + GG vs. AA | 2.022 (1.201–3.401) | 0.008 | 374.60 |
| Recessive | GG vs. AG + AA | 1.984 (0.326–12.079) | 0.457 | 380.97 |
| Overdominant | AG vs. AA + GG | 1.955 (1.154–3.311) | 0.013 | 375.42 |
| Additive | G | 1.888 (1.173–3.038) | 0.009 | 374.78 |
| Genotype/Allele | Periodontitis n = 151 n (%) | Control Group n = 368 n (%) | p-Value |
|---|---|---|---|
| rs3818292 | |||
| AA | 118 (78.1) 1 | 329 (89.4) 1 | 0.001 |
| AG | 31 (20.5) 2 | 39 (10.6) 2 | |
| GG | 2 (1.3) | 0 (0) | |
| Total | 151 (100.0) | 368 (100.0) | |
| A | 267 (88.4) | 697 (94.7) | 0.001 |
| G | 35 (11.6) | 39 (5.2) | |
| rs3758391 | |||
| CC | 74 (49.0) | 209 (56.8) | 0.187 |
| CT | 67 (44.4) | 144 (39.1) | |
| TT | 10 (6.6) | 15 (4.1) | |
| Total | 151 (100.0) | 368 (100.0) | |
| C | 215 (71.2) | 562 (76.4) | 0.081 |
| T | 87 (28.8) | 174 (23.6) | |
| rs7895833 | |||
| AA | 97 (64.2) 3 | 280 (76.1) 3 | 0.022 |
| AG | 51 (33.8) 4 | 84 (22.8) 4 | |
| GG | 3 (2.0) | 4 (1.1) | |
| Total | 151 (100.0) | 368 (100.0) | |
| A | 245 (81.1) | 644 (87.5) | 0.008 |
| G | 57 (18.9) | 92 (12.5) |
| Model | Genotype/Allele | OR (95% CI) | p-Value | AIC |
|---|---|---|---|---|
| Periodontitis | ||||
| SIRT1 rs3818292 | ||||
| Co-dominant | AG vs. AA GG vs. AA | 2.216 (1.322–3.714) 4.5 × 109 (0.000) | 0.003 1.0 | 616.12 |
| Dominant | AG + GG vs. AA | 2.359 (1.418–3.925) | 0.001 | 617.31 |
| Recessive | GG vs. AG + AA | 4.0 × 109 (0.000) | 1.0 | 622.95 |
| Overdominant | AG vs. AA + GG | 2.179 (1.301–3.650) | 0.003 | 619.43 |
| Additive | G | 2.408 (1.474–3.933) | 0.001 | 615.81 |
| SIRT1 rs3758391 | ||||
| Co-dominant | CT vs. CC TT vs. CC | 1.314 (0.887–1.946) 1.883 (0.810–4.374) | 0.173 0.141 | 626.63 |
| Dominant | CT + TT vs. CC | 1.368 (0.935–2.000) | 0.106 | 625.30 |
| Recessive | TT vs. CT + CC | 1.669 (0.732–3.803) | 0.223 | 626.48 |
| Overdominant | CT vs. CC + TT | 1.241 (0.846–1.820) | 0.270 | 626.70 |
| Additive | T | 1.340 (0.976–1.840 | 0.070 | 624.65 |
| SIRT1 rs7895833 | ||||
| Co-dominant | AG vs. AA GG vs. AA | 1.753 (1.154–2.661) 2.165 (0.476–9.846) | 0.008 0.318 | 622.50 |
| Dominant | AG + GG vs. AA | 1.771 (1.176–2.669) | 0.006 | 620.57 |
| Recessive | GG vs. AG + AA | 1.845 (0.408–8.342) | 0.427 | 627.30 |
| Overdominant | AG vs. AA + GG | 1.724 (1.138–2.614) | 0.010 | 621.44 |
| Additive | G | 1.688 (1.157–2.463 | 0.007 | 620.67 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kriaučiūnas, A.; Liutkevičienė, R.; Gedvilaitė, G.; Kaikarytė, K.; Vilkevičiūtė, A.; Gleiznys, D.; Pacauskienė, I.; Žekonis, G. Value of Serum Sirtuin-1 (SIRT1) Levels and SIRT1 Gene Variants in Periodontitis Patients. Medicina 2022, 58, 653. https://doi.org/10.3390/medicina58050653
Kriaučiūnas A, Liutkevičienė R, Gedvilaitė G, Kaikarytė K, Vilkevičiūtė A, Gleiznys D, Pacauskienė I, Žekonis G. Value of Serum Sirtuin-1 (SIRT1) Levels and SIRT1 Gene Variants in Periodontitis Patients. Medicina. 2022; 58(5):653. https://doi.org/10.3390/medicina58050653
Chicago/Turabian StyleKriaučiūnas, Albertas, Rasa Liutkevičienė, Greta Gedvilaitė, Kristė Kaikarytė, Alvita Vilkevičiūtė, Darius Gleiznys, Ingrida Pacauskienė, and Gediminas Žekonis. 2022. "Value of Serum Sirtuin-1 (SIRT1) Levels and SIRT1 Gene Variants in Periodontitis Patients" Medicina 58, no. 5: 653. https://doi.org/10.3390/medicina58050653
APA StyleKriaučiūnas, A., Liutkevičienė, R., Gedvilaitė, G., Kaikarytė, K., Vilkevičiūtė, A., Gleiznys, D., Pacauskienė, I., & Žekonis, G. (2022). Value of Serum Sirtuin-1 (SIRT1) Levels and SIRT1 Gene Variants in Periodontitis Patients. Medicina, 58(5), 653. https://doi.org/10.3390/medicina58050653







