Combined Effect of HPV and Several Gene SNPs in Laryngeal Cancer
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. Association of SNPs Distribution and Clinical-Pathological Characteristics in LSCC Patients
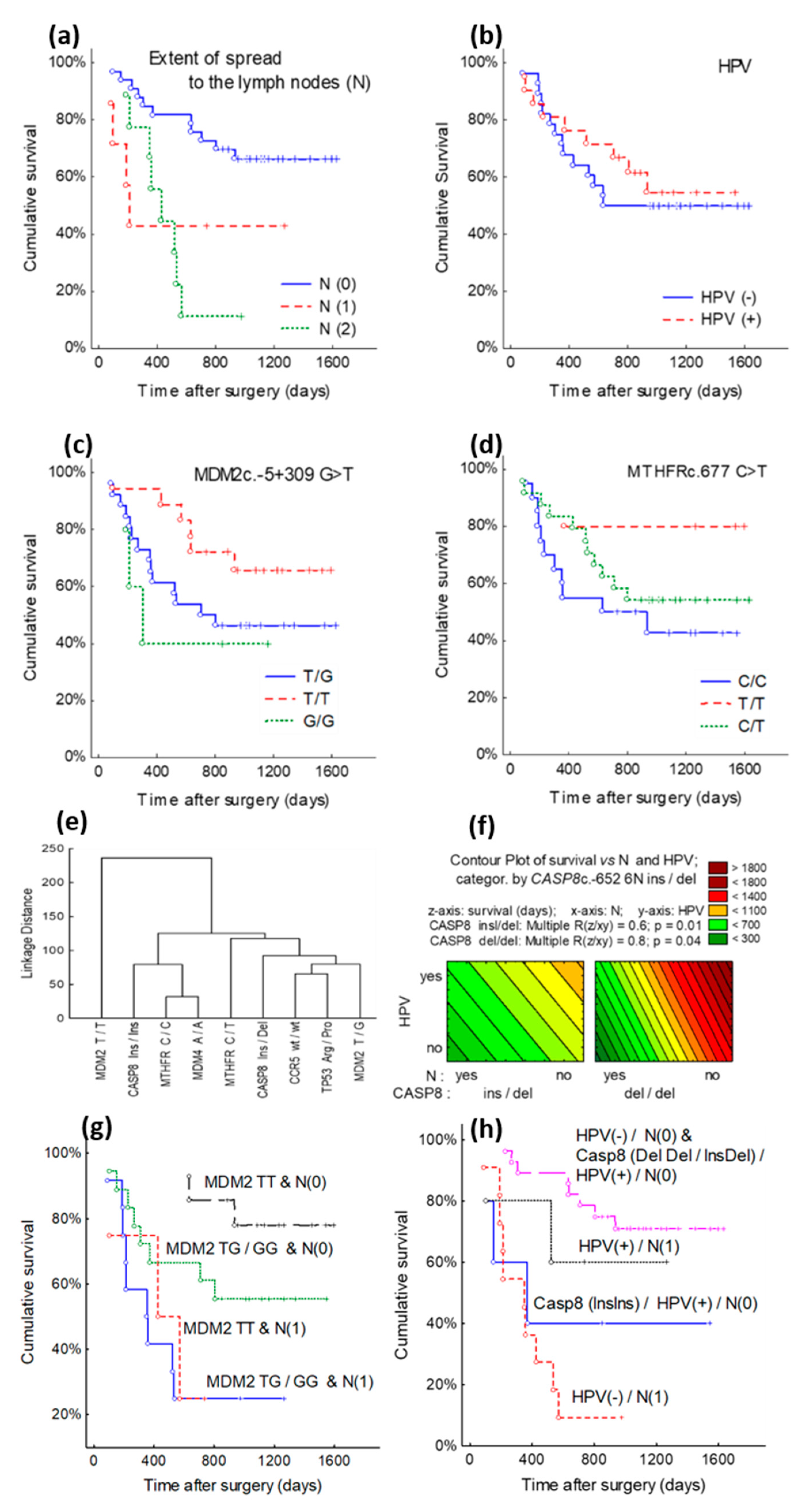
3.2. Survival Analysis according to TP53c.215 G > C (Arg72Pro), MDM2c.-5 + 309 G > T, MDM4c.1q32 A > C, MTHFRc.677 C > T, CASP8c.-652 6N ins/del, CCR5c.-Δ32 Genes SNPs
3.3. Cluster Analysis of TP53, MDM2, MDM4, MTHFR, CASP8, CCR5 Genes Polymorphic Variants and Clinical-Pathological Characteristics
3.4. Cox Model Analysis of TP53, MDM2, MDM4, MTHFR, CASP8, and CCR5 Genes Polymorphic Variants and Clinical-Pathological Characteristics
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Placa, J.R.; Bueno Rde, B.; Pinheiro, D.G.; Panepucci, R.A.; de Araújo, L.F.; Mamede, R.C.; Figueiredo, D.L.; Silva, W.A., Jr. Gene expression analysis of laryngeal squamous cell carcinoma. Genom. Data 2015, 5, 9–12. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.Y.; Lu, Z.M.; Luo, X.N.; Chen, L.S.; Ge, P.J.; Song, X.H.; Chen, S.H.; Wu, Y.L. Retrospective Analysis of Prognostic Factors in 205 Patients with Laryngeal Squamous Cell Carcinoma Who Underwent Surgical Treatment. PLoS ONE 2013, 8, e60157. [Google Scholar]
- Ahmadi, N.; Ahmadi, N.; Chan, M.V.; Huo, Y.R.; Sritharan, N.; Chin, R. Laryngeal Squamous Cell Carcinoma Survival in the Context of Human Papillomavirus: A Systematic Review and Meta-analysis. Cureus 2018, 10, e2234. [Google Scholar] [CrossRef]
- Sánchez Barrueco, A.; González Galán, F.; Lora Pablos, D.; Villacampa Aubá, J.M.; Ballestín Carcavilla, C.; Cenjor Español, C.; Almodóvar Álvarez, C. HPV in Larynx Squamous Cell Carcinoma: New Serotypes and Survival Study within 10-Year Follow-up. Otolaryngol. Head Neck Surg. 2017, 156, 677–682. [Google Scholar] [CrossRef]
- Quan, F.; Zhang, F.; Bai, Y.; Zhou, L.; Yang, H.; Li, B.; Jin, T.; Li, H.; Shao, Y. Association of genetic polymorphisms with laryngeal carcinoma prognosis in a Chinese population. Oncotarget 2017, 8, 10255–10263. [Google Scholar] [CrossRef][Green Version]
