Association of N-acetyltransferases 1 and 2 Polymorphisms with Susceptibility to Head and Neck Cancers—A Meta-Analysis, Meta-Regression, and Trial Sequential Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Identification of Articles
2.3. Eligibility Criteria
2.4. Data Summary
2.5. Quality Evaluation
2.6. Statistical Analysis
2.7. Primer Sequences
3. Results
3.1. Study Selection
3.2. Characteristics of Studies
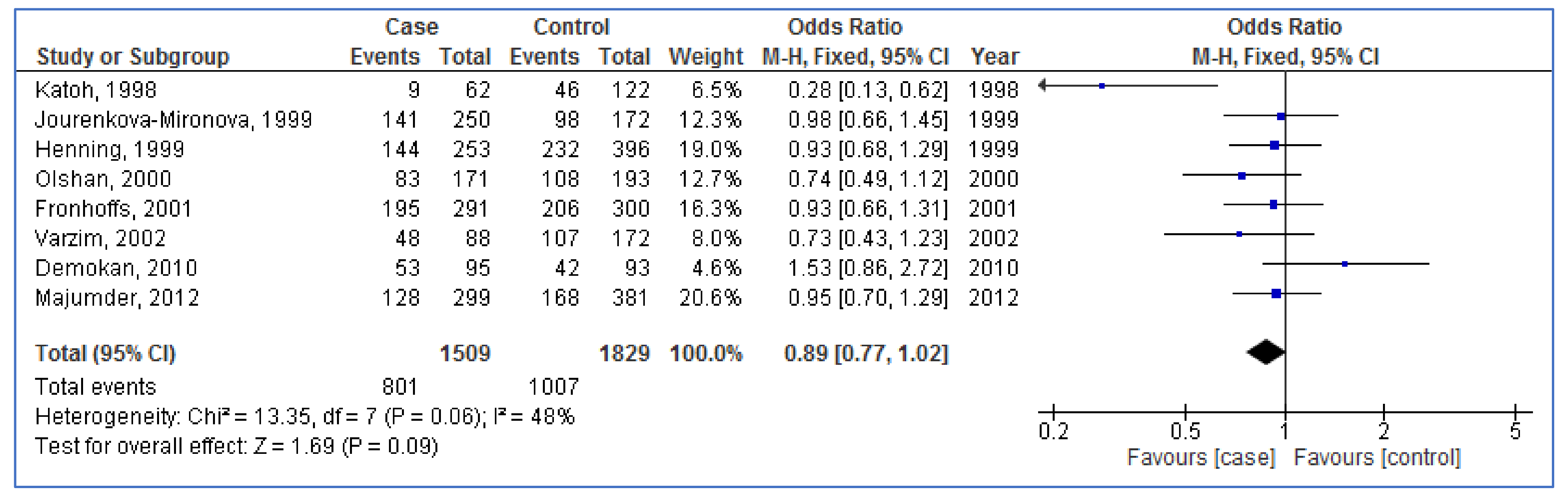
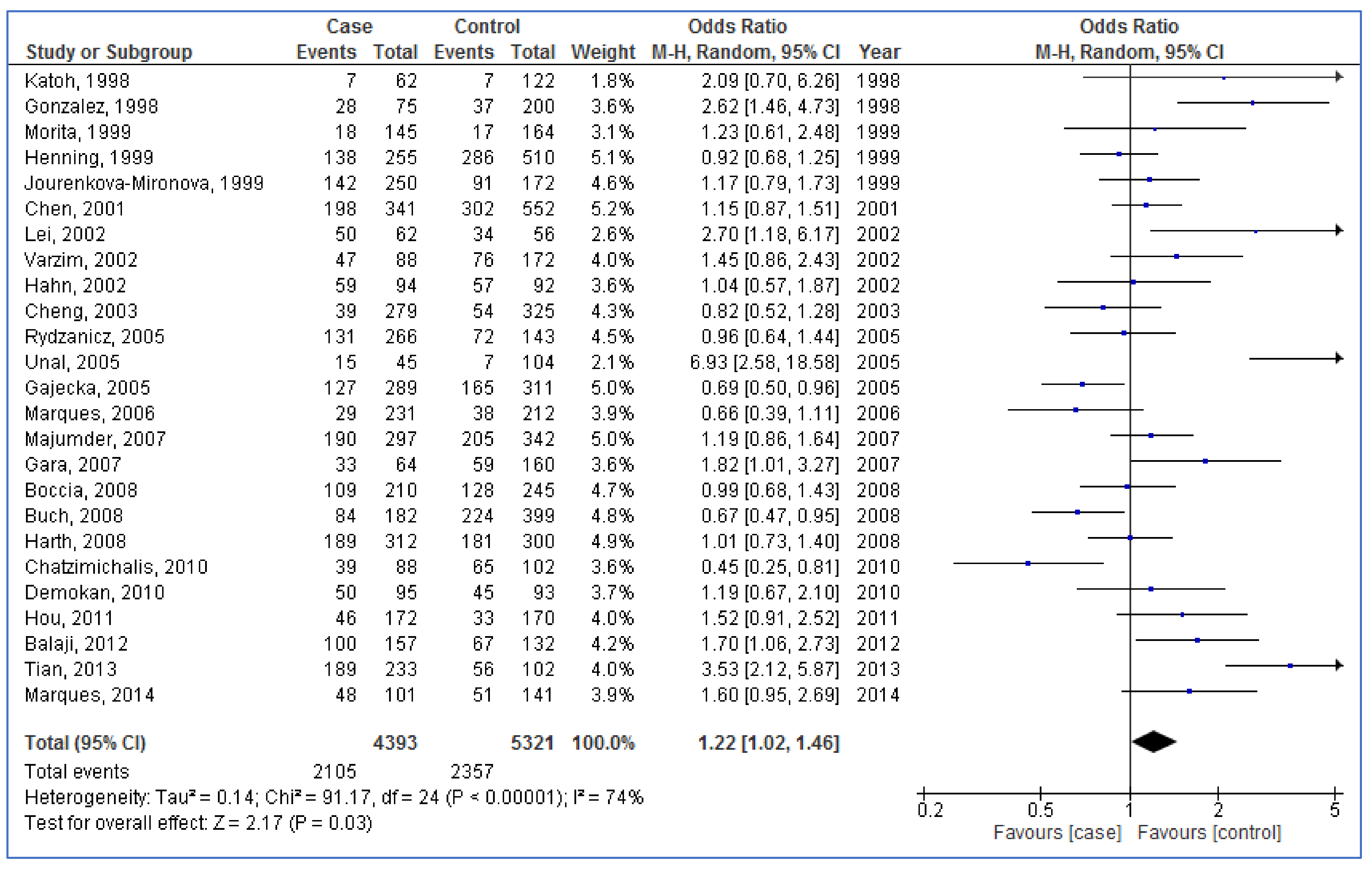
3.3. Pooled Analyses
3.4. Subgroup Analyses
3.5. Meta-Regression
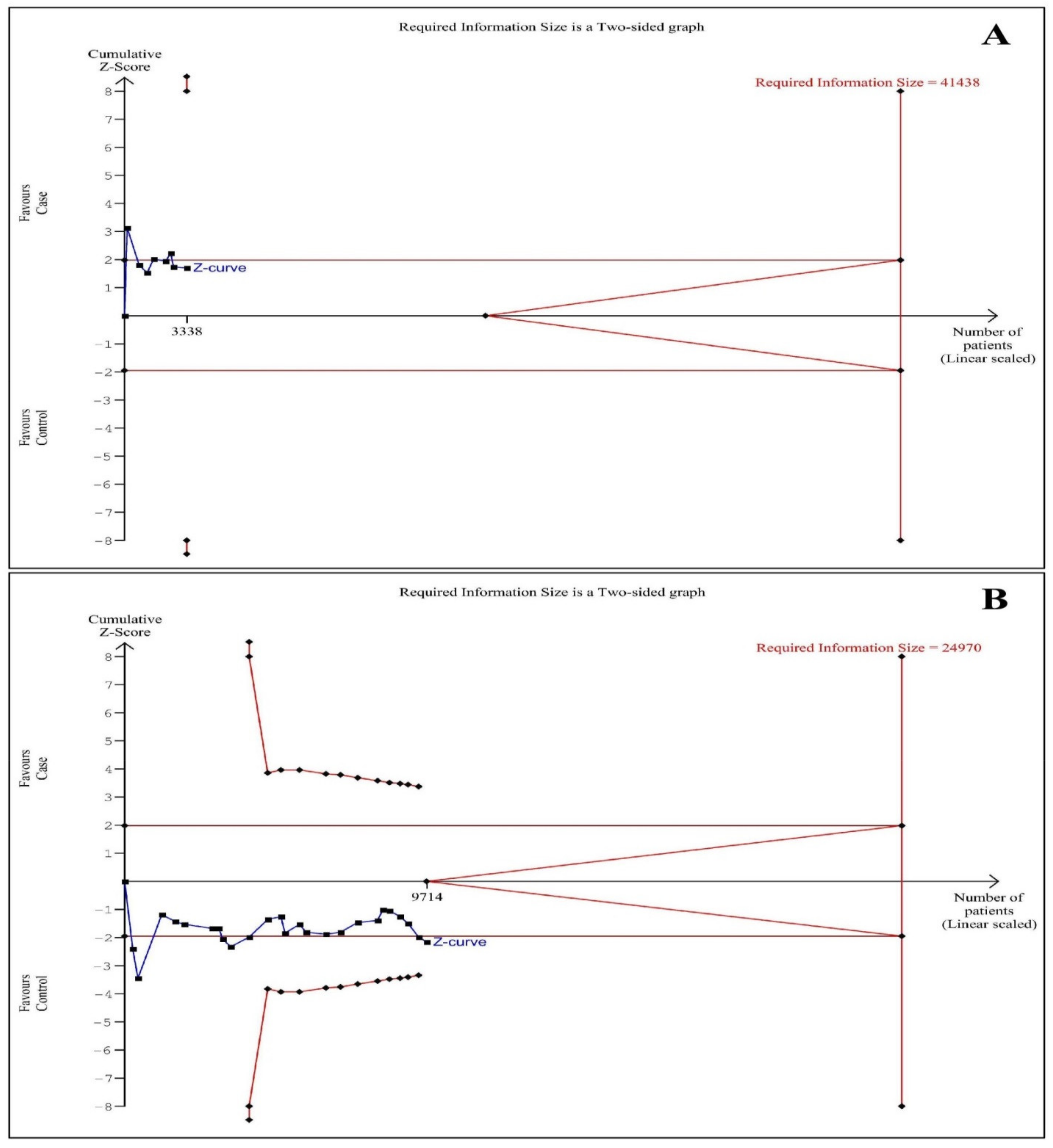
3.6. Trial Sequential Analysis
3.7. Sensitivity Analysis
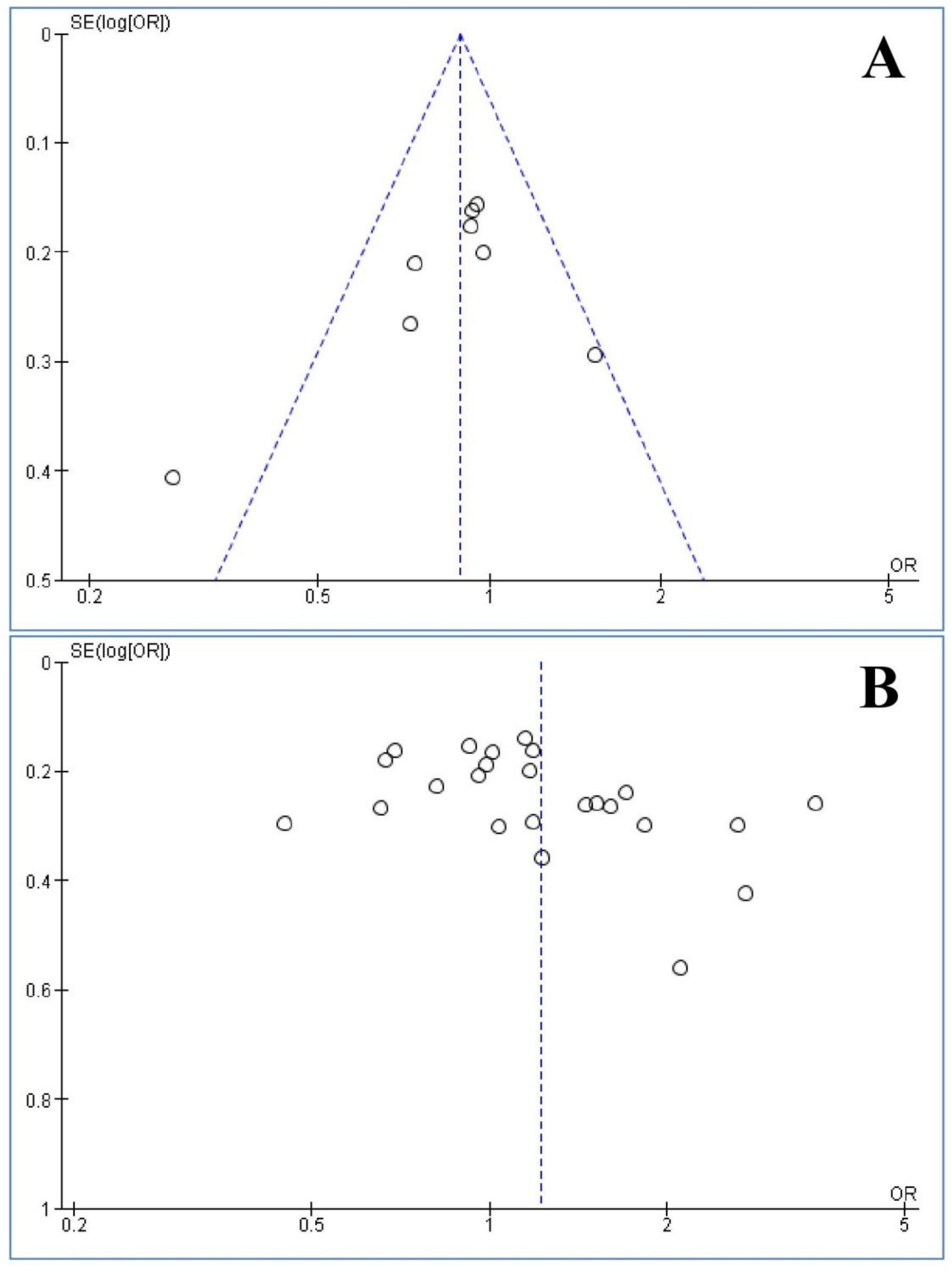
3.8. Publication Bias
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rezaei, F.; Mohammadi, H.; Heydari, M.; Sadeghi, M.; Mozaffari, H.R.; Khavid, A.; Godiny, M.; Brand, S.; M Dürsteler, K.; Brühl, A.B.; et al. Association between IL-8 (-251T/A) and IL-6 (-174G/C) Polymorphisms and Oral Cancer Susceptibility: A Systematic Review and Meta-Analysis. Medicina 2021, 57, 405. [Google Scholar] [CrossRef] [PubMed]
- Patterson, R.H.; Fischman, V.G.; Wasserman, I.; Siu, J.; Shrime, M.G.; Fagan, J.J.; Koch, W.; Alkire, B.C. Global burden of head and neck cancer: Economic consequences, health, and the role of surgery. Otolaryngol.–Head Neck Surg. 2020, 162, 296–303. [Google Scholar] [CrossRef]
- Vos, T.; Abajobir, A.A.; Abate, K.H.; Abbafati, C.; Abbas, K.M.; Abd-Allah, F.; Abdulkader, R.S.; Abdulle, A.M.; Abebo, T.A.; Abera, S.F. Global, regional, and national incidence, prevalence, and years lived with disability for 328 diseases and injuries for 195 countries, 1990–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet 2017, 390, 1211–1259. [Google Scholar] [CrossRef]
- Foreman, K.J.; Marquez, N.; Dolgert, A.; Fukutaki, K.; Fullman, N.; McGaughey, M.; Pletcher, M.A.; Smith, A.E.; Tang, K.; Yuan, C.-W. Forecasting life expectancy, years of life lost, and all-cause and cause-specific mortality for 250 causes of death: Reference and alternative scenarios for 2016—40 for 195 countries and territories. Lancet 2018, 392, 2052–2090. [Google Scholar] [CrossRef]
- Nigro, C.L.; Denaro, N.; Merlotti, A.; Merlano, M. Head and neck cancer: Improving outcomes with a multidisciplinary approach. Cancer Manag. Res. 2017, 9, 363. [Google Scholar] [CrossRef]
- Weinberger, P.M.; Yu, Z.; Haffty, B.G.; Kowalski, D.; Harigopal, M.; Brandsma, J.; Sasaki, C.; Joe, J.; Camp, R.L.; Rimm, D.L. Molecular classification identifies a subset of human papillomavirus–associated oropharyngeal cancers with favorable prognosis. J. Clin. Oncol. 2006, 24, 736–747. [Google Scholar] [CrossRef]
- Johnson, D.E.; Burtness, B.; Leemans, C.R.; Lui, V.W.Y.; Bauman, J.E.; Grandis, J.R. Head and neck squamous cell carcinoma. Nat. Rev. Dis. Primers 2020, 6, 1–22. [Google Scholar] [CrossRef]
- Mozaffari, H.R.; Rostamnia, M.; Sharifi, R.; Safaei, M.; Zavattaro, E.; Tadakamadla, S.K.; Imani, M.M.; Sadeghi, M.; Golshah, A.; Moradpoor, H. A PRISMA-compliant meta-analysis on association between X-ray repair cross complementing (XRCC1, XRCC2, and XRCC3) polymorphisms and oral cancer susceptibility. Gene 2021, 781, 145524. [Google Scholar] [CrossRef]
- Rezaei, F.; Doulatparast, D.; Sadeghi, M. Common polymorphisms of Interleukin-10 (-1082A/G,-592A/C, and-819C/T) in oral cancers: An updated meta-analysis. J. Interferon Cytokine Res. 2020, 40, 357–369. [Google Scholar] [CrossRef]
- Xia, S.; Wu, S.; Wang, M. The Association Between the XRCC1 Arg399Gln Polymorphism and the Risk of Head and Neck Cancer: An Updated Meta-Analysis Including 14586 Subjects. Technol. Cancer Res. Treat. 2021, 20, 15330338211033060. [Google Scholar] [CrossRef]
