How Dietary Factors Affect DNA Methylation: Lesson from Epidemiological Studies
Abstract
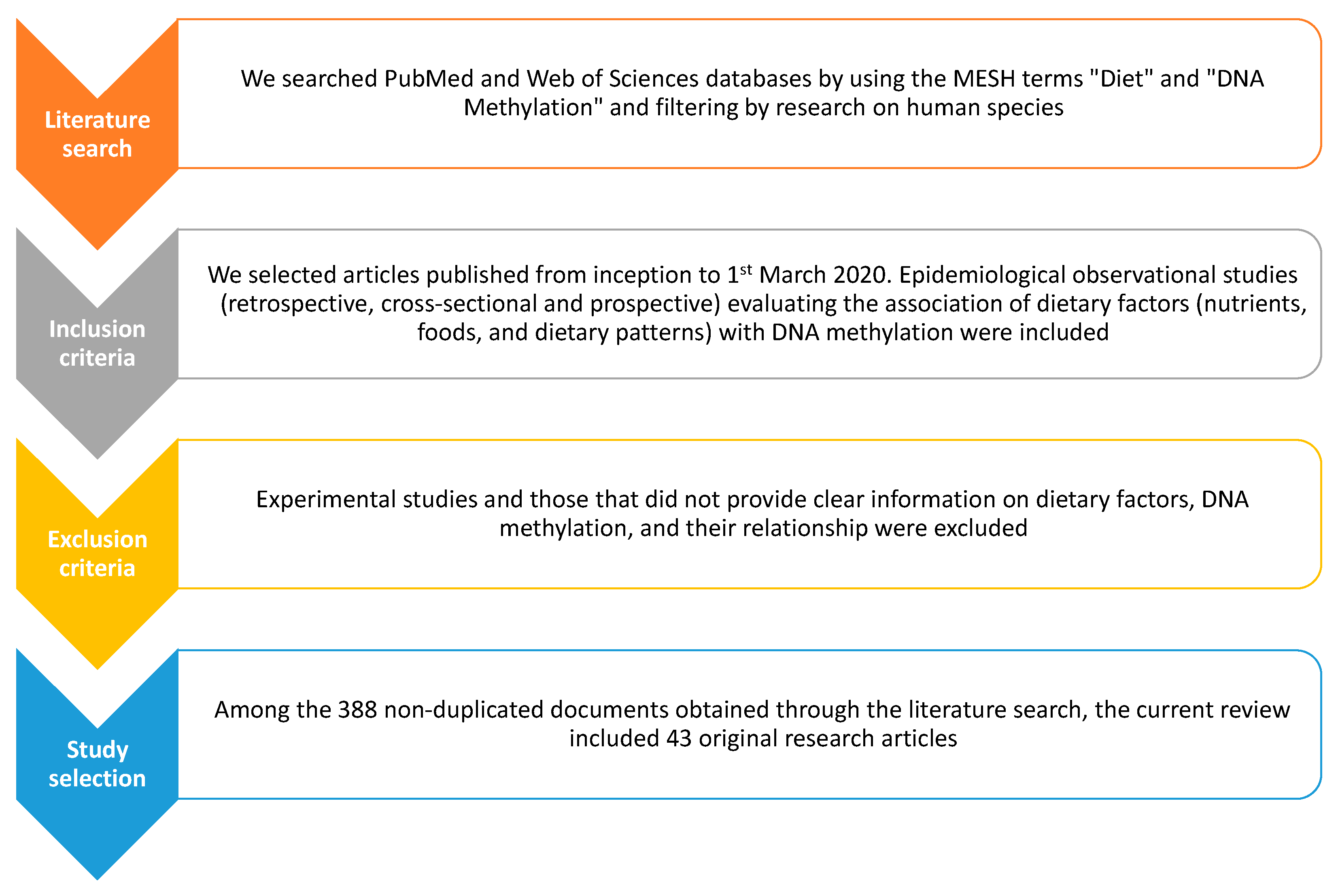
1. Introduction
2. Diet and DNA Methylation: The First Evidence from Cancer Research
2.1. Colon Cancer
2.2. Gastric Cancer
2.3. Breast Cancer
2.4. Other Cancers
3. The Effect of Diet on DNA Methylation in “Healthy” People
4. Prospects for Studying Obesity, Metabolic Disorders, and Cardiovascular Diseases
5. The Relationship between Dietary Factors and DNA Methylation in Mothers and Their Children
6. Methodological Pitfalls and Future Challenges
7. Discussion
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Sales, N.M.; Pelegrini, P.B.; Goersch, M.C. Nutrigenomics: Definitions and advances of this new science. J. Nutr. Metab. 2014, 2014, 202759. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.W.; Claycombe, K.J.; Martinez, J.A.; Friso, S.; Schalinske, K.L. Nutritional epigenomics: A portal to disease prevention. Adv. Nutr. 2013, 4, 530–532. [Google Scholar] [CrossRef] [PubMed]
- Wolffe, A.P.; Guschin, D. Review: Chromatin structural features and targets that regulate transcription. J. Struct. Biol. 2000, 129, 102–122. [Google Scholar] [CrossRef] [PubMed]
- Langemeijer, S.M.; Kuiper, R.P.; Berends, M.; Knops, R.; Aslanyan, M.G.; Massop, M.; Stevens-Linders, E.; van Hoogen, P.; van Kessel, A.G.; Raymakers, R.A.; et al. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat. Genet. 2009, 41, 838–842. [Google Scholar] [CrossRef]
- Auclair, G.; Weber, M. Mechanisms of DNA methylation and demethylation in mammals. Biochimie 2012, 94, 2202–2211. [Google Scholar] [CrossRef]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef]
- Jiang, M.H.; Fei, J.; Lan, M.S.; Lu, Z.P.; Liu, M.; Fan, W.W.; Gao, X.; Lu, D.R. Hypermethylation of hepatic Gck promoter in ageing rats contributes to diabetogenic potential. Diabetologia 2008, 51, 1525–1533. [Google Scholar] [CrossRef]
- Milagro, F.I.; Campión, J.; Cordero, P.; Goyenechea, E.; Gómez-Uriz, A.M.; Abete, I.; Zulet, M.A.; Martínez, J.A. A dual epigenomic approach for the search of obesity biomarkers: DNA methylation in relation to diet-induced weight loss. FASEB J. 2011, 25, 1378–1389. [Google Scholar] [CrossRef]
- Maier, S.; Olek, A. Diabetes: A candidate disease for efficient DNA methylation profiling. J. Nutr. 2002, 132, 2440S–2443S. [Google Scholar] [CrossRef]
- Chowdhury, S.; Erickson, S.W.; MacLeod, S.L.; Cleves, M.A.; Hu, P.; Karim, M.A.; Hobbs, C.A. Maternal genome-wide DNA methylation patterns and congenital heart defects. PLoS ONE 2011, 6, e16506. [Google Scholar] [CrossRef]
- Agodi, A.; Barchitta, M.; Quattrocchi, A.; Maugeri, A.; Vinciguerra, M. DAPK1 Promoter Methylation and Cervical Cancer Risk: A Systematic Review and a Meta-Analysis. PLoS ONE 2015, 10, e0135078. [Google Scholar] [CrossRef] [PubMed]
- Agodi, A.; Barchitta, M.; Maugeri, A.; Basile, G.; Zamboni, M.; Bernardini, G.; Corona, D.; Veroux, M. Unveiling the Role of DNA Methylation in Kidney Transplantation: Novel Perspectives toward Biomarker Identification. Biomed. Res. Int. 2019, 2019, 1602539. [Google Scholar] [CrossRef] [PubMed]
- Barchitta, M.; Quattrocchi, A.; Maugeri, A.; Vinciguerra, M.; Agodi, A. LINE-1 hypomethylation in blood and tissue samples as an epigenetic marker for cancer risk: A systematic review and meta-analysis. PLoS ONE 2014, 9, e109478. [Google Scholar] [CrossRef] [PubMed]
- Barchitta, M.; Quattrocchi, A.; Maugeri, A.; Canto, C.; La Rosa, N.; Cantarella, M.A.; Spampinato, G.; Scalisi, A.; Agodi, A. LINE-1 hypermethylation in white blood cell DNA is associated with high-grade cervical intraepithelial neoplasia. BMC Cancer 2017, 17, 601. [Google Scholar] [CrossRef] [PubMed]
- Barchitta, M.; Maugeri, A.; Li Destri, G.; Basile, G.; Agodi, A. Epigenetic Biomarkers in Colorectal Cancer Patients Receiving Adjuvant or Neoadjuvant Therapy: A Systematic Review of Epidemiological Studies. Int. J. Mol. Sci. 2019, 20, 3842. [Google Scholar] [CrossRef]
- Maugeri, A.; Barchitta, M.; Fallico, M.; Castellino, N.; Reibaldi, M.; Agodi, A. Characterization of SIRT1/DNMTs Functions and LINE-1 Methylation in Patients with Age-Related Macular Degeneration. J. Clin. Med. 2019, 8, 159. [Google Scholar] [CrossRef]
- Burris, H.H.; Baccarelli, A.A. Environmental epigenetics: From novelty to scientific discipline. J. Appl. Toxicol. 2014, 34, 113–116. [Google Scholar] [CrossRef]
- Ong, T.P.; Moreno, F.S.; Ross, S.A. Targeting the epigenome with bioactive food components for cancer prevention. J. Nutr. Nutr. 2011, 4, 275–292. [Google Scholar] [CrossRef]
- Piyathilake, C.J.; Badiga, S.; Kabagambe, E.K.; Azuero, A.; Alvarez, R.D.; Johanning, G.L.; Partridge, E.E. A dietary pattern associated with LINE-1 methylation alters the risk of developing cervical intraepithelial neoplasia. Cancer Prev. Res. 2012, 5, 385–392. [Google Scholar] [CrossRef]
- Ono, H.; Iwasaki, M.; Kuchiba, A.; Kasuga, Y.; Yokoyama, S.; Onuma, H.; Nishimura, H.; Kusama, R.; Ohnami, S.; Sakamoto, H.; et al. Association of dietary and genetic factors related to one-carbon metabolism with global methylation level of leukocyte DNA. Cancer Sci. 2012, 103, 2159–2164. [Google Scholar] [CrossRef]
- Zhang, F.F.; Santella, R.M.; Wolff, M.; Kappil, M.A.; Markowitz, S.B.; Morabia, A. White blood cell global methylation and IL-6 promoter methylation in association with diet and lifestyle risk factors in a cancer-free population. Epigenetics 2012, 7, 606–614. [Google Scholar] [CrossRef]
- Agodi, A.; Barchitta, M.; Quattrocchi, A.; Maugeri, A.; Canto, C.; Marchese, A.E.; Vinciguerra, M. Low fruit consumption and folate deficiency are associated with LINE-1 hypomethylation in women of a cancer-free population. Genes Nutr. 2015, 10, 480. [Google Scholar] [CrossRef] [PubMed]
- Moore, L.E.; Pfeiffer, R.M.; Poscablo, C.; Real, F.X.; Kogevinas, M.; Silverman, D.; García-Closas, R.; Chanock, S.; Tardón, A.; Serra, C.; et al. Genomic DNA hypomethylation as a biomarker for bladder cancer susceptibility in the Spanish Bladder Cancer Study: A case-control study. Lancet Oncol. 2008, 9, 359–366. [Google Scholar] [CrossRef]
- Choi, J.Y.; James, S.R.; Link, P.A.; McCann, S.E.; Hong, C.C.; Davis, W.; Nesline, M.K.; Ambrosone, C.B.; Karpf, A.R. Association between global DNA hypomethylation in leukocytes and risk of breast cancer. Carcinogenesis 2009, 30, 1889–1897. [Google Scholar] [CrossRef]
