New CagL Amino Acid Polymorphism Patterns of Helicobacter pylori in Peptic Ulcer and Non-Ulcer Dyspepsia
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. DNA Extraction
2.3. H. pylori Detection
2.4. H. pylori cagL Sequencing
2.5. Statistical Analysis
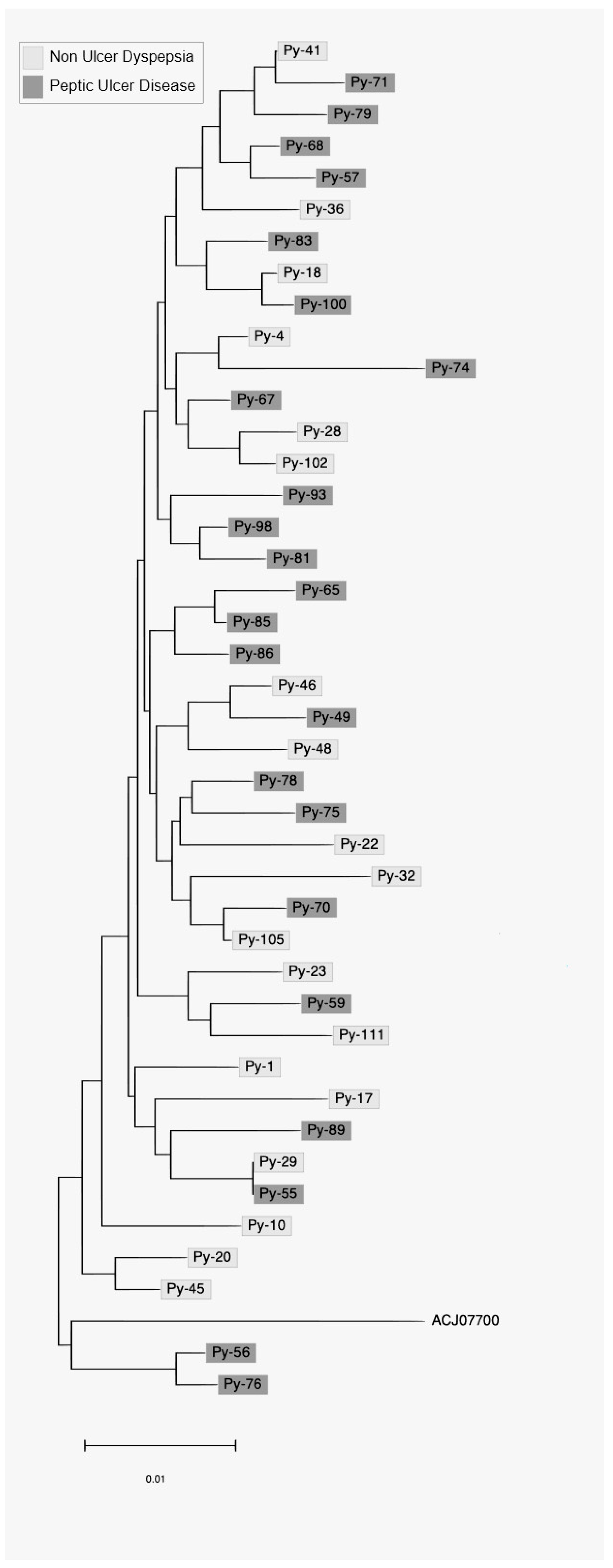
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rhee, K.-H.; Park, J.-S.; Cho, M.-J. Helicobacter Pylori: Bacterial Strategy for Incipient Stage and Persistent Colonization in Human Gastric Niches. Yonsei Med. J. 2014, 55, 1453–1466. [Google Scholar] [CrossRef] [PubMed]
- Sharndama, H.C.; Mba, I.E. Helicobacter Pylori: An up-to-Date Overview on the Virulence and Pathogenesis Mechanisms. Braz. J. Microbiol. 2022, 53, 33–50. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Liu, J.; Gong, Y.; Yuan, Y. Association of CagA EPIYA-D or EPIYA-C Phosphorylation Sites with Peptic Ulcer and Gastric Cancer Risks: A Meta-Analysis. Medicine 2017, 96, e6620. [Google Scholar] [CrossRef]
- Batista, S.A.; Rocha, G.A.; Rocha, A.M.; Saraiva, I.E.; Cabral, M.M.; Oliveira, R.C.; Queiroz, D.M. Higher Number of Helicobacter Pylori CagA EPIYA C Phosphorylation Sites Increases the Risk of Gastric Cancer, but Not Duodenal Ulcer. BMC Microbiol. 2011, 11, 61. [Google Scholar] [CrossRef] [PubMed]
- Kocazeybek, B.S.; Caliskan, R.; Erdamar Cetin, S.; Ergin, S.; Kuskucu, M.; Kepil, N.; Oyku Dinc, H.; Ziya Erzin, Y.; Saribas, S.; Bahar Tokman, H.; et al. Patterns of EPIYA Motifs among CagA-Positive Helicobacter Pylori Strains: A Case-Control Study in a Turkish Population with Eurasian Geographical Features. J. Med. Microbiol. 2015, 64, 1117–1123. [Google Scholar] [CrossRef]
- Demiryas, S.; Caliskan, R.; Saribas, S.; Akkus, S.; Gareayaghi, N.; Kirmusaoglu, S.; Kepil, N.; Dinc, H.; Dag, H.; Dagdeviren, E.; et al. The Association between CagL and CagA, VacAs-m, BabA Genes in Patients with Gastric Cancer, Duodenal Ulcer, and Non-Ulcer Dyspepsia Related to Helicobacter Pylori. Acta Gastroenterol. Belg. 2020, 83, 385–392. [Google Scholar]
- Odenbreit, S.; Püls, J.; Sedlmaier, B.; Gerland, E.; Fischer, W.; Haas, R. Translocation of Helicobacter Pylori CagA into Gastric Epithelial Cells by Type IV Secretion. Science 2000, 287, 1497–1500. [Google Scholar] [CrossRef]
- Bönig, T.; Olbermann, P.; Bats, S.H.; Fischer, W.; Josenhans, C. Systematic Site-Directed Mutagenesis of the Helicobacter Pylori CagL Protein of the Cag Type IV Secretion System Identifies Novel Functional Domains. Sci. Rep. 2016, 6, 38101. [Google Scholar] [CrossRef]
- Poppe, M.; Feller, S.M.; Römer, G.; Wessler, S. Phosphorylation of Helicobacter Pylori CagA by C-Abl Leads to Cell Motility. Oncogene 2007, 26, 3462–3472. [Google Scholar] [CrossRef]