- Morbini, P.; Benazzo, M. Human papillomavirus and head and neck carcinomas: Focus on evidence in the babel of published data. Acta Otorhinolaryngol. Ital. 2016, 36, 249–258. [Google Scholar]
- Hassumi-Fukasawa, M.K.; Miranda-Camargo, F.A.; Guimarães, M.C.; Simões, R.T.; Donadi, E.A.; Soares, C.P.; Soares, E.G. Possible implication of Mdm2 as a prognostic marker in invasive laryngeal carcinoma. Eur. Arch. Otorhinolaryngol. 2012, 269, 1795–1804. [Google Scholar] [CrossRef]
- Bychkov, V.A.; Nikitina, E.G.; Ibragimova, M.K.; Kaigorodova, E.V.; Choinzonov, E.L.; Litviakov, N.V. Comprehensive meta-analytical summary on human papillomavirus association with head and neck cancer. Exp. Oncol. 2016, 38, 68–72. [Google Scholar] [CrossRef]
- Chen, X.; Sturgis, E.M.; Lei, D.; Dahlstrom, K.; Wei, Q.; Li, G. Human papillomavirus seropositivity synergizes with MDM2 variants to increase the risk of oral squamous cell carcinoma. Cancer Res. 2010, 70, 7199–7208. [Google Scholar] [CrossRef]
- Tsimplaki, E.; Argyri, E.; Sakellaridis, A.; Kyrodimos, E.; Xesfyngi, D.; Panotopoulou, E. Oropharyngeal and laryngeal but not oral cancers are strongly associated with high-risk human papillomavirus in 172 Greek patients. J. Med. Virol. 2017, 89, 170–176. [Google Scholar] [CrossRef]
- Seifi, S.; Asvadi Kermani, I.; Dolatkhah, R.; Asvadi Kermani, A.; Sakhinia, E.; Asgarzadeh, M.; Dastgiri, S.; Ebrahimi, A.; Asghari Haggi, A.; Nadri, M.; et al. Prevalence of oral human papilloma virus in healthy individuals in the East Azerbaijan province of Iran. Iran. J. Public Health 2013, 42, 79–85. [Google Scholar]
- Nowińska, K.; Ciesielska, U.; Podhorska-Okołów, M.; Dzięgiel, P. The role of human papillomavirus in oncogenic transformation and its contribution to the etiology of precancerous lesions and cancer of the larynx: A review. Adv. Clin. Exp. Med. 2017, 26, 539–547. [Google Scholar] [CrossRef]
- Smith, E.M.; Rubenstein, L.M.; Hoffman, H.; Haugen, T.H.; Turek, L.P. Human papillomavirus, p16 and p53 expression associated with survival of head and neck cancer. Infect. Agent. Cancer 2010, 5, 4. [Google Scholar] [CrossRef]