- Zhang, F.F.; Morabia, A.; Carroll, J.; Gonzalez, K.; Fulda, K.; Kaur, M.; Vishwanatha, J.K.; Santella, R.M.; Cardarelli, R. Dietary patterns are associated with levels of global genomic DNA methylation in a cancer-free population. J. Nutr. 2011, 141, 1165–1171. [Google Scholar] [CrossRef] [PubMed]
- Ceccarelli, V.; Racanicchi, S.; Martelli, M.P.; Nocentini, G.; Fettucciari, K.; Riccardi, C.; Marconi, P.; Di Nardo, P.; Grignani, F.; Binaglia, L.; et al. Eicosapentaenoic acid demethylates a single CpG that mediates expression of tumor suppressor CCAAT/enhancer-binding protein delta in U937 leukemia cells. J. Biol. Chem. 2011, 286, 27092–27102. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, A.; Dangat, K.; Kale, A.; Sable, P.; Chavan-Gautam, P.; Joshi, S. Effects of altered maternal folic acid, vitamin B12 and docosahexaenoic acid on placental global DNA methylation patterns in Wistar rats. PLoS ONE 2011, 6, e17706. [Google Scholar] [CrossRef]
- Kiec-Wilk, B.; Polus, A.; Mikolajczyk, M.; Mathers, J.C. Beta-carotene and arachidonic acid induced DNA methylation and the regulation of pro-chemotactic activity of endothelial cells and its progenitors. J. Physiol. Pharmacol. 2007, 58, 757–766. [Google Scholar]
- Zam, W.; Khadour, A. Impact of Phytochemicals and Dietary Patterns on Epigenome and Cancer. Nutr. Cancer 2017, 69, 184–200. [Google Scholar] [CrossRef]
- Maugeri, A.; Mazzone, M.G.; Giuliano, F.; Vinciguerra, M.; Basile, G.; Barchitta, M.; Agodi, A. Curcumin Modulates DNA Methyltransferase Functions in a Cellular Model of Diabetic Retinopathy. Oxid. Med. Cell Longev. 2018, 2018, 5407482. [Google Scholar] [CrossRef]
- Maugeri, A.; Barchitta, M.; Mazzone, M.G.; Giuliano, F.; Basile, G.; Agodi, A. Resveratrol Modulates SIRT1 and DNMT Functions and Restores LINE-1 Methylation Levels in ARPE-19 Cells under Oxidative Stress and Inflammation. Int. J. Mol. Sci. 2018, 19, 2118. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Kelly, T.K.; Jones, P.A. Epigenetics in cancer. Carcinogenesis 2010, 31, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Van Engeland, M.; Weijenberg, M.P.; Roemen, G.M.; Brink, M.; de Bruïne, A.P.; Goldbohm, R.A.; van den Brandt, P.A.; Baylin, S.B.; de Goeij, A.F.; Herman, J.G. Effects of dietary folate and alcohol intake on promoter methylation in sporadic colorectal cancer: The Netherlands cohort study on diet and cancer. Cancer Res. 2003, 63, 3133–3137. [Google Scholar] [PubMed]
- Nan, H.M.; Song, Y.J.; Yun, H.Y.; Park, J.S.; Kim, H. Effects of dietary intake and genetic factors on hypermethylation of the hMLH1 gene promoter in gastric cancer. World J. Gastroenterol. 2005, 11, 3834–3841. [Google Scholar] [CrossRef] [PubMed]
- Yuasa, Y.; Nagasaki, H.; Akiyama, Y.; Sakai, H.; Nakajima, T.; Ohkura, Y.; Takizawa, T.; Koike, M.; Tani, M.; Iwai, T.; et al. Relationship between CDX2 gene methylation and dietary factors in gastric cancer patients. Carcinogenesis 2005, 26, 193–200. [Google Scholar] [CrossRef]
- Kraunz, K.S.; Hsiung, D.; McClean, M.D.; Liu, M.; Osanyingbemi, J.; Nelson, H.H.; Kelsey, K.T. Dietary folate is associated with p16(INK4A) methylation in head and neck squamous cell carcinoma. Int. J. Cancer 2006, 119, 1553–1557. [Google Scholar] [CrossRef]
- Slattery, M.L.; Curtin, K.; Sweeney, C.; Levin, T.R.; Potter, J.; Wolff, R.K.; Albertsen, H.; Samowitz, W.S. Diet and lifestyle factor associations with CpG island methylator phenotype and BRAF mutations in colon cancer. Int. J. Cancer 2007, 120, 656–663. [Google Scholar] [CrossRef]
- Mas, S.; Lafuente, M.J.; Crescenti, A.; Trias, M.; Ballesta, A.; Molina, R.; Zheng, S.; Wiencke, J.K.; Lafuente, A. Lower specific micronutrient intake in colorectal cancer patients with tumors presenting promoter hypermethylation in p16(INK4a), p4(ARF) and hMLH1. Anticancer Res. 2007, 27, 1151–1156. [Google Scholar]
- De Vogel, S.; Bongaerts, B.W.; Wouters, K.A.; Kester, A.D.; Schouten, L.J.; de Goeij, A.F.; de Bruïne, A.P.; Goldbohm, R.A.; van den Brandt, P.A.; van Engeland, M.; et al. Associations of dietary methyl donor intake with MLH1 promoter hypermethylation and related molecular phenotypes in sporadic colorectal cancer. Carcinogenesis 2008, 29, 1765–1773. [Google Scholar] [CrossRef]
- Schernhammer, E.S.; Giovannucci, E.; Kawasaki, T.; Rosner, B.; Fuchs, C.S.; Ogino, S. Dietary folate, alcohol and B vitamins in relation to LINE-1 hypomethylation in colon cancer. Gut 2010, 59, 794–799. [Google Scholar] [CrossRef]
- Curtin, K.; Samowitz, W.S.; Ulrich, C.M.; Wolff, R.K.; Herrick, J.S.; Caan, B.J.; Slattery, M.L. Nutrients in folate-mediated, one-carbon metabolism and the risk of rectal tumors in men and women. Nutr. Cancer 2011, 63, 357–366. [Google Scholar] [CrossRef] [PubMed]
- Tao, M.H.; Mason, J.B.; Marian, C.; McCann, S.E.; Platek, M.E.; Millen, A.; Ambrosone, C.; Edge, S.B.; Krishnan, S.S.; Trevisan, M.; et al. Promoter methylation of E-cadherin, p16, and RAR-β(2) genes in breast tumors and dietary intake of nutrients important in one-carbon metabolism. Nutr. Cancer 2011, 63, 1143–1150. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Gammon, M.D.; Jefferson, E.; Zhang, Y.; Cho, Y.H.; Wetmur, J.G.; Teitelbaum, S.L.; Bradshaw, P.T.; Terry, M.B.; Garbowski, G.; et al. The influence of one-carbon metabolism on gene promoter methylation in a population-based breast cancer study. Epigenetics 2011, 6, 1276–1283. [Google Scholar] [CrossRef]
- Zhang, Y.W.; Eom, S.Y.; Yim, D.H.; Song, Y.J.; Yun, H.Y.; Park, J.S.; Youn, S.J.; Kim, B.S.; Kim, Y.D.; Kim, H. Evaluation of the relationship between dietary factors, CagA-positive Helicobacter pylori infection, and RUNX3 promoter hypermethylation in gastric cancer tissue. World J. Gastroenterol. 2013, 19, 1778–1787. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Yang, C.; Yang, L.; Qi, C.; Tian, S.; Han, Y.; Dou, Y.; Ma, Y.; Tian, D.; Zheng, Y. Association of roasting meat intake with the risk of esophageal squamous cell carcinoma of Kazakh Chinese via affecting promoter methylation of p16 gene. Asia Pac. J. Clin. Nutr. 2014, 23, 488–497. [Google Scholar] [CrossRef]
- Nishihara, R.; Wang, M.; Qian, Z.R.; Baba, Y.; Yamauchi, M.; Mima, K.; Sukawa, Y.; Kim, S.A.; Inamura, K.; Zhang, X.; et al. Alcohol, one-carbon nutrient intake, and risk of colorectal cancer according to tumor methylation level of IGF2 differentially methylated region. Am. J. Clin. Nutr. 2014, 100, 1479–1488. [Google Scholar] [CrossRef] [PubMed]