- Wu, T.; Zhang, Z.T.; Li, L.; Liu, R.Y.; Bei, B.T. Correlation between hypoxia-inducible factor-1α C1772T/G1790A polymorphisms and head and neck cancer risk: A meta-analysis. World J. Surg. Oncol. 2021, 19, 210. [Google Scholar] [CrossRef] [PubMed]
- Vineis, P. Epidemiology of cancer from exposure to arylamines. Environ. Health Perspect. 1994, 102, 7–10. [Google Scholar] [PubMed]
- Bartsch, H.; Malaveille, C.; Friesen, M.; Kadlubar, F.; Vineis, P. Black (air-cured) and blond (flue-cured) tobacco cancer risk IV: Molecular dosimetry studies implicate aromatic amines as bladder carcinogens. Eur. J. Cancer 1993, 29, 1199–1207. [Google Scholar] [CrossRef]
- Hein, D.W.; Doll, M.A.; Rustan, T.D.; Gray, K.; Feng, Y.; Ferguson, R.J.; Grant, D.M. Metabolic activation and deactivation of arylamine carcinogens by recombinant human NAT1 and polymorphic NAT2 acetyltransferases. Carcinogenesis 1993, 14, 1633–1638. [Google Scholar] [CrossRef]
- Hein, D.W.; Doll, M.A.; Fretland, A.J.; Leff, M.A.; Webb, S.J.; Xiao, G.H.; Devanaboyina, U.-S.; Nangju, N.A.; Feng, Y. Molecular genetics and epidemiology of the NAT1 and NAT2 acetylation polymorphisms. Cancer Epidemiol. Prev. Biomark. 2000, 9, 29–42. [Google Scholar]
- Vatsis, K.P.; Weber, W.W. Structural heterogeneity of Caucasian N-acetyltransferase at the NAT1 gene locus. Arch. Biochem. Biophys. 1993, 301, 71–76. [Google Scholar] [CrossRef]
- Feki-Tounsi, M.; Khlifi, R.; Louati, I.; Fourati, M.; Mhiri, M.-N.; Hamza-Chaffai, A.; Rebai, A. Polymorphisms in XRCC1, ERCC2, and ERCC3 DNA repair genes, CYP1A1 xenobiotic metabolism gene, and tobacco are associated with bladder cancer susceptibility in Tunisian population. Environ. Sci. Pollut. Res. 2017, 24, 22476–22484. [Google Scholar] [CrossRef]
- Imam, H.; Imam, T.; Abbas, S.Z.; Ismail, M.; Muhammad, S.A. Association study of NAT2 gene polymorphism and risk of oral cancer in Southern Punjab, Pakistan. J. Pak. Med Assoc. 2021, 71, 1954–1958. [Google Scholar] [PubMed]
- Sabbagh, N.; Delaporte, E.; Marez, D.; Lo-Guidice, J.-M.; Piette, F.; Broly, F. NAT2 genotyping and efficacy of sulfasalazine in patients with chronic discoid lupus erythematosus. Pharmacogenetics 1997, 7, 131–135. [Google Scholar] [CrossRef]
- Wang, L.; Minchin, R.F.; Butcher, N.J. Arylamine N-acetyltransferase 1 protects against reactive oxygen species during glucose starvation: Role in the regulation of p53 stability. PLoS ONE 2018, 13, e0193560. [Google Scholar] [CrossRef]
- Minchin, R.F. Acetylation of p-aminobenzoylglutamate, a folic acid catabolite, by recombinant human arylamine N-acetyltransferase and U937 cells. Biochem. J. 1995, 307, 1–3. [Google Scholar] [CrossRef][Green Version]
- Boccia, S.; Cadoni, G.; Sayed-Tabatabaei, F.A.; Volante, M.; Arzani, D.; De Lauretis, A.; Cattel, C.; Almadori, G.; Van Duijn, C.M.; Paludetti, G. CYP1A1, CYP2E1, GSTM1, GSTT1, EPHX1 exons 3 and 4, and NAT2 polymorphisms, smoking, consumption of alcohol and fruit and vegetables and risk of head and neck cancer. J. Cancer Res. Clin. Oncol. 2008, 134, 93–100. [Google Scholar] [CrossRef]
- Henning, S.; Cascorbi, I.; Münchow, B.; Jahnke, V.; Roots, I. Association of arylamine N-acetyltransferases NAT1 and NAT2 genotypes to laryngeal cancer risk. Pharmacogenetics 1999, 9, 103–111. [Google Scholar] [CrossRef]
- Zhang, K.; Gao, L.; Wu, Y.; Chen, J.; Lin, C.; Liang, S.; Su, J.; Ye, J.; He, X. NAT1 polymorphisms and cancer risk: A systematic review and meta-analysis. Int. J. Clin. Exp. Med. 2015, 8, 9177. [Google Scholar] [PubMed]