- Liu, L.; Wu, C.; Wang, Y.; Zhong, R.; Duan, S.; Wei, S.; Lin, S.; Zhang, X.; Tan, W.; Yu, D.; et al. Combined Effect of Genetic Polymorphisms in P53, P73 and MDM2 on Non-small Cell Lung Cancer Survival. J. Thorac. Oncol. 2011, 6, 1793–1800. [Google Scholar] [CrossRef]
- Wang, Z.; Sturgis, E.M.; Zhang, Y.; Huang, Z.; Zhou, Q.; Wei, Q.; Li, G. Combined p53-related genetic variants together with HPV infection increase oral cancer risk. Int. J. Cancer 2012, 131, E251–E258. [Google Scholar] [CrossRef]
- Bojesen, S.E.; Nordestgaard, B.G. The common germline Arg72Pro polymorphism of p53 and increased longevity in humans. Cell Cycle 2008, 7, 158–163. [Google Scholar] [CrossRef]
- Denaro, N.; Lo Nigro, C.; Natoli, G.; Russi, E.G.; Adamo, V.; Merlano, M.C. The Role of p53 and MDM2 in Head and Neck Cancer. ISRN Otolaryngol. 2011, 2011, 931813. [Google Scholar] [CrossRef][Green Version]
- Zhou, J.; Liu, F.; Zhang, D.; Chen, B.; Li, Q.; Zhou, L.; Lu, L.M.; Tao, L. Significance of MDM2-309 polymorphisms and induced corresponding plasma MDM2 levels in susceptibility to laryngeal squamous cell carcinoma. DNA Cell Biol. 2014, 33, 88–94. [Google Scholar] [CrossRef]
- Ruttkay-Nedecky, B.; Jimenez, A.M.; Nejdl, L.; Chudobova, D.; Gumulec, J.; Masarik, M.; Adam, V.; Kizek, R. Relevance of infection with human papillomavirus: The role of the p53 tumor suppressor protein and E6/E7 zinc finger proteins. Int. J. Oncol. 2013, 43, 1754–1762. [Google Scholar] [CrossRef]
- Vivenza, D.; Gasco, M.; Monteverde, M.; Lattanzio, L.; Syed, N.; Colantonio, I.; Denaro, N.; Natoli, G.; Comino, A.; Russi, E.; et al. MDM2 309 polymorphism predicts outcome in platinum-treated locally advanced head and neck cancer. Oral. Oncol. 2012, 48, 602–607. [Google Scholar] [CrossRef]