- Pirouzpanah, S.; Taleban, F.A.; Mehdipour, P.; Atri, M. Association of folate and other one-carbon related nutrients with hypermethylation status and expression of RARB, BRCA1, and RASSF1A genes in breast cancer patients. J. Mol. Med. 2015, 93, 917–934. [Google Scholar] [CrossRef] [PubMed]
- Mehta, R.S.; Song, M.; Nishihara, R.; Drew, D.A.; Wu, K.; Qian, Z.R.; Fung, T.T.; Hamada, T.; Masugi, Y.; da Silva, A.; et al. Dietary Patterns and Risk of Colorectal Cancer: Analysis by Tumor Location and Molecular Subtypes. Gastroenterology 2017, 152, 1944–1953.e1941. [Google Scholar] [CrossRef]
- Ferrari, A.; Torrezan, G.T.; Carraro, D.M.; Aguiar Junior, S. Association of Folate and Vitamins Involved in the 1-Carbon Cycle with Polymorphisms in the Methylenetetrahydrofolate Reductase Gene (MTHFR) and Global DNA Methylation in Patients with Colorectal Cancer. Nutrients 2019, 11, 1368. [Google Scholar] [CrossRef]
- Van Engeland, M.; Derks, S.; Smits, K.M.; Meijer, G.A.; Herman, J.G. Colorectal cancer epigenetics: Complex simplicity. J. Clin. Oncol. 2011, 29, 1382–1391. [Google Scholar] [CrossRef]
- Stidley, C.A.; Picchi, M.A.; Leng, S.; Willink, R.; Crowell, R.E.; Flores, K.G.; Kang, H.; Byers, T.; Gilliland, F.D.; Belinsky, S.A. Multivitamins, folate, and green vegetables protect against gene promoter methylation in the aerodigestive tract of smokers. Cancer Res. 2010, 70, 568–574. [Google Scholar] [CrossRef]
- Leng, S.; Picchi, M.A.; Kang, H.; Wu, G.; Filipczak, P.T.; Juri, D.E.; Zhang, X.; Gauderman, W.J.; Gilliland, F.D.; Belinsky, S.A. Dietary Nutrient Intake, Ethnicity, and Epigenetic Silencing of Lung Cancer Genes Detected in Sputum in New Mexican Smokers. Cancer Prev. Res. 2018, 11, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Mandaviya, P.R.; Joehanes, R.; Brody, J.; Castillo-Fernandez, J.E.; Dekkers, K.F.; Do, A.N.; Graff, M.; Hänninen, I.K.; Tanaka, T.; de Jonge, E.A.L.; et al. Association of dietary folate and vitamin B-12 intake with genome-wide DNA methylation in blood: A large-scale epigenome-wide association analysis in 5841 individuals. Am. J. Clin. Nutr. 2019, 110, 437–450. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.F.; Cardarelli, R.; Carroll, J.; Fulda, K.G.; Kaur, M.; Gonzalez, K.; Vishwanatha, J.K.; Santella, R.M.; Morabia, A. Significant differences in global genomic DNA methylation by gender and race/ethnicity in peripheral blood. Epigenetics 2011, 6, 623–629. [Google Scholar] [CrossRef] [PubMed]
- Perng, W.; Villamor, E.; Shroff, M.R.; Nettleton, J.A.; Pilsner, J.R.; Liu, Y.; Diez-Roux, A.V. Dietary intake, plasma homocysteine, and repetitive element DNA methylation in the Multi-Ethnic Study of Atherosclerosis (MESA). Nutr. Metab. Cardiovasc. Dis. 2014, 24, 614–622. [Google Scholar] [CrossRef]
- Barchitta, M.; Maugeri, A.; Quattrocchi, A.; Barone, G.; Mazzoleni, P.; Catalfo, A.; De Guidi, G.; Iemmolo, M.G.; Crimi, N.; Agodi, A. Mediterranean Diet and Particulate Matter Exposure Are Associated With LINE-1 Methylation: Results From a Cross-Sectional Study in Women. Front. Genet. 2018, 9, 514. [Google Scholar] [CrossRef] [PubMed]
- Barchitta, M.; Maugeri, A.; Magnano San Lio, R.; Favara, G.; La Rosa, M.C.; La Mastra, C.; Quattrocchi, A.; Agodi, A. Dietary Patterns are Associated with Leukocyte LINE-1 Methylation in Women: A Cross-Sectional Study in Southern Italy. Nutrients 2019, 11, 1843. [Google Scholar] [CrossRef]
- Nicodemus-Johnson, J.; Sinnott, R.A. Fruit and Juice Epigenetic Signatures Are Associated with Independent Immunoregulatory Pathways. Nutrients 2017, 9, 752. [Google Scholar] [CrossRef]
- Marques-Rocha, J.L.; Milagro, F.I.; Mansego, M.L.; Mourão, D.M.; Martínez, J.A.; Bressan, J. LINE-1 methylation is positively associated with healthier lifestyle but inversely related to body fat mass in healthy young individuals. Epigenetics 2016, 11, 49–60. [Google Scholar] [CrossRef]
- Shimazu, T.; Asada, K.; Charvat, H.; Kusano, C.; Otake, Y.; Kakugawa, Y.; Watanabe, H.; Gotoda, T.; Ushijima, T.; Tsugane, S. Association of gastric cancer risk factors with DNA methylation levels in gastric mucosa of healthy Japanese: A cross-sectional study. Carcinogenesis 2015, 36, 1291–1298. [Google Scholar] [CrossRef]
- Hermsdorff, H.H.; Mansego, M.L.; Campión, J.; Milagro, F.I.; Zulet, M.A.; Martínez, J.A. TNF-alpha promoter methylation in peripheral white blood cells: Relationship with circulating TNFα, truncal fat and n-6 PUFA intake in young women. Cytokine 2013, 64, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Carraro, J.C.; Hermsdorff, H.H.; Mansego, M.L.; Zulet, M.; Milagro, F.I.; Bressan, J.; Martínez, J.A. Higher Fruit Intake Is Related to TNF-α Hypomethylation and Better Glucose Tolerance in Healthy Subjects. Lifestyle Genom. 2016, 9, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Lopez, O.; Samblas, M.; Milagro, F.I.; Zulet, M.A.; Mansego, M.L.; Riezu-Boj, J.I.; Martinez, J.A. Association of low dietary folate intake with lower CAMKK2 gene methylation, adiposity, and insulin resistance in obese subjects. Nutr. Res. 2018, 50, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Lopez, O.; Riezu-Boj, J.I.; Milagro, F.I.; Martinez, J.A.; Project, M. Dopamine gene methylation patterns are associated with obesity markers and carbohydrate intake. Brain Behav. 2018, 8, e01017. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Uriz, A.M.; Goyenechea, E.; Campión, J.; de Arce, A.; Martinez, M.T.; Puchau, B.; Milagro, F.I.; Abete, I.; Martínez, J.A.; Lopez de Munain, A. Epigenetic patterns of two gene promoters (TNF-α and PON) in stroke considering obesity condition and dietary intake. J. Physiol. Biochem. 2014, 70, 603–614. [Google Scholar] [CrossRef]
- Heijmans, B.T.; Tobi, E.W.; Stein, A.D.; Putter, H.; Blauw, G.J.; Susser, E.S.; Slagboom, P.E.; Lumey, L.H. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc. Natl. Acad. Sci. USA 2008, 105, 17046–17049. [Google Scholar] [CrossRef]
- Tobi, E.W.; Lumey, L.H.; Talens, R.P.; Kremer, D.; Putter, H.; Stein, A.D.; Slagboom, P.E.; Heijmans, B.T. DNA methylation differences after exposure to prenatal famine are common and timing- and sex-specific. Hum. Mol. Genet. 2009, 18, 4046–4053. [Google Scholar] [CrossRef]
- Tobi, E.W.; Goeman, J.J.; Monajemi, R.; Gu, H.; Putter, H.; Zhang, Y.; Slieker, R.C.; Stok, A.P.; Thijssen, P.E.; Müller, F.; et al. DNA methylation signatures link prenatal famine exposure to growth and metabolism. Nat. Commun. 2014, 5, 5592. [Google Scholar] [CrossRef]