- Zheng, Y.; Li, Y.; Teng, Y.; Zhang, Z.; Cao, X. Association of NAT2 phenotype with risk of head and neck carcinoma: A meta-analysis. Oncol. Lett. 2012, 3, 429–434. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, X.-L.; Ling, J.-J.; Zhou, Y.; Zhao, H.-Y.; Song, Y.-F.; Tan, Y.-H. NAT2 polymorphisms with oral carcinoma susceptibility: A meta-analysis. Mol. Biol. Rep. 2012, 39, 8813–8819. [Google Scholar] [CrossRef] [PubMed]
- Ying, X.-J.; Dong, P.; Shen, B.; Wang, J.; Wang, S.; Wang, G. Possible association of NAT2 polymorphism with laryngeal cancer risk: An evidence-based meta-analysis. J. Cancer Res. Clin. Oncol. 2011, 137, 1661–1667. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Xiang, Z.; Hao, R.; Li, R.; Zhu, Y. N-acetyltransferase 2 genetic variants confer the susceptibility to head and neck carcinoma: Evidence from 23 case–control studies. Tumor Biol. 2014, 35, 3585–3595. [Google Scholar] [CrossRef] [PubMed]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. Int. J. Surg. 2010, 8, 336–341. [Google Scholar] [CrossRef]
- Wells, G.A.; Shea, B.; O’Connell, D.; Peterson, J.; Welch, V.; Losos, M.; Tugwell, P. The Newcastle-Ottawa Scale (NOS) for Assessing the Quality of Nonrandomised Studies in Meta-Analyses. 2000. Available online: http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp (accessed on 12 January 2016).
- DerSimonian, R.; Laird, N. Meta-analysis in clinical trials revisited. Contemp. Clin. Trials 2015, 45, 139–145. [Google Scholar] [CrossRef]
- Mantel, N.; Haenszel, W. Statistical aspects of the analysis of data from retrospective studies of disease. J. Natl. Cancer Inst. 1959, 22, 719–748. [Google Scholar]
- Imberger, G.; Thorlund, K.; Gluud, C.; Wetterslev, J. False-positive findings in Cochrane meta-analyses with and without application of trial sequential analysis: An empirical review. BMJ Open 2016, 6, e011890. [Google Scholar] [CrossRef] [PubMed]
- Wetterslev, J.; Jakobsen, J.C.; Gluud, C. Trial sequential analysis in systematic reviews with meta-analysis. BMC Med. Res. Methodol. 2017, 17, 1–18. [Google Scholar] [CrossRef]
- Katoh, T.; Kaneko, S.; Boissy, R.; Watson, M.; Ikemura, K.; Bell, D.A. A pilot study testing the association between N-acetyltransferases 1 and 2 and risk of oral squamous cell carcinoma in Japanese people. Carcinogenesis 1998, 19, 1803–1807. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Ricks, S.; Doody, D.R.; Fitzgibbons, E.D.; Porter, P.L.; Schwartz, S.M. N-Acetyltransferase 2 polymorphisms, cigarette smoking and alcohol consumption, and oral squamous cell cancer risk. Carcinogenesis 2001, 22, 1993–1999. [Google Scholar] [CrossRef][Green Version]
- Chatzimichalis, M.; Xenellis, J.; Tzagaroulakis, A.; Sarof, P.; Banis, K.; Gazouli, M.; Bibas, A. GSTT1, GSTM1, GSTM3 and NAT2 polymorphisms in laryngeal squamous cell carcinoma in a Greek population. J. Laryngol. Otol. 2010, 124, 318–323. [Google Scholar] [CrossRef]
- Demokan, S.; Suoglu, Y.; Gözeler, M.; Demir, D.; Dalay, N. N-acetyltransferase 1 and 2 gene sequence variants and risk of head and neck cancer. Mol. Biol. Rep. 2010, 37, 3217–3226. [Google Scholar] [CrossRef]
- Fronhoffs, S.; Brüning, T.; Ortiz-Pallardo, E.; Bröde, P.; Koch, B.; Harth, V.; Sachinidis, A.; Bolt, H.M.; Herberhold, C.; Vetter, H. Real-time PCR analysis of the N-acetyltransferase NAT1 allele* 3,* 4,* 10,* 11,* 14 and* 17 polymorphism in squamous cell cancer of head and neck. Carcinogenesis 2001, 22, 1405–1412. [Google Scholar] [CrossRef] [PubMed]