- Bond, G.L.; Hu, W.; Bond, E.E.; Robins, H.; Lutzker, S.G.; Arva, N.C.; Bargonetti, J.; Bartel, F.; Taubert, H.; Wuerl, P.; et al. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell 2004, 119, 591–602. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Sturgis, E.M.; Liu, Z.; Wang, L.E.; Wei, Q.; Li, G. Modifying effect of MDM4 variants on risk of HPV16-associated squamous cell carcinoma of oropharynx. Cancer 2012, 118, 1684–1692. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.J.; Luo, Y.J.; Shi, Z.Y.; Xu, X.L.; Yao, G.L.; Liu, R.P.; Zhao, H. The associations between MDM4 gene polymorphisms and cancer risk. Oncotarget 2016, 7, 55611–55623. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.M.; Deng, M.H.; Chen, W.; Zeng, X.T.; Luo, J. MTHFR C677T gene polymorphism and head and neck cancer risk: A meta-analysis based on 23 publications. Dis. Mark. 2015, 2015, 681313. [Google Scholar] [CrossRef]
- Anders, Q.S.; Stur, E.; Agostini, L.P.; Garcia, F.M.; Reis, R.S.; Santos, J.A.; Mendes, S.O.; Maia, L.L.; Peterle, G.T.; Stange, V.; et al. MTHFR C677T and A1298C polymorphisms as predictors of radiotherapy response in head and neck squamous cell carcinoma. Genet. Mol. Res. 2015, 14, 13105–13109. [Google Scholar] [CrossRef]
- Zhuo, X.; Ye, H.; Li, Q.; Xiang, Z.; Zhang, X. Is MDM2 SNP309 Variation a Risk Factor for Head and Neck Carcinoma? An Updated Meta-Analysis Based on 11,552 Individuals. Medicine 2016, 95, e2948. [Google Scholar] [CrossRef]
- Ying, Y.; Xu, J.; Qi, Y.; Zhang, M.; Yang, Y. CASP8 rs3834129 (-652 6N insertion/deletion) Polymorphism and Colorectal Cancer Susceptibility: An Updated Meta-Analysis. J. Cancer 2018, 9, 4166–4171. [Google Scholar] [CrossRef]
- Li, C.; Lu, J.; Liu, Z.; Wang, L.E.; Zhao, H.; El-Naggar, A.K.; Sturgis, E.M.; Wei, Q. The sixnucleotide deletion/insertion variant in the CASP8 promoter region is inversely associated with risk of squamous cell carcinoma of the head and neck. Cancer Prev. Res. 2010, 3, 246–253. [Google Scholar] [CrossRef]
- Chen, D.; Ma, T.; Liu, X.W.; Liu, Z. CASP-8 -652 6N ins/del polymorphism and cancer risk: Aliterature-based systematic HuGE review and meta-analysis. Exp. Ther. Med. 2012, 4, 762–770. [Google Scholar] [CrossRef][Green Version]