- Boeke, C.E.; Baccarelli, A.; Kleinman, K.P.; Burris, H.H.; Litonjua, A.A.; Rifas-Shiman, S.L.; Tarantini, L.; Gillman, M. Gestational intake of methyl donors and global LINE-1 DNA methylation in maternal and cord blood: Prospective results from a folate-replete population. Epigenetics 2012, 7, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Taylor, R.M.; Smith, R.; Collins, C.E.; Mossman, D.; Wong-Brown, M.W.; Chan, E.C.; Evans, T.J.; Attia, J.R.; Smith, T.; Butler, T.; et al. Methyl-Donor and Cofactor Nutrient Intakes in the First 2-3 Years and Global DNA Methylation at Age 4: A Prospective Cohort Study. Nutrients 2018, 10, 273. [Google Scholar] [CrossRef]
- Haggarty, P.; Hoad, G.; Campbell, D.M.; Horgan, G.W.; Piyathilake, C.; McNeill, G. Folate in pregnancy and imprinted gene and repeat element methylation in the offspring. Am. J. Clin. Nutr. 2013, 97, 94–99. [Google Scholar] [CrossRef] [PubMed]
- Rijlaarsdam, J.; Cecil, C.A.; Walton, E.; Mesirow, M.S.; Relton, C.L.; Gaunt, T.R.; McArdle, W.; Barker, E.D. Prenatal unhealthy diet, insulin-like growth factor 2 gene (IGF2) methylation, and attention deficit hyperactivity disorder symptoms in youth with early-onset conduct problems. J. Child Psychol. Psychiatry 2017, 58, 19–27. [Google Scholar] [CrossRef]
- Pauwels, S.; Ghosh, M.; Duca, R.C.; Bekaert, B.; Freson, K.; Huybrechts, I.; AS Langie, S.; Koppen, G.; Devlieger, R.; Godderis, L. Dietary and supplemental maternal methyl-group donor intake and cord blood DNA methylation. Epigenetics 2017, 12, 1–10. [Google Scholar] [CrossRef] [PubMed]
- McCullough, L.E.; Miller, E.E.; Calderwood, L.E.; Shivappa, N.; Steck, S.E.; Forman, M.R.; A Mendez, M.; Maguire, R.; Fuemmeler, B.F.; Kollins, S.H.; et al. Maternal inflammatory diet and adverse pregnancy outcomes: Circulating cytokines and genomic imprinting as potential regulators? Epigenetics 2017, 12, 688–697. [Google Scholar] [CrossRef] [PubMed]
- Willett, W. Nutritional Epidemiology; Oxford University Press: Oxford, UK, 2012. [Google Scholar]
- Maugeri, A.; Barchitta, M. A Systematic Review of Ecological Momentary Assessment of Diet: Implications and Perspectives for Nutritional Epidemiology. Nutrients 2019, 11, 2696. [Google Scholar] [CrossRef]
- Barchitta, M.; Maugeri, A.; Quattrocchi, A.; Agrifoglio, O.; Scalisi, A.; Agodi, A. The Association of Dietary Patterns with High-Risk Human Papillomavirus Infection and Cervical Cancer: A Cross-Sectional Study in Italy. Nutrients 2018, 10, 469. [Google Scholar] [CrossRef] [PubMed]
- Maugeri, A.; Hruskova, J.; Jakubik, J.; Hlinomaz, O.; Medina-Inojosa, J.R.; Barchitta, M.; Agodi, A.; Vinciguerra, M. How dietary patterns affect left ventricular structure, function and remodelling: Evidence from the Kardiovize Brno 2030 study. Sci. Rep. 2019, 9, 19154. [Google Scholar] [CrossRef]
- Maugeri, A.; Barchitta, M.; Favara, G.; La Rosa, M.C.; La Mastra, C.; Magnano San Lio, R.; Agodi, A. Maternal Dietary Patterns Are Associated with Pre-Pregnancy Body Mass Index and Gestational Weight Gain: Results from the "Mamma & Bambino" Cohort. Nutrients 2019, 11, 1308. [Google Scholar] [CrossRef]
- Barchitta, M.; Maugeri, A.; Agrifoglio, O.; Favara, G.; La Mastra, C.; La Rosa, M.C.; Magnano San Lio, R.; Agodi, A. Dietary patterns and school performance: Evidence from a sample of adolescents in Sicily, Italy. Ann. Ig 2019, 31, 72–80. [Google Scholar]
- Maugeri, A.; Hruskova, J.; Jakubik, J.; Kunzova, S.; Sochor, O.; Barchitta, M.; Agodi, A.; Bauerova, H.; Medina-Inojosa, J.R.; Vinciguerra, M. Dietary antioxidant intake decreases carotid intima media thickness in women but not in men: A cross-sectional assessment in the Kardiovize study. Free Radic. Biol. Med. 2019, 131, 274–281. [Google Scholar] [CrossRef]
- Barchitta, M.; Maugeri, A.; La Mastra, C.; Rosa, M.C.; Favara, G.; Lio, R.M.S.; Agodi, A. Dietary Antioxidant Intake and Human Papillomavirus Infection: Evidence from a Cross-Sectional Study in Italy. Nutrients 2020, 12, 1384. [Google Scholar] [CrossRef] [PubMed]
- Atkins, J.L.; Whincup, P.H.; Morris, R.W.; Lennon, L.T.; Papacosta, O.; Wannamethee, S.G. Dietary patterns and the risk of CVD and all-cause mortality in older British men. Br. J. Nutr. 2016, 116, 1246–1255. [Google Scholar] [CrossRef] [PubMed]
- Bouwland-Both, M.I.; Steegers-Theunissen, R.P.; Vujkovic, M.; Lesaffre, E.M.; Mook-Kanamori, D.O.; Hofman, A.; Lindemans, J.; Russcher, H.; Jaddoe, V.W.; Steegers, E.A. A periconceptional energy-rich dietary pattern is associated with early fetal growth: The Generation R study. BJOG 2013, 120, 435–445. [Google Scholar] [CrossRef] [PubMed]
- Hearty, A.P.; Gibney, M.J. Comparison of cluster and principal component analysis techniques to derive dietary patterns in Irish adults. Br. J. Nutr. 2009, 101, 598–608. [Google Scholar] [CrossRef]
- Heidemann, C.; Scheidt-Nave, C.; Richter, A.; Mensink, G.B. Dietary patterns are associated with cardiometabolic risk factors in a representative study population of German adults. Br. J. Nutr. 2011, 106, 1253–1262. [Google Scholar] [CrossRef]
- Laird, P.W. Principles and challenges of genomewide DNA methylation analysis. Nat. Rev. Genet. 2010, 11, 191–203. [Google Scholar] [CrossRef]
- Harrison, A.; Parle-McDermott, A. DNA methylation: A timeline of methods and applications. Front. Genet. 2011, 2, 74. [Google Scholar] [CrossRef]
- Zhang, N. Epigenetic modulation of DNA methylation by nutrition and its mechanisms in animals. Anim. Nutr. 2015, 1, 144–151. [Google Scholar] [CrossRef]
- Kadayifci, F.Z.; Zheng, S.; Pan, Y.X. Molecular Mechanisms Underlying the Link between Diet and DNA Methylation. Int. J. Mol. Sci. 2018, 19, 4055. [Google Scholar] [CrossRef]
- Bollati, V.; Schwartz, J.; Wright, R.; Litonjua, A.; Tarantini, L.; Suh, H.; Sparrow, D.; Vokonas, P.; Baccarelli, A. Decline in genomic DNA methylation through aging in a cohort of elderly subjects. Mech. Ageing Dev. 2009, 130, 234–239. [Google Scholar] [CrossRef]
- Bollati, V.; Galimberti, D.; Pergoli, L.; Dalla Valle, E.; Barretta, F.; Cortini, F.; Scarpini, E.; Bertazzi, P.A.; Baccarelli, A. DNA methylation in repetitive elements and Alzheimer disease. Brain Behav. Immun. 2011, 25, 1078–1083. [Google Scholar] [CrossRef] [PubMed]