- Gajecka, M.; Rydzanicz, M.; Jaskula-Sztul, R.; Kujawski, M.; Szyfter, W.; Szyfter, K. CYP1A1, CYP2D6, CYP2E1, NAT2, GSTM1 and GSTT1 polymorphisms or their combinations are associated with the increased risk of the laryngeal squamous cell carcinoma. Mutat. Res. Fundam. Mol. Mech. Mutagenesis 2005, 574, 112–123. [Google Scholar] [CrossRef]
- Gonzalez, M.; Alvarez, V.; Pello, M.; Menendez, M.; Suarez, C.; Coto, E. Genetic polymorphism of N-acetyltransferase-2, glutathione S-transferase-M1, and cytochromes P450IIE1 and P450IID6 in the susceptibility to head and neck cancer. J. Clin. Pathol. 1998, 51, 294–298. [Google Scholar] [CrossRef]
- Hahn, M.; Hagedorn, G.; Kuhlisch, E.; Schackert, H.K.; Eckelt, U. Genetic polymorphisms of drug-metabolizing enzymes and susceptibility to oral cavity cancer. Oral Oncol. 2002, 38, 486–490. [Google Scholar] [CrossRef]
- Harth, V.; Schäfer, M.; Abel, J.; Maintz, L.; Neuhaus, T.; Besuden, M.; Primke, R.; Wilkesmann, A.; Thier, R.; Vetter, H. Head and neck squamous-cell cancer and its association with polymorphic enzymes of xenobiotic metabolism and repair. J. Toxicol. Environ. Health Part A 2008, 71, 887–897. [Google Scholar] [CrossRef]
- Jourenkova-Mironova, N.; Wikman, H.; Bouchardy, C.; Mitrunen, K.; Dayer, P.; Benhamou, S.; Hirvonen, A. Role of arylamine N-acetyltransferase 1 and 2 (NAT1 and NAT2) genotypes in susceptibility to oral/pharyngeal and laryngeal cancers. Pharm. Genom. 1999, 9, 533–537. [Google Scholar]
- Rydzanicz, M.; Wierzbicka, M.; Gajęcka, M.; Szyfter, W.; Szyfter, K. The impact of genetic factors on the incidence of multiple primary tumors (MPT) of the head and neck. Cancer Lett. 2005, 224, 263–278. [Google Scholar] [CrossRef] [PubMed]
- Ünal, M.; Tamer, L.; Akbaş, Y.; Pata, Y.S.; Vayisoǧlu, Y.; Deǧirmenci, U.; Çamdeviren, H. Genetic polymorphism of N-acetyltransferase 2 in the susceptibility to laryngeal squamous cell carcinoma. Head Neck J. Sci. Spec. Head Neck 2005, 27, 1056–1060. [Google Scholar] [CrossRef]
- Varzim, G.; Monteiro, E.; Silva, R.; Pinheiro, C.; Lopes, C. Polymorphisms of arylamine N-acetyltransferase (NAT1 and NAT2) and larynx cancer susceptibility. ORL 2002, 64, 206–212. [Google Scholar] [CrossRef]
- Balaji, L.; Krishna, B.S.; Bhaskar, L. An unlikely role for the NAT2 genotypes and haplotypes in the oral cancer of south Indians. Arch. Oral Biol. 2012, 57, 513–518. [Google Scholar] [CrossRef]
- Cheng, Y.-J.; Chien, Y.-C.; Hildesheim, A.; Hsu, M.-M.; Chen, I.-H.; Chuang, J.; Chang, J.; Ma, Y.D.; Luo, C.-T.; Hsu, W.-L. No association between genetic polymorphisms of CYP1A1, GSTM1, GSTT1, GSTP1, NAT2, and nasopharyngeal carcinoma in Taiwan. Cancer Epidemiol. Prev. Biomark. 2003, 12, 179–180. [Google Scholar]
- Hou, Y.-Y.; Ou, H.-L.; Chu, S.-T.; Wu, P.-C.; Lu, P.-J.; Chi, C.-C.; Leung, K.-W.; Lee, C.-Y.; Wu, P.-H.; Hsiao, M. NAT2 slow acetylation haplotypes are associated with the increased risk of betel quid–related oral and pharyngeal squamous cell carcinoma. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endodontol. 2011, 112, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Lei, D.; Pan, X.; Guo, C.; Xu, F.; Zhang, L.; Liu, D.; Luan, X. Relationship between polymorphism of N-acetyltransferase 2 and genetic susceptibility to laryngeal carcinoma. Zhonghua Zhong Liu Za Zhi [Chin. J. Oncol.] 2002, 24, 154–156. [Google Scholar]
- Majumder, M.; Ghosh, S.; Roy, B. Association between polymorphisms at N-acetyltransferase 1 (NAT1) & risk of oral leukoplakia & cancer. Indian J. Med Res. 2012, 136, 605. [Google Scholar]
- Majumder, M.; Sikdar, N.; Ghosh, S.; Roy, B. Polymorphisms at XPD and XRCC1 DNA repair loci and increased risk of oral leukoplakia and cancer among NAT2 slow acetylators. Int. J. Cancer 2007, 120, 2148–2156. [Google Scholar] [CrossRef] [PubMed]