- Santos, E.U.; Lima, G.D.; Oliveira Mde, L.; Heráclio Sde, A.; Silva, H.D.; Crovella, S.; Maia Mde, M.; Souza, P.R. CCR2 and CCR5 genes polymorphisms in women with cervical lesions from Pernambuco, Northeast Region of Brazil: A case-control study. Mem. Inst. Oswaldo Cruz 2016, 111, 174–180. [Google Scholar] [CrossRef]
- Zheng, B.; Wiklund, F.; Gharizadeh, B.; Sadat, M.; Gambelunghe, G.; Hallmans, G.; Dillner, J.; Wallin, K.L.; Ghaderi, M. Genetic polymorphism of chemokine receptors CCR2 and CCR5 in Swedish cervical cancer patients. Anticancer Res. 2006, 26, 3669–3674. [Google Scholar]
- Hütter, G.; Neumann, M.; Nowak, D.; Klein, S.; Klüter, H.; Hofmann, W.K. The effect of the CCR5-Δ32 deletion on global gene expression considering immune response and inflammation. J. Inflamm. 2011, 8, 29. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Lu, Z.M.; Guo, M.; Wu, Q.J.; Chen, K.N.; Xing, H.P.; Mei, Q.; Ke, Y. p53 codon 72 polymorphism (C/G) and the risk of human papillomavirus-associated carcinomas in China. Cancer 2002, 95, 2571–2576. [Google Scholar] [CrossRef] [PubMed]
- Horikawa, Y.; Nadaoka, J.; Saito, M.; Kumazawa, T.; Inoue, T.; Yuasa, T.; Tsuchiya, N.; Nishiyama, H.; Ogawa, O.; Habuchi, T. Clinical implications of the MDM2 SNP309 and p53 Arg72Pro polymorphisms in transitional cell carcinoma of the bladder. Oncol. Rep. 2008, 20, 49. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liu, J.; Tang, X.; Li, M.; Lu, C.; Shi, J.; Zhou, L.; Yuan, Q.; Yang, M. Functional MDM4 rs4245739 genetic variant, alone and in combination with P53 Arg72Pro polymorphism, contributes to breast cancer susceptibility. Breast Cancer Res. Treat. 2013, 140, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Gama, R.R.; Carvalho, A.L.; Longatto Filho, A.; Scorsato, A.P.; López, R.V.; Rautava, J.; Syrjänen, S.; Syrjänen, K. Detection of human papillomavirus in laryngeal squamous cell carcinoma: Systematic review and meta-analysis. Laryngoscope 2016, 126, 885–893. [Google Scholar] [CrossRef]