- Carraro, J.C.; Mansego, M.L.; Milagro, F.I.; Chaves, L.O.; Vidigal, F.C.; Bressan, J.; Martínez, J.A. LINE-1 and inflammatory gene methylation levels are early biomarkers of metabolic changes: Association with adiposity. Biomarkers 2016, 21, 625–632. [Google Scholar] [CrossRef] [PubMed]
- Delgado-Cruzata, L.; Zhang, W.; McDonald, J.A.; Tsai, W.Y.; Valdovinos, C.; Falci, L.; Wang, Q.; Crew, K.D.; Santella, R.M.; Hershman, D.L.; et al. Dietary modifications, weight loss, and changes in metabolic markers affect global DNA methylation in Hispanic, African American, and Afro-Caribbean breast cancer survivors. J. Nutr. 2015, 145, 783–790. [Google Scholar] [CrossRef] [PubMed]
- Carreira, P.E.; Richardson, S.R.; Faulkner, G.J. L1 retrotransposons, cancer stem cells and oncogenesis. FEBS J. 2014, 281, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Beck, S.; Rakyan, V.K. The methylome: Approaches for global DNA methylation profiling. Trends Genet. 2008, 24, 231–237. [Google Scholar] [CrossRef]
- Champagne, F.A. Epigenetic influence of social experiences across the lifespan. Dev. Psychobiol. 2010, 52, 299–311. [Google Scholar] [CrossRef]
- Galea, S.; Uddin, M.; Koenen, K. The urban environment and mental disorders: Epigenetic links. Epigenetics 2011, 6, 400–404. [Google Scholar] [CrossRef]
- Agodi, A.; Barchitta, M.; Valenti, G.; Marzagalli, R.; Frontini, V.; Marchese, A.E. Increase in the prevalence of the MTHFR 677 TT polymorphism in women born since 1959: Potential implications for folate requirements. Eur. J. Clin. Nutr. 2011, 65, 1302–1308. [Google Scholar] [CrossRef]
- Agodi, A.; Quattrocchi, A.; Maugeri, A.; Barchitta, M. The link between MTHFR C677T polymorphism, folate metabolism and global DNA methylation: A literature review. In Methylenetetrahydrofolate Reductase (MTHFR) in Health and Disease; Evans, R., Ed.; Nova Science Publishers: New York, NY, USA, 2015; pp. 71–82. [Google Scholar]
- Cabo, R.; Hernes, S.; Slettan, A.; Haugen, M.; Ye, S.; Blomhoff, R.; Mansoor, M.A. Effect of genetic polymorphisms involved in folate metabolism on the concentration of serum folate and plasma total homocysteine (p-tHcy) in healthy subjects after short-term folic acid supplementation: A randomized, double blind, crossover study. Genes Nutr. 2015, 10, 456. [Google Scholar] [CrossRef]
- Agodi, A.; Barchitta, M.; Cipresso, R.; Marzagalli, R.; La Rosa, N.; Caruso, M.; Castiglione, M.G.; Travali, S. Distribution of p53, GST, and MTHFR polymorphisms and risk of cervical intraepithelial lesions in sicily. Int. J. Gynecol. Cancer 2010, 20, 141–146. [Google Scholar] [CrossRef]
- Simone, B.; Mazzucco, W.; Gualano, M.R.; Agodi, A.; Coviello, D.; Dagna Bricarelli, F.; Dallapiccola, B.; Di Maria, E.; Federici, A.; Genuardi, M.; et al. The policy of public health genomics in Italy. Health Policy 2013, 110, 214–219. [Google Scholar] [CrossRef]
- Robertson, K.D.; Wolffe, A.P. DNA methylation in health and disease. Nat. Rev. Genet. 2000, 1, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Qu, J.; Liu, G.H.; Belmonte, J.C.I. The ageing epigenome and its rejuvenation. Nat. Rev. Mol. Cell Biol. 2020, 21, 137–150. [Google Scholar] [CrossRef]
- Zhang, Y.; Kutateladze, T.G. Diet and the epigenome. Nat. Commun. 2018, 9, 3375. [Google Scholar] [CrossRef] [PubMed]
- Agodi, A.; Barchitta, M.; Grillo, A.; Sciacca, S. Detection of genetically modified DNA sequences in milk from the Italian market. Int. J. Hyg. Environ. Health 2006, 209, 81–88. [Google Scholar] [CrossRef]
- Mitchell, C.; Schneper, L.M.; Notterman, D.A. DNA methylation, early life environment, and health outcomes. Pediatr. Res. 2016, 79, 212–219. [Google Scholar] [CrossRef] [PubMed]
- Borghesan, M.; Fusilli, C.; Rappa, F.; Panebianco, C.; Rizzo, G.; Oben, J.A.; Mazzoccoli, G.; Faulkes, C.; Pata, I.; Agodi, A.; et al. DNA Hypomethylation and Histone Variant macroH2A1 Synergistically Attenuate Chemotherapy-Induced Senescence to Promote Hepatocellular Carcinoma Progression. Cancer Res. 2016, 76, 594–606. [Google Scholar] [CrossRef] [PubMed]
- Barchitta, M.; Maugeri, A.; Quattrocchi, A.; Agrifoglio, O.; Agodi, A. The Role of miRNAs as Biomarkers for Pregnancy Outcomes: A Comprehensive Review. Int. J. Genom. 2017, 2017, 8067972. [Google Scholar] [CrossRef] [PubMed]
- Lo Re, O.; Maugeri, A.; Hruskova, J.; Jakubik, J.; Kucera, J.; Bienertova-Vasku, J.; Oben, J.A.; Kubala, L.; Dvorakova, A.; Ciz, M.; et al. Obesity-induced nucleosome release predicts poor cardio-metabolic health. Clin. Epigenet. 2019, 12, 2. [Google Scholar] [CrossRef]
- Agodi, A.; Barchitta, M.; Quattrocchi, A.; Marchese, A.E.; Boffetta, P. Folate deficiency is not associated with increased mitochondrial genomic instability: Results from dietary intake and lymphocytic mtDNA 4977-bp deletion in healthy young women in Italy. Mutagenesis 2014, 29, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Barchitta, M.; Quattrocchi, A.; Adornetto, V.; Marchese, A.E.; Agodi, A. Tumor necrosis factor-alpha -308 G>A polymorphism, adherence to Mediterranean diet, and risk of overweight/obesity in young women. Biomed. Res. Int. 2014, 2014, 742620. [Google Scholar] [CrossRef]
- Barchitta, M.; Maugeri, A.; La Rosa, M.C.; Magnano San Lio, R.; Favara, G.; Panella, M.; Cianci, A.; Agodi, A. Single Nucleotide Polymorphisms in Vitamin D Receptor Gene Affect Birth Weight and the Risk of Preterm Birth: Results From the "Mamma & Bambino" Cohort and A Meta-Analysis. Nutrients 2018, 10, 1172. [Google Scholar] [CrossRef]
- Maugeri, A.; Medina-Inojosa, J.R.; Kunzova, S.; Agodi, A.; Barchitta, M.; Sochor, O.; Lopez-Jimenez, F.; Geda, Y.E.; Vinciguerra, M. Sleep Duration and Excessive Daytime Sleepiness Are Associated with Obesity Independent of Diet and Physical Activity. Nutrients 2018, 10, 1219. [Google Scholar] [CrossRef]
- Maugeri, A.; Barchitta, M.; Fiore, V.; Rosta, G.; Favara, G.; La Mastra, C.; La Rosa, M.C.; Magnano San Lio, R.; Agodi, A. Determinants of Adherence to the Mediterranean Diet: Findings from a Cross-Sectional Study in Women from Southern Italy. Int. J. Environ. Res. Public Health 2019, 16, 2963. [Google Scholar] [CrossRef]
- Maugeri, A.; Jana, H.; Jakubik, J.; Barchitta, M.; Lo Re, O.; Kunzova, S.; Medina-Inojosa, J.R.; Agodi, A.; Sciacca, S.; Vinciguerra, M. Independent effects of hypertension and obesity on left ventricular mass and geometry: Evidence from the Cardiovision 2030 study. J. Clin. Med. 2019, 8, 370. [Google Scholar] [CrossRef] [PubMed]