- Morita, S.; Yano, M.; Tsujinaka, T.; Akiyama, Y.; Taniguchi, M.; Kaneko, K.; Miki, H.; Fujii, T.; Yoshino, K.; Kusuoka, H. Genetic polymorphisms of drug-metabolizing enzymes and susceptibility to head-and-neck squamous-cell carcinoma. Int. J. Cancer 1999, 80, 685–688. [Google Scholar] [CrossRef]
- Tian, S.; Zhang, J.; Yuan, X.; Huang, M.; Guo, Z.; Chen, F.; Li, Q.; Guan, Z. The association between genetic polymorphisms of NAT1, NAT2 and susceptibility to laryngeal squamous carcinoma (LSCC) in Han population in Guangdong China. J. Modern Oncol. 2013, 6, 1213–1218. [Google Scholar]
- Buch, S.C.; Nazar-Stewart, V.; Weissfeld, J.L.; Romkes, M. Case–control study of oral and oropharyngeal cancer in whites and genetic variation in eight metabolic enzymes. Head Neck J. Sci. Spec. Head Neck 2008, 30, 1139–1147. [Google Scholar] [CrossRef]
- Gara, S.; Abdennebi, M.; Chatti, S.; Touati, S.; Ladgham, A.; Guemira, F. Association of NAT2 gene substitution mutation T341C with increased risk for head and neck cancer in Tunisia. Acta Oncol. 2007, 46, 834–837. [Google Scholar] [CrossRef] [PubMed]
- Marques, C.F.; Koifman, S.; Koifman, R.J.; Boffetta, P.; Brennan, P.; Hatagima, A. Influence of CYP1A1, CYP2E1, GSTM3 and NAT2 genetic polymorphisms in oral cancer susceptibility: Results from a case-control study in Rio de Janeiro. Oral Oncol. 2006, 42, 632–637. [Google Scholar] [CrossRef]
- Marques, C.R.; Da Silva, T.M.; De Albuquerque, D.M.; Chaves, M.S.; Marques Filho, M.F.; Oliveira, J.S.; Di Pietro, G.; Sousa, S.M.B.; Simoes, A.L.; Rios-Santos, F. NAT2, XRCC1 and hOGG1 polymorphisms, cigarette smoking, alcohol consumption and risk of upper aerodigestive tract cancer. Anticancer. Res. 2014, 34, 3217–3224. [Google Scholar]
- Olshan, A.F.; Weissler, M.C.; Watson, M.A.; Bell, D.A. GSTM1, GSTT1, GSTP1, CYP1A1, and NAT1 polymorphisms, tobacco use, and the risk of head and neck cancer. Cancer Epidemiol. Prev. Biomark. 2000, 9, 185–191. [Google Scholar]
- Richardson, M.; Garner, P.; Donegan, S. Interpretation of subgroup analyses in systematic reviews: A tutorial. Clin. Epidemiol. Glob. Health 2019, 7, 192–198. [Google Scholar] [CrossRef]
- Hickman, D.; Risch, A.; Buckle, V.; Spurr, N.; Jeremiah, S.; McCarthy, A.; Sim, E. Chromosomal localization of human genes for arylamine N-acetyltransferase. Biochem. J. 1994, 297, 441–445. [Google Scholar] [CrossRef] [PubMed]
- Moslehi, R.; Chatterjee, N.; Church, T.R.; Chen, J.; Yeager, M.; Weissfeld, J.; Hein, D.W.; Hayes, R.B. Cigarette smoking, N-acetyltransferase genes and the risk of advanced colorectal adenoma. Future Med. 2006, 7, 819–829. [Google Scholar] [CrossRef] [PubMed]
- Cascorbi, I.; Roots, I.; Brockmöller, J. Association of NAT1 and NAT2 polymorphisms to urinary bladder cancer: Significantly reduced risk in subjects with NAT1* 10. Cancer Res. 2001, 61, 5051–5056. [Google Scholar] [PubMed]
- Chen, J.; Stampfer, M.J.; Hough, H.L.; Garcia-Closas, M.; Willett, W.C.; Hennekens, C.H.; Kelsey, K.T.; Hunter, D.J. A prospective study of N-acetyltransferase genotype, red meat intake, and risk of colorectal cancer. Cancer Res. 1998, 58, 3307–3311. [Google Scholar]
- Badawi, A.F.; Hirvonen, A.; Bell, D.A.; Lang, N.P.; Kadlubar, F.F. Role of aromatic amine acetyltransferases, NAT1 and NAT2, in carcinogen-DNA adduct formation in the human urinary bladder. Cancer Res. 1995, 55, 5230–5237. [Google Scholar]