- Mallen-St Clair, J.; Alani, M.; Wang, M.B.; Srivatsan, E.S. Human papillomavirus inoropharyngeal cancer: The changing face of a disease. Biochim. Biophys Acta 2016, 1866, 141–150. [Google Scholar]
- Nakashima, M.; Kondo, S.; Shimizu, Y.; Wakisaka, N.; Murono, S.; Furukawa, M.; Yoshizaki, T. Impact of MDM2 single nucleotide polymorphism on tumor onset in head and neck squamous cell carcinoma. Acta Otolaryngol. 2008, 128, 808–813. [Google Scholar] [CrossRef]
- Spence, T.; Bruce, J.; Yip, K.W.; Liu, F.F. HPV Associated Head and Neck Cancer. Cancers 2016, 8, 75. [Google Scholar] [CrossRef]
- Chen, W.C.; Chuang, H.C.; Lin, Y.T.; Huang, C.C.; Chien, C.Y. Clinical impact of human papillomavirus in laryngeal squamous cell carcinoma: A retrospective study. Peer J. 2017, 5, e3395. [Google Scholar] [CrossRef]
- Vermorken, J.B.; Psyrri, A.; Mesía, R.; Peyrade, F.; Beier, F.; de Blas, B.; Celik, I.; Licitra, L. Impact of tumor HPV status on outcome in patients with recurrent and/or metastatic squamous cell. carcinoma of the head and neck receiving chemotherapy with or without cetuximab: Retrospective analysis of the phase III EXTREME trial. Ann. Oncol. 2014, 25, 801–807. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Concha-Benavente, F.; Shayan, G.; Srivastava, R.M.; Gibson, S.P.; Wang, L.; Gooding, W.E.; Ferris, R.L. STING activation enhances cetuximab-mediated NK cell activation and DC maturation and correlates with HPV+ status in head and neck cancer. Oral. Oncol. 2018, 78, 186–193. [Google Scholar] [CrossRef] [PubMed]
- Pan, C.; Issaeva, N.; Yarbrough, W.G. HPV-driven oropharyngeal cancer: Current knowledge of molecular biology and mechanisms of carcinogenesis. Cancers Head Neck 2018, 3, 12. [Google Scholar] [CrossRef] [PubMed]
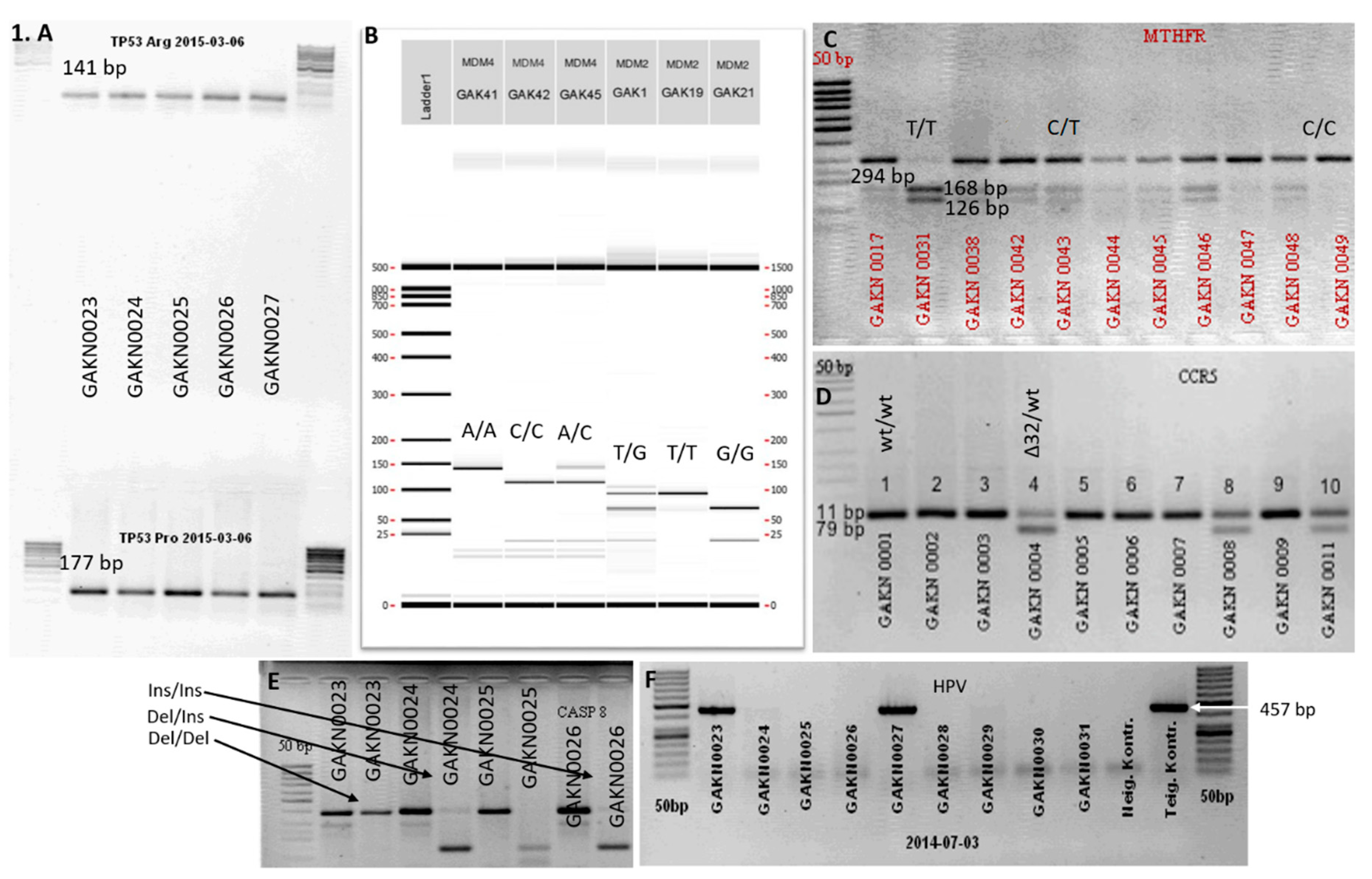
| Gene | SNP | Primer | Amplification Products (bp) | Authors |
|---|---|---|---|---|
| TP53 | Arg F | 5’→TCCCCCTTGCCGTCCCAA→3’ | 141 bp | [30] |
| Arg R | 5’→CTGGTGCAGGGGCCACGC→3’ | |||
| Pro F | 5’→GCCAGAGGCTGCTCCCCCC→3’ | 177 bp | ||
| Pro R | 5’→CGTGCAAGTCACAGACTT→3’ | |||