- Tammer, I.; Brandt, S.; Hartig, R.; König, W.; Backert, S. Activation of Abl by Helicobacter Pylori: A Novel Kinase for CagA and Crucial Mediator of Host Cell Scattering. Gastroenterology 2007, 132, 1309–1319. [Google Scholar] [CrossRef]
- Higashi, H.; Tsutsumi, R.; Fujita, A.; Yamazaki, S.; Asaka, M.; Azuma, T.; Hatakeyama, M. Biological Activity of the Helicobacter Pylori Virulence Factor CagA Is Determined by Variation in the Tyrosine Phosphorylation Sites. Proc. Natl. Acad. Sci. USA 2002, 99, 14428–14433. [Google Scholar] [CrossRef] [PubMed]
- Barden, S.; Lange, S.; Tegtmeyer, N.; Conradi, J.; Sewald, N.; Backert, S.; Niemann, H.H. A Helical RGD Motif Promoting Cell Adhesion: Crystal Structures of the Helicobacter Pylori Type IV Secretion System Pilus Protein CagL. Structure 2013, 21, 1931–1941. [Google Scholar] [CrossRef] [PubMed]
- Conradi, J.; Huber, S.; Gaus, K.; Mertink, F.; Royo Gracia, S.; Strijowski, U.; Backert, S.; Sewald, N. Cyclic RGD Peptides Interfere with Binding of the Helicobacter Pylori Protein CagL to Integrins AVβ3 and A5β1. Amino Acids 2012, 43, 219–232. [Google Scholar] [CrossRef] [PubMed]
- Wiedemann, T.; Hofbaur, S.; Tegtmeyer, N.; Huber, S.; Sewald, N.; Wessler, S.; Backert, S.; Rieder, G. Helicobacter Pylori CagL Dependent Induction of Gastrin Expression via a Novel Avβ5-Integrin–Integrin Linked Kinase Signalling Complex. Gut 2012, 61, 986–996. [Google Scholar] [CrossRef] [PubMed]
- Tegtmeyer, N.; Hartig, R.; Delahay, R.M.; Rohde, M.; Brandt, S.; Conradi, J.; Takahashi, S.; Smolka, A.J.; Sewald, N.; Backert, S. A Small Fibronectin-Mimicking Protein from Bacteria Induces Cell Spreading and Focal Adhesion Formation*. J. Biol. Chem. 2010, 285, 23515–23526. [Google Scholar] [CrossRef] [PubMed]
- Wiedemann, T.; Hofbaur, S.; Loell, E.; Rieder, G. A C-Terminal Coiled-Coil Region of CagL Is Responsible for Helicobacter Pylori-Induced Il-8 Expression. Eur. J. Microbiol. Immunol. (Bp) 2016, 6, 186–196. [Google Scholar] [CrossRef] [PubMed]
- Conradi, J.; Tegtmeyer, N.; Woźna, M.; Wissbrock, M.; Michalek, C.; Gagell, C.; Cover, T.L.; Frank, R.; Sewald, N.; Backert, S. An RGD Helper Sequence in CagL of Helicobacter Pylori Assists in Interactions with Integrins and Injection of CagA. Front. Cell Infect. Microbiol. 2012, 2, 70. [Google Scholar] [CrossRef] [PubMed]
- Barden, S.; Niemann, H.H. Adhesion of Several Cell Lines to Helicobacter Pylori CagL Is Mediated by Integrin AVβ6 via an RGDLXXL Motif. J. Mol. Biol. 2015, 427, 1304–1315. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.M.; Choi, Y.H.; Sudhanva, M.S.; Devakumar, S.; Lee, K.H.; Cha, J.-H.; Lee, S.H. Crystal Structure of CagL from Helicobacter Pylori K74 Strain. Biochem. Biophys. Res. Commun. 2015, 460, 964–970. [Google Scholar] [CrossRef]