| The First Author, Publication Year | Country | Ethnicity | Control Source | Number | Mean Year | Male Percentage | Type of Tumor | Genotyping Method | Quality Score | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | Case | Control | Case | Control | |||||||
| Gonzalez, 1998 [41] | Spain | Caucasian | PB | 75 | 200 | 58.7 | 45 | 100 | 75 | Oral, pharyngeal, laryngeal | PCR-RFLP | 7 |
| Katoh, 1998 [35] | Japan | Asian | HB | 62 | 122 | 61.7 | 62.4 | 64.5 | 61.5 | Oral | PCR-RFLP | 7 |
| Henning, 1999 [23] | Germany | Caucasian | HB | 255 | 510 | 61.4 | NA | 90.6 | NA | Laryngeal | PCR | 7 |
| Jourenkova-Mironova, 1999 [44] | France | Caucasian | HB | 250 | 172 | 54.4 | 54.9 | 96 | 94.8 | Oral, pharyngeal, laryngeal | PCR-RFLP | 7 |
| Morita, 1999 [54] | Japan | Asian | PB | 145 | 164 | 59.0 | 49.8 | 86.9 | 62.2 | Oral, pharyngeal, laryngeal | PCR | 7 |
| Olshan, 2000 [60] | USA | Mixed | HB | 171 | 193 | 59.5 | 56.8 | 81.3 | 59.1 | Oral, pharyngeal, laryngeal | PCR | 7 |
| Chen, 2001 [36] | USA | Caucasian | PB | 341 | 552 | NA | NA | 70.4 | 71.6 | Oral | PCR-RFLP | 9 |
| Fronhoffs, 2001 [39] | Germany | Caucasian | HB | 291 | 300 | 59.8 | 47.1 | 80.1 | 58 | Oral, pharyngeal, laryngeal | RT-PCR | 6 |
| Hahn, 2002 [42] | Germany | Caucasian | PB | 94 | 92 | 61.5 | 45.1 | 65.9 | 51.1 | Oral | PCR-RFLP | 7 |
| Lei, 2002 [51] | China | Asian | HB | 62 | 56 | 60.2 | 58.2 | NA | NA | Laryngeal | PCR-RFLP | 7 |
| Varzim, 2002 [47] | Portugal | Caucasian | PB | 88 | 172 | 62.8 | 43.0 | 94.3 | 72.7 | Laryngeal | PCR-RFLP | 7 |
| Cheng, 2003 [49] | Taiwan | Asian | HB | 279 | 325 | NA | NA | NA | NA | Pharyngeal | PCR-RFLP | 6 |
| Gajecka, 2005 [40] | Poland | Caucasian | HB | 289 | 311 | 57.9 | 45.9 | 100 | 100 | Laryngeal | PCR-RFLP | 8 |
| Rydzanicz, 2005 [45] | Poland | Caucasian | HB | 266 | 143 | 61.6 | 53.1 | 95.1 | 100 | Oral, pharyngeal, laryngeal | PCR-RFLP | 8 |
| Unal, 2005 [46] | Turkey | Caucasian | HB | 45 | 104 | 53.5 | 50.0 | 93.3 | 65.4 | Laryngeal | PCR-RFLP | 7 |
| Marques, 2006 [58] | Brazil | Mixed | HB | 231 | 212 | 56.6 | 55.3 | 83.5 | 79.2 | Oral | PCR-RFLP | 8 |
| Gara, 2007 [57] | Tunisia | Mixed | HB | 64 | 160 | 50.7 | 53.6 | 65.6 | 45 | Oral, pharyngeal, laryngeal | PCR-RFLP | 7 |
| Majumder, 2007 [53] | India | Asian | HB | 297 | 342 | NA | NA | NA | NA | Oral | PCR-RFLP | 6 |
| Boccia, 2008 [22] | Italy | Caucasian | HB | 210 | 245 | 63.6 | 63.3 | 71.4 | 72.2 | Oral, pharyngeal, laryngeal | PCR-RFLP | 8 |
| Buch, 2008 [56] | USA | Mixed | PB | 182 | 399 | 58.7 | 58.7 | 87.4 | 75.7 | Oral | PCR-RFLP | 9 |
| Harth, 2008 [43] | Germany | Caucasian | HB | 312 | 300 | 59.7 | 47.2 | 80.4 | 58.7 | Oral, pharyngeal, laryngeal | PCR-RFLP | 6 |
| Chatzimichalis, 2010 [37] | Greece | Caucasian | HB | 88 | 102 | 66.5 | 62.5 | 87.5 | 74.5 | Laryngeal | PCR-RFLP | 8 |
| Demokan, 2010 [38] | Turkey | Caucasian | PB | 95 | 93 | 59.6 | 53.3 | 86.3 | 52.7 | Oral, pharyngeal, laryngeal | PCR | 8 |
| Hou, 2011 [50] | China | Asian | PB | 172 | 170 | 49.6 | 49.6 | 100 | 100 | Oral, pharyngeal | PCR-RFLP and Taqman | 9 |
| Balaji, 2012 [48] | India | Asian | HB | 157 | 132 | 53.1 | 55.1 | 54.8 | 34.8 | Oral | Taqman | 7 |
| Majumder, 2012 [52] | India | Asian | HB | 299 | 381 | NA | NA | NA | NA | Oral | PCR | 6 |
| Tian, 2013 [55] | China | Asian | PB | 233 | 102 | 60.0 | 60.0 | NA | NA | Laryngeal | PCR | 8 |
| Marques, 2014 [59] | Brazil | Mixed | PB | 101 | 141 | NA | NA | NA | NA | Oral, pharyngeal, laryngeal | PCR-RFLP | 7 |
| Author, Year | NAT1 | |||
|---|---|---|---|---|
| Case | Control | |||
| Slow | Rapid | Slow | Rapid | |
| Katoh, 1998 [35] | 9 | 53 | 46 | 76 |
| Henning, 1999 [23] | 144 | 109 | 232 | 164 |
| Jourenkova-Mironova, 1999 [44] | 141 | 109 | 98 | 74 |