| MDM2 | F | 5’→TTCGGAGGTCTCCGCGGGAGTTCAG→3’ | 89 bp, 64 bp, 25 bp | [31] |
| R | 5’→TGCGATCATCCGGACCTCCCGCGTC→3’ | |||
| MDM4 | F | 5’→AAGACTAAAGAAGGCTGGGG→3’ | 134 bp, 111 bp, 23 bp | [32] |
| R | 5′→TTCAAATAATGTGGCAAGTGACC→3’ | |||
| MTHFR | F | 5’→CCTTGAACAGGTGGAGGCCAG→3’, | 294 bp, 168 bp, 126 bp | [33] |
| R | 5’→GCGGTGAGAGTGGGGTGGAG→3’ | |||
| CASP8 | F | 5’→AGTGAAAACTTCTCCCATGGCCTC→3’ | 139 bp, 291 bp, 396 bp | [34] |
| R | 5’→GATTGATACTGGCACAGTATACTTACC→3’ | |||
| Ins | 5’→GTAATTCTTGCTCTGCCAAGCTG→3’; | |||
| Del | 5’→CCAAGGTCACGCAGCTAGTAAG→3’ | |||
| CCR5 | F | 5’→ACCTGCAGCTCTCATTTTCC→3’ | 111 bp, 79 bp | [28] |
| R | 5’→GCAGATGACCATGACAAGCA→3’ | |||
| HPV | MY09 | 5′→CGT-CCA-AAA-GGA-AAC-TGA-GC→3′ | 450 bp | [35] |
| MY11 | 5′→GCA-CAG-GGA-CAT-AAC-AAT-GG→3′ |
| HPV Infection | p | Stage | p | N | p | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | N (%) | + (n = 21) | − (n = 28) | I–II (n = 9) | III–IV (n = 40) | 0 (n = 33) | 1–2 (n = 16) | |||
| TP53 Arg/Arg Arg/Pro Pro/Pro | 0 (0) 49 (100) 0 (0) | 0 (0) 21 (100) 0 (0) | 0 (0) 28 (100) 0 (0) | - | 0 (0) 9 (100) 0 (0) | 0 (0) 40 (100) 0 (0) | - | - 33 (100) - | - 16 (100) - | - |
| MDM2 T/T T/G G/G | 18 (36.7) 26 (53.1) 5 (10.2) | 7 (33.3) 13 (61.9) 1 (4.8) | 11 (37.5) 13 (43.7) 4 (18.8) | 0.42 | 3 (33.3) 6 (66.7) 0 (0) | 15 (37.5) 20 (50.0) 5 (12.5) | 0.46 | 14 (42.4) 16 (48.5) 3 (9.1) | 4 (25.0) 10 (62.5) 2 (12.5) | 0.24 |
| MDM4 A/A A/C C/C | 33 (67.3) 15 (30.6) 1 (2.1) | 14 (66.7) 7 (33.3) 0 (0) | 19 (67.9) 8 (28.6) 1 (3.6) | 0.97 | 7 (77.8) 2 (22.2) 0 (0) | 26 (65.0) 13 (32.5) 1 (2.5) | 0.70 | 23 (69.7) 10 (30.3) - | 10 (62.5) 5 (31.3) 1 (6.2) | 0.74 |
| MTHFR C > T C/C C/T T/T | 20 (40.8) 24 (48.9) 5 (10.2) | 8 (38.0) 9 (42.9) 4 (19.1) | 12 (42.9) 15 (53.6) 1 (3.6) | 0.21 | 2 (22.2) 7 (77.8) 0 (0) | 18 (45.0) 17 (42.5) 5 (12.5) | 0.14 | 13 (39.4) 15 (45.5) 5 (15.2) | 7 (43.8) 9 (56.2) - | 0.44 |
| CASP8 Ins/ins Ins/del Del/del | 16 (32.7) 24 (49.0) 9 (18.3) | 6 (28.6) 13 (61.9) 2 (9.5) | 10 (35.7) 11 (39.3) 7 (25.0) | 0.22 | 2 (22.2) 6 (66.7) 1 (11.1) | 14 (35.0) 18 (45.0) 8 (20.0) | 0.50 | 13 (39.4) 16 (48.5) 4 (12.1) | 3 (18.8) 8 (50.0) 5 (31.2) | 0.20 |