- Maugeri, A.; Barchitta, M.; Agrifoglio, O.; Favara, G.; La Mastra, C.; La Rosa, M.C.; Magnano San Lio, R.; Panella, M.; Cianci, A.; Agodi, A. The impact of social determinants and lifestyles on dietary patterns during pregnancy: Evidence from the “Mamma & Bambino” study. Ann. Ig 2019, 31, 81–89. [Google Scholar] [PubMed]
- Maugeri, A.; Barchitta, M.; Kunzova, S.; Bauerova, H.; Agodi, A.; Vinciguerra, M. The association of social and behavioral factors with dietary risks in adults: Evidence from the Kardiovize Brno 2030 study. Nutr. Metab. Cardiovasc. Dis. 2020. [Google Scholar] [CrossRef] [PubMed]
- Maugeri, A.; Kunzova, S.; Medina-Inojosa, J.R.; Agodi, A.; Barchitta, M.; Homolka, M.; Kiacova, N.; Bauerova, H.; Sochor, O.; Lopez-Jimenez, F.; et al. Association between eating time interval and frequency with ideal cardiovascular health: Results from a random sample Czech urban population. Nutr. Metab. Cardiovasc. Dis. 2018. [Google Scholar] [CrossRef] [PubMed]
- Kunzova, S.; Maugeri, A.; Medina-Inojosa, J.; Lopez-Jimenez, F.; Vinciguerra, M.; Marques-Vidal, P. Determinants of Metabolic Health across Body Mass Index Categories in Central Europe: A Comparison Between Swiss and Czech Populations. Front. Public Health 2020, 8, 108. [Google Scholar] [CrossRef]
- Lodato, F.; Araújo, J.; Barros, H.; Lopes, C.; Agodi, A.; Barchitta, M.; Ramos, E. Caffeine intake reduces sleep duration in adolescents. Nutr. Res. 2013, 33, 726–732. [Google Scholar] [CrossRef]
| First Author and Year of Publication | Country | Study Design | Study Population | Dietary Factors | DNA Methylation Markers | Sample Type | DNA Methylation Method | Main Findings |
|---|---|---|---|---|---|---|---|---|
| van Engeland et al. 2003 [33] | The Netherlands | Cross-sectional | 122 patients with colorectal cancer | Alcohol and folate | APC, p14, p16, MLH1, MGMT, and RASSF1A | Tumor biopsy | Methylation Specific PCR | For each gene, prevalence of promoter hypermethylation was higher in patients with low folate intake and high alcohol consumption. The number of patients with at least one gene methylated was higher in the low folate intake/high alcohol intake group than their counterparts |
| Nan et al. 2004 [34] | Korea | Case Control | 110 patients with gastric cancer and 220 age- and sex-matched controls | Foods, calories, nutrients, vitamins, and minerals | MLH1 | Tumor biopsy | Methylation Specific PCR | Alcohol consumption was associated with higher odds of MLH1 hypermethylation. High consumption of vegetables and low consumption of potato were associated with higher odds of MLH1 hypermethylation |
| Yuasa et al. 2005 [35] | Japan | Cross-sectional | 73 patients with gastric cancer | Foods and nutrients | CDX2, p16 and MLH1 | Tumor biopsy | Methylation Specific PCR | Among men, consumption of green tea and cruciferous vegetables was negatively correlated with CDX2 methylation |
| Kraunz et al. 2006 [36] | USA | Cross-sectional | 242 patients with head and neck squamous cell carcinoma | Folate | p16 | Tumor biopsy | Methylation Specific PCR | Low intake of folate was associated with higher odds of p16 methylation |
| Slattery et al. 2006 [37] | USA | Case Control | 1154 patients with colon cancer and 1256 controls | Cruciferous vegetables, alcohol, folate, vitamins B6 and B12, methionine, and fiber | CIMP | Tumor biopsy | Methylation Specific PCR | Alcohol consumption was associated with higher odds of CIMP-low. The intake of fiber was associated with CIMP status |
| Mas et al. 2007 [38] | Spain | Case Control | 120 patients with colorectal cancer and 296 controls | Nutrients | p16, p14 and MLH1 | Tumor biopsy | Methylation Specific PCR | Patients with low intake of folate, vitamin A, vitamin B1, potassium and iron showed lower p16 methylation than controls. Patients with low vitamin A intake showed lower p14 and MLH1 methylation |
| De Vogel et al. 2008 [39] | The Netherlands | Cross-sectional | 648 patients with colorectal cancer | Folate, vitamin B2 and vitamin B6, methionine and alcohol | MLH1 | Tumor biopsy | Methylation Specific PCR | Intakes of folate, vitamin B2, methionine and alcohol were not associated with MLH1 hypermethylation. Among men, intake of vitamin B6 was associated with MLH1 hypermethylation |
| Schernhammer et al. 2009 [40] | USA | Cross-sectional | 609 patients with colon cancer | Alcohol, folate and B vitamins | LINE-1 | Tumor biopsy | Pyrosequencing | Participants with higher folate intake were less likely to exhibit LINE-1 hypomethylation. Alcohol consumption was positively associated with LINE-1 hypomethylation |
| Curtin et al. 2011 [41] | USA | Case Control | 951 patients with rectal cancer and 1205 controls | Folate, riboflavin, vitamins B6 andB12, and methionine | CIMP | Tumor biopsy | Methylation Specific PCR | Women with higher folate intake had lower odds of CIMP+ phenotype. Men with higher folate intake had higher odds of CIMP+ tumor |
| Tao et al. 2011 [42] | USA | Cross-sectional | 1170 women with breast cancer | One-carbon-related micronutrients and compounds | E-cadherin, p16, and RAR-β | Tumor biopsy | Methylation Specific PCR | Dietary intake of folate, vitamins B2, B6, B12, and methionine was not associated with methylation of E- cadherin, p16, and RAR-β |
| Xu et al. 2011 [43] | USA | Cross-sectional | 851 women with breast cancer | One-carbon-related micronutrients and compounds | 13 breast cancer-related genes | Tumor biopsy | Methylation Specific PCR and Methyight assay | Intake of B2 and B6 correlated with promoter methylation status in 3 out of the 13 breast cancer genes evaluated. Both positive (hypermethylation) and inverse (hypomethylation) associations were observed |
| Piyathilake et al. 2012 [19] | USA | Cross-sectional | 319 women with abnormal cervical cytology | Dietary patterns | LINE-1 | Blood | Pyrosequencing | Women with healthy dietary pattern were more likely to have higher LINE-1 methylation than those who adhered to an unhealthy dietary pattern |
| Zhang et al. 2013 [44] | South Korea | Cross-sectional | 184 patients with gastric cancer | Calories, foods, nutrients, vitamins and minerals | RUNX3 | Tumor biopsy | Methylation Specific PCR | High consumption of eggs was associated with higher odds of RUNX3 methylation. High consumption of nuts was associated with higher odds of RUNX3 methylation in patients with Helicobacter pylori infection. High consumption of fruits and high intake of carbohydrate, vitamin B1, and vitamin E was associated with lower odds of RUNX3 methylation |