| Olshan, 2000 [60] | 83 | 88 | 108 | 85 |
| Fronhoffs, 2001 [39] | 195 | 96 | 206 | 94 |
| Varzim, 2002 [47] | 48 | 40 | 107 | 65 |
| Demokan, 2010 [38] | 53 | 42 | 42 | 51 |
| Majumder, 2012 [52] | 128 | 171 | 168 | 213 |
| Author, Year | NAT2 | |||
| Case | Control | |||
| Slow | Rapid | Slow | Rapid | |
| Gonzalez, 1998 [41] | 28 | 47 | 37 | 163 |
| Katoh, 1998 [35] | 7 | 55 | 7 | 115 |
| Henning, 1999 [23] | 138 | 117 | 286 | 224 |
| Jourenkova-Mironova, 1999 [44] | 142 | 108 | 91 | 81 |
| Morita, 1999 [54] | 18 | 127 | 17 | 147 |
| Chen, 2001 [36] | 198 | 143 | 302 | 250 |
| Hahn, 2002 [42] | 59 | 35 | 57 | 35 |
| Lei, 2002 [51] | 50 | 12 | 34 | 22 |
| Varzim, 2002 [47] | 47 | 41 | 76 | 96 |
| Cheng, 2003 [49] | 39 | 240 | 54 | 271 |
| Gajecka, 2005 [40] | 127 | 162 | 165 | 146 |
| Rydzanicz, 2005 [45] | 131 | 135 | 72 | 71 |
| Unal, 2005 [46] | 15 | 30 | 7 | 97 |
| Marques, 2006 [58] | 29 | 202 | 38 | 174 |
| Gara, 2007 [57] | 33 | 31 | 59 | 101 |
| Majumder, 2007 [53] | 190 | 107 | 205 | 137 |
| Boccia, 2008 [22] | 109 | 101 | 128 | 117 |
| Buch, 2008 [56] | 84 | 98 | 224 | 175 |
| Harth, 2008 [43] | 189 | 123 | 181 | 119 |
| Chatzimichalis, 2010 [37] | 39 | 49 | 65 | 37 |
| Demokan, 2010 [38] | 50 | 45 | 45 | 48 |
| Hou, 2011 [50] | 46 | 126 | 33 | 137 |
| Balaji, 2012 [48] | 100 | 57 | 67 | 65 |
| Tian, 2013 [55] | 189 | 44 | 56 | 46 |
| Marques, 2014 [59] | 48 | 53 | 51 | 90 |
| Polymorphism | Variable (N) | OR | 95% CI | p-Value | I2 | Pheterogeneity |
|---|---|---|---|---|---|---|
| NAT1 | Overall (8) | 0.89 | 0.77, 1.02 | 0.09 | 48% | 0.06 |
| Ethnicity | ||||||
| Caucasian (5) | 0.96 | 0.80, 1.15 | 0.64 | 0% | 0.45 | |
| Asian (2) | 0.55 | 0.17, 1.80 | 0.32 | 87% | 0.005 | |
| Control source | ||||||
| Hospital-based (6) | 0.87 | 0.74, 1.01 | 0.06 | 46% | 0.10 | |
| Population-based (2) | 1.05 | 0.51, 2.17 | 0.90 | 72% | 0.06 | |
| Sample size | ||||||
| ≥200 (6) | 0.90 | 0.77, 1.04 | 0.15 | 0% | 0.87 | |
| <200 (2) | 0.67 | 0.13, 3.56 | 0.64 | 91% | 0.0007 | |
| Genotyping method | ||||||
| PCR (4) | 0.94 | 0.79, 1.14 | 0.54 | 26% | 0.26 | |
| PCR-RFLP (3) | 0.64 | 0.34, 1.18 | 0.15 | 74% | 0.02 | |
| Tumor type | ||||||
| Oral (2) | 0.55 | 0.17, 1.80 | 0.32 | 87% | 0.005 | |
| Laryngeal (2) | 0.87 | 0.67, 1.15 | 0.33 | 0% | 0.43 | |
| NAT2 | Overall (25) | 1.22 | 1.02, 1.46 | 0.03 | 74% | <0.00001 |
| Ethnicity | ||||||
| Caucasian (13) | 1.10 | 0.89, 1.37 | 0.38 | 71% | <0.0001 | |
| Asian (8) | 1.60 | 1.13, 2.26 | 0.008 | 69% | 0.002 | |
| Mixed (4) | 1.04 | 0.61, 1.77 | 0.89 | 79% | 0.003 | |
| Control source | ||||||
| Hospital-based (15) | 1.10 | 0.88, 1.37 | 0.39 | 71% | <0.0001 | |
| Population-based (10) | 1.41 | 1.04, 1.92 | 0.03 | 75% | <0.0001 | |
| Sample size | ||||||
| ≥200 (20) | 1.19 | 1.00, 1.42 | 0.05 | 70% | <0.00001 | |
| <200 (5) | 1.49 | 0.68, 3.29 | 0.32 | 85% | <0.0001 | |
| Genotyping method | ||||||
| PCR (4) | 1.47 | 0.77, 2.78 | 0.24 | 85% | 0.0002 | |
| PCR-RFLP (19) | 1.14 | 0.93, 1.39 | 0.21 | 72% | <0.00001 | |
| Tumor type | ||||||
| Oral (7) | 1.05 | 0.80,1.38 | 0.72 | 62% | 0.01 | |
| Pharyngeal (2) | 0.82 | 0.54, 1.24 | 0.35 | 0% | 0.96 | |
| Laryngeal (8) | 1.48 | 0.88, 2.51 | 0.14 | 88% | <0.00001 |
| Polymorphism | Variable | Point Estimate | Standard Error | Lower Limit | Upper Limit | Z-Value | p-Value | |
|---|---|---|---|---|---|---|---|---|
| NAT1 | Publication year | Slope | 0.01830 | 0.01361 | −0.00837 | 0.04497 | 1.34462 | 0.17875 |
| Intercept | −36.77098 | 27.26207 | −90.20365 | 16.66169 | −1.34880 | 0.17740 | ||