- Rizzato, C.; Torres, J.; Plummer, M.; Muñoz, N.; Franceschi, S.; Camorlinga-Ponce, M.; Fuentes-Pananá, E.M.; Canzian, F.; Kato, I. Variations in Helicobacter Pylori Cytotoxin-Associated Genes and Their Influence in Progression to Gastric Cancer: Implications for Prevention. PLoS ONE 2012, 7, e29605. [Google Scholar] [CrossRef]
- Shukla, S.K.; Prasad, K.N.; Tripathi, A.; Jaiswal, V.; Khatoon, J.; Ghsohal, U.C.; Krishnani, N.; Husain, N. Helicobacter PyloricagL Amino Acid Polymorphisms and Its Association with Gastroduodenal Diseases. Gastric Cancer 2013, 16, 435–439. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yeh, Y.-C.; Chang, W.-L.; Yang, H.-B.; Cheng, H.-C.; Wu, J.-J.; Sheu, B.-S.H. Pylori CagL Amino Acid Sequence Polymorphism Y58E59 Induces a Corpus Shift of Gastric Integrin A5β1 Related with Gastric Carcinogenesis. Mol. Carcinog. 2011, 50, 751–759. [Google Scholar] [CrossRef] [PubMed]
- Yeh, Y.-C.; Cheng, H.-C.; Yang, H.-B.; Chang, W.-L.; Sheu, B.-S.H. Pylori CagL-Y58/E59 Prime Higher Integrin A5β1 in Adverse PH Condition to Enhance Hypochlorhydria Vicious Cycle for Gastric Carcinogenesis. PLoS ONE 2013, 8, e72735. [Google Scholar] [CrossRef] [PubMed]
- Cherati, M.R.; Shokri-Shirvani, J.; Karkhah, A.; Rajabnia, R.; Nouri, H.R. Helicobacter Pylori CagL Amino Acid Polymorphism D58E59 Pave the Way toward Peptic Ulcer Disease While N58E59 Is Associated with Gastric Cancer in North of Iran. Microb. Pathog. 2017, 107, 413–418. [Google Scholar] [CrossRef] [PubMed]
- Canzian, F.; Rizzato, C.; Obazee, O.; Stein, A.; Flores-Luna, L.; Camorlinga-Ponce, M.; Mendez-Tenorio, A.; Vivas, J.; Trujillo, E.; Jang, H.; et al. Genetic Polymorphisms in the Cag Pathogenicity Island of Helicobacter Pylori and Risk of Stomach Cancer and High-Grade Premalignant Gastric Lesions. Int. J. Cancer 2020, 147, 2437–2445. [Google Scholar] [CrossRef]
- Román-Román, A.; Martínez-Santos, V.I.; Castañón-Sánchez, C.A.; Albañil-Muñoz, A.J.; González-Mendoza, P.; Soto-Flores, D.G.; Martínez-Carrillo, D.N.; Fernández-Tilapa, G. CagL Polymorphisms D58/K59 Are Predominant in Helicobacter Pylori Strains Isolated from Mexican Patients with Chronic Gastritis. Gut Pathog. 2019, 11, 5. [Google Scholar] [CrossRef]
- Gorrell, R.J.; Zwickel, N.; Reynolds, J.; Bulach, D.; Kwok, T. Helicobacter Pylori CagL Hypervariable Motif: A Global Analysis of Geographical Diversity and Association With Gastric Cancer. J. Infect. Dis. 2016, 213, 1927–1931. [Google Scholar] [CrossRef][Green Version]