| Chen et al. 2014 [45] | China | Case Control | 90 patients with esophageal squamous cell carcinoma and 60 healthy adults | Roast meat | p16 | Esophageal mucosa tissue | Pyrosequencing | Consumption of roast meat was positively associated with p16 methylation among cases. No association was evident among healthy subjects |
| Nishihara et al. 2014 [46] | USA | Cross-sectional | 993 patients with colorectal cancer from the Nurses’ Health Study and the Health Professionals’ Follow-up Study | Alcohol, vitamin B6, vitamin B12, folate, and methionine | IGF2 | Tumor biopsy | Pyrosequencing | Consumption of >15 g alcohol/d was associated with higher risk of colorectal cancer with lower IGF2 methylation levels. The association of vitamin B-6, vitamin B-12, and folate intakes with cancer risk did not significantly differ according to IGF2 methylation level |
| Pirouzpanah et al. 2015 [47] | Iran | Cross-sectional | 149 women with breast cancer | Folate, vitamins B2, B6, B12, and methionine | RAR-β, BRCA1 and RASSF1A | Tumor biopsy | Methylation Specific PCR | Intake of folate and vitamin B12 was negatively associated with RAR-β and BRCA1 methylation. Intake of riboflavin and pyridoxine was positively associated with RAR-β methylation |
| Mehta et al. 2017 [48] | USA | Cross-sectional | 1285 patients with colorectal cancer from the Health Professionals’ Follow-up Study and the Nurses’ Health Study | Dietary patterns | CIMP | Tumor biopsy | Pyrosequencing | Adherence to the western dietary pattern, characterized by red and processed meats, high-fat dairy products, refined grains, and desserts, was associated with CIMP-low phenotype |
| Ferrari et al. 2019 [49] | Brazil | Cross-sectional | 189 patients with colon or rectal adenocarcinoma | Alcohol, folate, vitamins B2, B6, and B12, choline, betaine, methionine, energy, carbohydrate, protein, and lipid | Global DNA methylation | Tumor biopsy and blood | Enzyme-linked immunosorbent assay | No association between dietary intakes and global DNA methylation was evident |
| First Author and Year of Publication | Country | Study Design | Study Population | Dietary Factors | DNA Methylation Markers | Sample Type | DNA Methylation Method | Main Findings |
|---|---|---|---|---|---|---|---|---|
| Stidley et al. 2010 [51] | USA | Cross-sectional | 1101 smokers from the Lovelace Smokers Cohort | Total and animal fat, vitamin C, vitamin E, folate, carotene, alpha carotene, beta-carotene, lycopene, lutein and zeaxanthin, and retinol | p16, MGMT, DAPK, RASSF1A, PAX5 α, PAX5 β, GATA4 and GATA5 | Sputum | Methylation Specific PCR | High intake of folate was associated with lower DNA methylation |
| Zhang et al. 2011 [54] | USA | Cross-sectional | 161 cancer-free individuals | Folate, vitamins B12 and B6, riboflavin and methionine | LINE-1 | Blood | Pyrosequencing | No association between intake of nutrients in one-carbon metabolism and LINE-1 methylation |
| Ono et al. 2012 [20] | Japan | Cross-sectional | 384 healthy women | Folate and vitamins B2, B6, and B12 | Global DNA methylation | Blood | Methylight assay | Folate intake was negatively associated with global DNA methylation |
| Zhang et al. 2012 [21] | USA | Cross-sectional | 180 cancer-free individuals | Folate and dietary patterns | IL-6 and LINE-1 | Blood | Pyrosequencing | Folate intake was positively associated with LINE-1 methylation |
| Perng et al. 2014 [55] | USA | Cross-sectional | 1002 participants of the Multi-Ethnic Study of Atherosclerosis study | Folate, vitamins B12 and B6, zinc, and methionine | LINE-1 and Alu | Blood | Pyrosequencing | Intake of methyl-donor micronutrients was not associated with DNA methylation |
| Agodi et al. 2015 [22] | Italy | Cross-sectional | 177 healthy women | Mediterranean Diet and folate | LINE-1 | Blood | Pyrosequencing | Women with low consumption of fruit and those with folate deficiency were more likely to exhibit LINE-1 hypomethylation |
| Shimazu et al. 2015 [60] | Japan | Cross-sectional | 281 subjects without cancer and with no history of treatment against Helicobacter pylori infection | Green/yellow vegetables, fruit and salt | miR-124a-3, EMX1 and NKX6-1 | Gastric mucosa | Methylation Specific PCR | Intake of green/yellow vegetables was negatively associated with methylation of miR-124a-3 |
| Marques-Rocha et al. 2016 [59] | Brazil | Cross-sectional | 156 subjects without metabolic disease, chronic inflammation, hydric balance disorders, changes in body composition and problems in nutrient absorption or metabolism | Energy and nutrients | LINE-1, TNF-α and IL-6 | Blood | Methylation-sensitive high-resolution melting analysis | Individuals with higher LINE-1 methylation had higher daily intakes of calories, iron and riboflavin, and lower intakes of copper, niacin and thiamin |
| Nicodemus-Johnson et al. 2017 [58] | USA | Prospective | 2148 Caucasian individuals from the Framingham Heart Study Offspring cohort | Fruits and juices | Genomic methylation profile | Blood | Infinium Illumina Human Methylation 450 k BeadChip arrays | There were 5221 and 5434 CpG sites associated with the intake of fruit and juice, respectively |
| Barchitta et al. 2018 [56] | Italy | Cross-sectional | 299 healthy women | Mediterranean diet | LINE-1 | Blood | Pyrosequencing | Adherence to the Mediterranean diet was positively associated with LINE-1 methylation level |
| Leng et al. 2018 [52] | Mexico | Prospective | 327 Hispanics and 1502 non-Hispanic White smokers from the Lovelace Smokers Cohort | Nutrients | 12 tumor suppressor genes | Sputum | Methylation Specific PCR | Intake of vitamin A, folate, and vitamin D was negatively associated with DNA methylation levels. Intake of saturated fat was positively associated with DNA methylation levels |
| Barchitta et al. 2019 [57] | Italy | Cross-sectional | 349 healthy women | Foods and dietary patterns | LINE-1 | Blood | Pyrosequencing | Consumption of whole-meal bread, cereals, fish, fruit, raw and cooked vegetables, legumes, soup, potatoes, fries, rice and pizza were positively correlated with LINE-1 methylation. LINE-1 methylation level increased with increasing adherence to a prudent dietary pattern |