| Sample size | Slope | 0.00027 | 0.00045 | −0.00060 | 0.00115 | 0.61240 | 0.54027 | |
| Intercept | −0.25993 | 0.24912 | −0.74819 | 0.22833 | −1.04340 | 0.29676 | ||
| Mean age of cases | Slope | −0.01179 | 0.03248 | −0.07546 | 0.05186 | −0.36300 | 0.71660 | |
| Intercept | 0.57037 | 1.93376 | −3.21972 | 4.36047 | 0.29496 | 0.76803 | ||
| Mean age of controls | Slope | −0.02263 | 0.03624 | −0.09365 | 0.04839 | −0.62459 | 0.53224 | |
| Intercept | 1.17938 | 2.13386 | −3.00290 | 5.36167 | 0.55270 | 0.58047 | ||
| Male percentage of cases | Slope | −0.01131 | 0.01256 | −0.03593 | 0.01331 | −0.90074 | 0.36773 | |
| Intercept | 0.86738 | 1.11137 | −1.31087 | 3.04562 | 0.78046 | 0.43512 | ||
| Male percentage of controls | Slope | −0.00268 | 0.00617 | −0.01478 | 0.00942 | −0.43474 | 0.066375 | |
| Intercept | 0.03230 | 0.43459 | −0.81948 | 0.88409 | 0.07433 | 0.94074 | ||
| NAT2 | Publication year | Slope | 0.00944 | 0.01016 | −0.01047 | 0.02934 | 0.092942 | 0.35267 |
| Intercept | −18.82284 | 20.36308 | −58.73373 | 21.08806 | −0.92436 | 0.35530 | ||
| Sample size | Slope | −0.00080 | 0.00020 | −0.00120 | −0.00040 | −3.91239 | 0.00009 | |
| Intercept | 0.50882 | 0.11300 | 0.28733 | 0.73030 | 4.50265 | 0.00001 | ||
| Mean age of cases | Slope | −0.04050 | 0.01356 | −0.06706 | −0.01393 | −2.098776 | 0.00281 | |
| Intercept | 2.47888 | 0.80007 | 0.91077 | 4.04699 | 3.09832 | 0.00195 | ||
| Mean age of controls | Slope | −0.00438 | 0.00889 | −0.02180 | 0.01305 | −0.49203 | 0.62270 | |
| Intercept | 0.34691 | 0.47403 | −0.58217 | 1.27600 | 0.73184 | 0.46427 | ||
| Male percentage of cases | Slope | −0.0629 | 0.00393 | −0.01399 | 0.00141 | −1.60201 | 0.10915 | |
| Intercept | 0.57366 | 0.33428 | −0.08152 | 1.22884 | 1.71610 | 0.08614 | ||
| Male percentage of controls | Slope | −0.00785 | 0.00289 | −0.01351 | −0.00219 | −2.71989 | 0.00653 | |
| Intercept | 0.64373 | 0.22152 | 0.20956 | 1.07790 | 2.90598 | 0.00366 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohammadi, H.; Roochi, M.M.; Sadeghi, M.; Garajei, A.; Heidar, H.; Ghaderi, B.; Tadakamadla, J.; Meybodi, A.A.; Dallband, M.; Mostafavi, S.; et al. Association of N-acetyltransferases 1 and 2 Polymorphisms with Susceptibility to Head and Neck Cancers—A Meta-Analysis, Meta-Regression, and Trial Sequential Analysis. Medicina 2021, 57, 1095. https://doi.org/10.3390/medicina57101095
Mohammadi H, Roochi MM, Sadeghi M, Garajei A, Heidar H, Ghaderi B, Tadakamadla J, Meybodi AA, Dallband M, Mostafavi S, et al. Association of N-acetyltransferases 1 and 2 Polymorphisms with Susceptibility to Head and Neck Cancers—A Meta-Analysis, Meta-Regression, and Trial Sequential Analysis. Medicina. 2021; 57(10):1095. https://doi.org/10.3390/medicina57101095
Chicago/Turabian StyleMohammadi, Hady, Mehrnoush Momeni Roochi, Masoud Sadeghi, Ata Garajei, Hosein Heidar, Bayazid Ghaderi, Jyothi Tadakamadla, Ali Aghaie Meybodi, Mohsen Dallband, Sarton Mostafavi, and et al. 2021. "Association of N-acetyltransferases 1 and 2 Polymorphisms with Susceptibility to Head and Neck Cancers—A Meta-Analysis, Meta-Regression, and Trial Sequential Analysis" Medicina 57, no. 10: 1095. https://doi.org/10.3390/medicina57101095
APA StyleMohammadi, H., Roochi, M. M., Sadeghi, M., Garajei, A., Heidar, H., Ghaderi, B., Tadakamadla, J., Meybodi, A. A., Dallband, M., Mostafavi, S., Mostafavi, M., Salehi, M., Sadeghi-Bahmani, D., & Brand, S. (2021). Association of N-acetyltransferases 1 and 2 Polymorphisms with Susceptibility to Head and Neck Cancers—A Meta-Analysis, Meta-Regression, and Trial Sequential Analysis. Medicina, 57(10), 1095. https://doi.org/10.3390/medicina57101095