| CCR5 wt/wt wt/Δ32 Δ32/Δ32 | 39 (79.6) 10 (20.4) 0 (0) | 19 (90.5) 2 (9.5) 0 (0) | 20 (71.4) 8 (28.6) 0 (0) | 0.16 | 6 (66.7) 3 (33.3) 0 (0) | 33 (82.5) 7 (17.5) 0 (0) | 0.74 | 26 (78.8) 7 (21.2) - | 13 (81.3) 3 (18.7) - | 0.11 |
| No = 49 | No = 49 | No = 49 | ||||||||
| Cluster | I | II | III | ||||||
|---|---|---|---|---|---|---|---|---|---|
| SNP | MDM2 T/T | MDM2 T/G | TP53 Arg/Pro | CASP8 ins/del | MTHFR C/T | CCR5 wt/wt | MDM4 A/A | CASP8 ins/ins | MTHFR C/C |
| % * | 33 | 54 | 47 | 54 | 46 | 49 | 48 | 38 | 55 |
| M ** (IQR) | 601 (170) | 312 (289) | 353 (350) | 428 (330) | 522 (360.5) | 369 (358) | 250 (197) | 210 (228) | 229 (166) |
| Variable | HR * | 95% CI | p |
|---|---|---|---|
| Multivariate analysis for single effects | |||
| HPV(-)/N1/CASP8 Ins/Ins and HPV(+)/N(0) | 5.49 | 2.34–12.86 | <0.001 |
| MDM2 T/T | 0.25 | 0.07–0.85 | 0.026 |
| Univariate analysis for combined effect | |||
| HPV(-)/N1/CASP8 Ins/Ins and HPV(+)/N(0) | 4.49 | 1.87–10.8 | 0.001 |
| MDM2 T/T | 0.36 | 0.10–1.27 | 0.112 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stumbrytė-Kaminskienė, A.; Gudlevičienė, Ž.; Dabkevičienė, D.; Mackevičienė, I. Combined Effect of HPV and Several Gene SNPs in Laryngeal Cancer. Medicina 2020, 56, 81. https://doi.org/10.3390/medicina56020081
Stumbrytė-Kaminskienė A, Gudlevičienė Ž, Dabkevičienė D, Mackevičienė I. Combined Effect of HPV and Several Gene SNPs in Laryngeal Cancer. Medicina. 2020; 56(2):81. https://doi.org/10.3390/medicina56020081
Chicago/Turabian StyleStumbrytė-Kaminskienė, Aušra, Živilė Gudlevičienė, Daiva Dabkevičienė, and Irina Mackevičienė. 2020. "Combined Effect of HPV and Several Gene SNPs in Laryngeal Cancer" Medicina 56, no. 2: 81. https://doi.org/10.3390/medicina56020081
APA StyleStumbrytė-Kaminskienė, A., Gudlevičienė, Ž., Dabkevičienė, D., & Mackevičienė, I. (2020). Combined Effect of HPV and Several Gene SNPs in Laryngeal Cancer. Medicina, 56(2), 81. https://doi.org/10.3390/medicina56020081