- Choi, Y.H.; Lai, J.; Kim, M.-A.; Kim, A.; Kim, J.; Su, H.; Ge, L.; Cha, J.-H. CagL Polymorphisms between East Asian and Western Helicobacter Pylori Are Associated with Different Abilities to Induce IL-8 Secretion. J. Microbiol. 2021, 59, 763–770. [Google Scholar] [CrossRef]
- Saez, J.; Belda, S.; Santibáñez, M.; Rodríguez, J.C.; Sola-Vera, J.; Galiana, A.; Ruiz-García, M.; Brotons, A.; López-Girona, E.; Girona, E.; et al. Real-Time PCR for Diagnosing Helicobacter Pylori Infection in Patients with Upper Gastrointestinal Bleeding: Comparison with Other Classical Diagnostic Methods. J. Clin. Microbiol. 2012, 50, 3233–3237. [Google Scholar] [CrossRef]
- Yadegar, A.; Mohabati Mobarez, A.; Zali, M.R. Genetic Diversity and Amino Acid Sequence Polymorphism in Helicobacter Pylori CagL Hypervariable Motif and Its Association with Virulence Markers and Gastroduodenal Diseases. Cancer Med. 2019, 8, 1619–1632. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A Multiple Sequence Alignment Editor and Analysis Workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- Tegtmeyer, N.; Lind, J.; Schmid, B.; Backert, S. Helicobacter Pylori CagL Y58/E59 Mutation Turns-Off Type IV Secretion-Dependent Delivery of CagA into Host Cells. PLoS ONE 2014, 9, e97782. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tafreshi, M.; Zwickel, N.; Gorrell, R.J.; Kwok, T. Preservation of Helicobacter Pylori CagA Translocation and Host Cell Proinflammatory Responses in the Face of CagL Hypervariability at Amino Acid Residues 58/59. PLoS ONE 2015, 10, e0133531. [Google Scholar] [CrossRef] [PubMed]
- Özbey, D. Gastroduodenal Patolojilerle Helicobacter Pylori CagL (Sitotoksinle ilişkili Gen L) Polimorfizmi ilişkisi. Master’s Thesis, Istanbul University Cerrahpaşa, Istanbul, Turkey, 2020. [Google Scholar]
- Raei, N.; Latifi-Navid, S.; Zahri, S. Helicobacter Pylori Cag Pathogenicity Island CagL and Orf17 Genotypes Predict Risk of Peptic Ulcerations but Not Gastric Cancer in Iran. Asian Pac. J. Cancer Prev. 2015, 16, 6645–6650. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, H.; Iwamoto, A.; Tanahashi, T.; Okada, R.; Yamamoto, K.; Nishiumi, S.; Yoshida, M.; Azuma, T. Genetic Variants of Helicobacter Pylori Type IV Secretion System Components CagL and CagI and Their Association with Clinical Outcomes. Gut Pathog. 2017, 9, 21. [Google Scholar] [CrossRef]
| CagL Amino Acid Polymorphisms | PUD † (n = 23) | NUD ‡ (n = 19) | p Value |
|---|---|---|---|
| 22D/F | 23/0 | 19/0 | >0.05 |
| 32S/N | 6/17 | 4/15 | >0.05 |
| 35Q/K | 9/14 | 9/10 | >0.05 |
| 41V/A/T | 18/2/3 | 17/1/1 | >0.05 |
| 56A/T | 1/22 | 1/18 | >0.05 |
| 58D/N | 21/2 | 16/3 | >0.05 |
| 59K/E | 13/10 | 12/7 | >0.05 |
| 60M/I | 23/0 | 17/2 | >0.05 |
| 62E/Q | 23/0 | 18/1 | >0.05 |