| Mandaviya et al. 2019 [53] | Netherlands, Italy, Finland, USA, UK | Prospective | 5841 participants with no history of cancer from 10 cohorts | Folate and vitamin B-12 | Genomic methylation profile | Blood | Infinium Illumina Human Methylation 450 k BeadChip arrays | 74 folate-associated DMRs, of which 73 were negatively associated with folate intake. The most significant folate-associated DMR was a 400-base pair (bp) spanning region annotated to the LGALS3BP gene |
| Zhang et al. 2011 [25] | USA | Cross-sectional | 149 cancer-free individuals | Dietary patterns | LINE-1 | Blood | Pyrosequencing | Adherence to a prudent dietary pattern was associated with a lower prevalence of LINE-1 hypomethylation. No association between the Western dietary pattern and LINE-1 methylation was evident |
| First Author and Year of Publication | Country | Study Design | Study Population | Dietary Factors | DNA Methylation Markers | Sample Type | DNA Methylation Method | Main Findings |
|---|---|---|---|---|---|---|---|---|
| Hermsdorff et al. 2013 [61] | Spain | Cross-sectional | 40 normal-weight women | Energy and fat | TNF-α | Blood | Epityper Methylation Analysis | Women with high truncal fat showed lower TNF-α methylation than those with lower truncal adiposity. Intake of n-6 fatty acid was negatively associated with TNF-α methylation |
| Gomez-Uriz et al. 2014 [65] | Spain | Case Control | 12 patients with a first episode of parenchymal ischemic stroke and 12 patients with non-vascular neurological disorder | Nutrients and indexes of quality of diet | TNF-α and PON | Blood | Matrix-Assisted Laser Desorption/Ionization-Time Of Flight (MALDI-TOF) mass spectrometry | TNF-α methylation was related to lipid intake and dietary indexes in non-stroke patients. PON methylation was related to energy intake and quality of the diet |
| Carraro et al. 2016 [62] | Spain | Cross-sectional | 40 normal-weight healthy women | Fruit and Healthy Eating Index | TNF-α | Blood | MALDI-TOF mass spectrometry | Healthy eating index was negatively associated with TNF-α methylation level. A higher intake of fruits was associated with lower TNF-α methylation |
| Ramos-Lopez et al. 2017 [63] | Spain | Cross-sectional | 47 obese adults from the Metabolic Syndrome Reduction in Navarra, Spain trial | Folate | Genomic methylation profile | Blood | Infinium Illumina Human Methylation 450 k BeadChip arrays | A total of 51 CpGs were associated with folate intake, including one located in the CAMKK2 gene. Folate deficiency was related to lower CAMKK2 methylation |
| Ramos-Lopez et al. 2019 [64] | Spain | Cross-sectional | 247 adults from the Methyl Epigenome Network Association project | Energy, carbohydrates, protein, and fat | Genomic methylation profile | Blood | Infinium Illumina Human Methylation 450 k BeadChip arrays | Methylation of SLC18A1, SLC6A3, and SLC6A3 correlated with total energy consumption and carbohydrate intake |
| First Author and Year of Publication | Country | Study Design | Study Population | Dietary Factors | DNA Methylation Markers | Sample Type | DNA Methylation Method | Main Findings |
|---|---|---|---|---|---|---|---|---|
| Boeke et al. 2012 [69] | USA | Prospective | 830 mother–child pairs | Vitamin B12, betaine, choline, folate, cadmium, zinc and iron | LINE-1 | Maternal and infant cord blood | Pyrosequencing | No association of maternal intake of methyl donor nutrients with maternal and cord blood methylation. Periconceptional betaine intake was inversely associated with cord blood methylation; dietary cadmium was positively associated with first trimester methylation and inversely with cord blood methylation |
| Haggarty et al. 2013 [71] | United Kingdom | Prospective | 913 mother–child pairs | Folate intake | PEG3, IGF2, small nuclear ribonucleoprotein polypeptide N, and LINE-1 | Infant cord blood | Pyrosequencing | Folate intake was positively associated with IGF2 methylation and negatively with PEG3 and LINE-1 methylation in the offspring |
| McCullough et al. 2017 [74] | USA | Prospective | 338 mother–child pairs from the NEST cohort | Dietary inflammatory potential | IGF2, H19, MEG3, MEG3-IG, PEG3, MEST, SGCE/PEG10, NNAT, PLAGL1 | Infant cord blood | Pyrosequencing | Pro-inflammatory diets increased cytokine levels, but no association between dietary inflammatory potential and DNA methylation was evident |
| Pauwels et al. 2017 [73] | Belgium | Prospective | 115 mother–child pairs from the Maternal Nutrition and Offspring’s Epigenome study | Betaine, choline, folate, and methionine | Global DNA methylation and RXRA, LEP, DNMT1, and IGF2 | Infant cord blood | Liquid chromatography–tandem mass spectrometry and Pyrosequencing | Before pregnancy, intakes of betaine and methionine were positively associated with DNMT and LEP methylation. In the second trimester, methyl group donor intake was negatively associated with LEP and DNMT methylation. In the last trimester, intake of choline and folate was positively associated with DNMT methylation and negatively with RXRA methylation |
| Rijlaarsdam et al. 2017 [72] | UK | Prospective | 346 mother–child pairs from the Avon Longitudinal Study of Parents and Children | Dietary patterns | IGF2 | Infant cord blood and blood at 7 years | Infinium Illumina Human Methylation 450 k BeadChip arrays | Maternal diet high in fat and carbohydrates before pregnancy was positively associated with IGF2 methylation at birth |
| Taylor et al. 2017 [70] | Australia | Prospective | 73 children from the WATCH study | Methionine, folate, vitamins B2, B6 and B12 and choline | Global DNA methylation | Buccal cells | Enzyme-linked immunosorbent assay | No association between one-carbon metabolism nutrient intake and global DNA methylation levels was evident |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maugeri, A.; Barchitta, M. How Dietary Factors Affect DNA Methylation: Lesson from Epidemiological Studies. Medicina 2020, 56, 374. https://doi.org/10.3390/medicina56080374
Maugeri A, Barchitta M. How Dietary Factors Affect DNA Methylation: Lesson from Epidemiological Studies. Medicina. 2020; 56(8):374. https://doi.org/10.3390/medicina56080374
Chicago/Turabian StyleMaugeri, Andrea, and Martina Barchitta. 2020. "How Dietary Factors Affect DNA Methylation: Lesson from Epidemiological Studies" Medicina 56, no. 8: 374. https://doi.org/10.3390/medicina56080374
APA StyleMaugeri, A., & Barchitta, M. (2020). How Dietary Factors Affect DNA Methylation: Lesson from Epidemiological Studies. Medicina, 56(8), 374. https://doi.org/10.3390/medicina56080374