| 84A/T | 1/22 | 0/19 | >0.05 |
| 114I/M | 23/0 | 19/0 | >0.05 |
| 122K/N | 19/4 | 15/4 | >0.05 |
| 134I/V | 22/1 | 15/4 | >0.05 |
| 154E/Q/K | 22/0/1 | 18/1/0 | >0.05 |
| 171A/T/V | 20/2/1 | 18/1/0 | >0.05 |
| 172S/P | 1/22 | 0/19 | >0.05 |
| 174I/V | 1/22 | 0/19 | >0.05 |
| 175T/I | 1/22 | 1/18 | >0.05 |
| 194R/K | 2/21 | 2/17 | >0.05 |
| 200Q/H | 22/1 | 18/1 | >0.05 |
| 203V/I | 0/23 | 1/18 | >0.05 |
| 206N/S | 0/23 | 1/18 | >0.05 |
| 210E/K | 1/22 | 2/17 | >0.05 |
| 223R/Q | 0/23 | 1/18 | >0.05 |
| CagLHM Amino Acid Sequences | Total n = 42 (%) | PUD † n = 23 (%) | NUD ‡ n = 19 (%) | p Value |
|---|---|---|---|---|
| NEIGQ | 25 (59.52) | 13 (56.5) | 12 (63.1) | >0.05 |
| NKIGQ | 11 (26.19) | 8 (34.8) | 3 (15.8) | >0.05 |
| DKIGQ | 4 (9.52) | 2 (8.7) | 2 (10.5) | >0.05 |
| NKMGQ | 1 (2.38) | 0 (0) | 1 (5.3) | >0.05 |
| DKMGE | 1 (2.38) | 0 (0) | 1 (5.3) | >0.05 |
| Patterns | CagL Amino Acid Polymorphism Combinations |
|---|---|
| Pattern 1 | K35/N122/V134/T175/R194/E210 |
| Pattern 2 | V41/I134 |
| Pattern 3 | V41/K122/A171/I174 |
| Patterns | PUD † n = 23 (%) | NUD ‡ n = 19 (%) | p Value | Odds Ratio | 95% CI ‡‡ |
|---|---|---|---|---|---|
| 1 | 6 (26.1) | 0 (0) | 0.02 | 1.353 | 1.061–1.725 |
| 2 | 0 (0) | 4 (17.4) | 0.03 | 1.26 | 1.004–1.597 |
| 3 | 0 (0) | 4 (17.4) | 0.03 | 1.26 | 1.004–1.597 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caliskan, R.; Polat Sari, S.; Ercan, B.; Peker, K.D.; Omac Sonmez, M.; Akgul, O.; Sapmaz, B.; Soylu, A.; Adas, G.T.; Oner, Y.A.; et al. New CagL Amino Acid Polymorphism Patterns of Helicobacter pylori in Peptic Ulcer and Non-Ulcer Dyspepsia. Medicina 2022, 58, 1738. https://doi.org/10.3390/medicina58121738
Caliskan R, Polat Sari S, Ercan B, Peker KD, Omac Sonmez M, Akgul O, Sapmaz B, Soylu A, Adas GT, Oner YA, et al. New CagL Amino Acid Polymorphism Patterns of Helicobacter pylori in Peptic Ulcer and Non-Ulcer Dyspepsia. Medicina. 2022; 58(12):1738. https://doi.org/10.3390/medicina58121738
Chicago/Turabian StyleCaliskan, Reyhan, Silva Polat Sari, Bahadir Ercan, Kivanc Derya Peker, Mehtap Omac Sonmez, Ozer Akgul, Burcu Sapmaz, Aliye Soylu, Gokhan Tolga Adas, Yasar Ali Oner, and et al. 2022. "New CagL Amino Acid Polymorphism Patterns of Helicobacter pylori in Peptic Ulcer and Non-Ulcer Dyspepsia" Medicina 58, no. 12: 1738. https://doi.org/10.3390/medicina58121738
APA StyleCaliskan, R., Polat Sari, S., Ercan, B., Peker, K. D., Omac Sonmez, M., Akgul, O., Sapmaz, B., Soylu, A., Adas, G. T., Oner, Y. A., & Yuksel Mayda, P. (2022). New CagL Amino Acid Polymorphism Patterns of Helicobacter pylori in Peptic Ulcer and Non-Ulcer Dyspepsia. Medicina, 58(12), 1738. https://doi.org/10.3390/medicina58121738







