In-Depth Bioinformatic Study of the CLDN16 Gene and Protein: Prediction of Subcellular Localization to Mitochondria
Abstract
1. Introduction
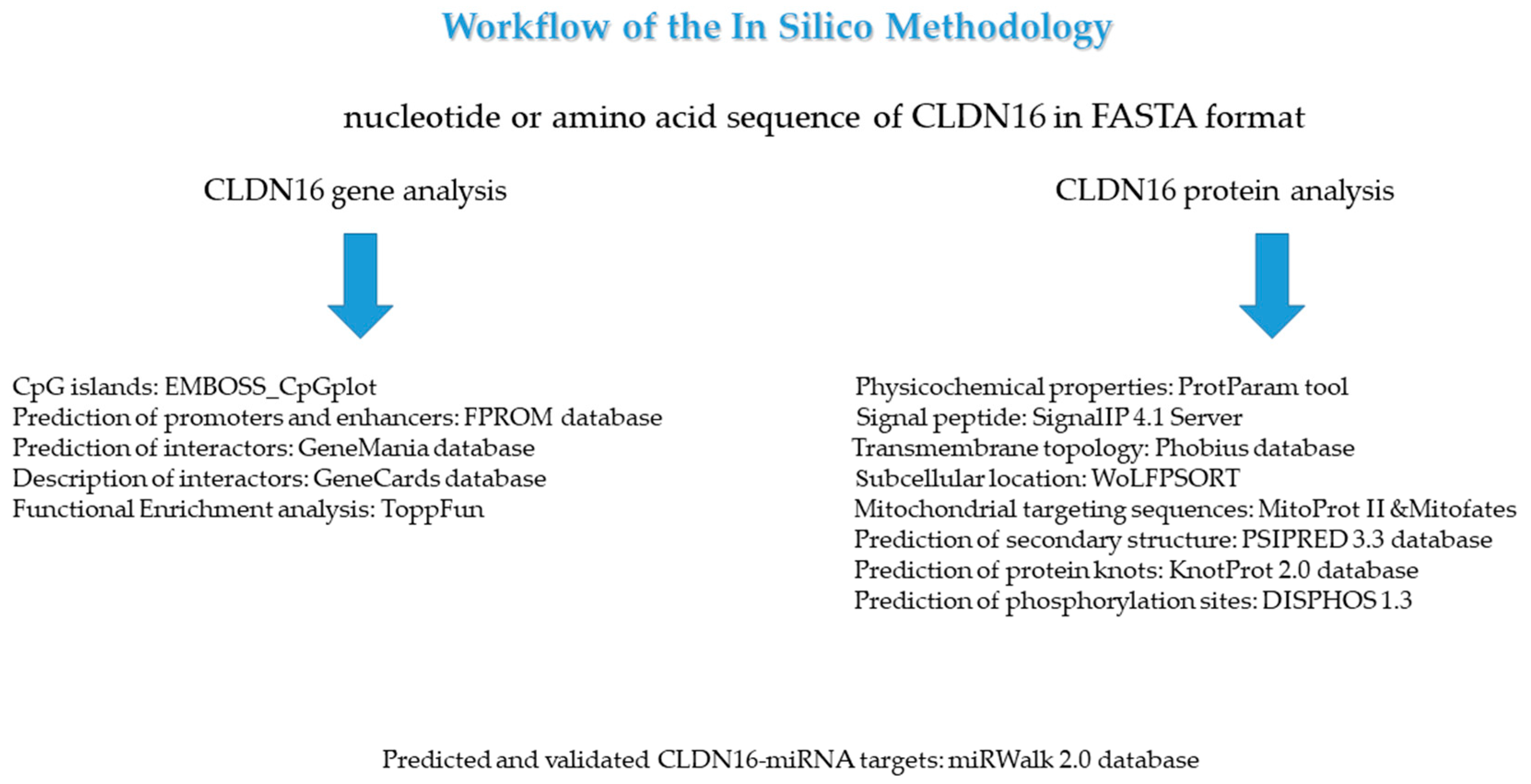
2. Materials and Methods
3. Results
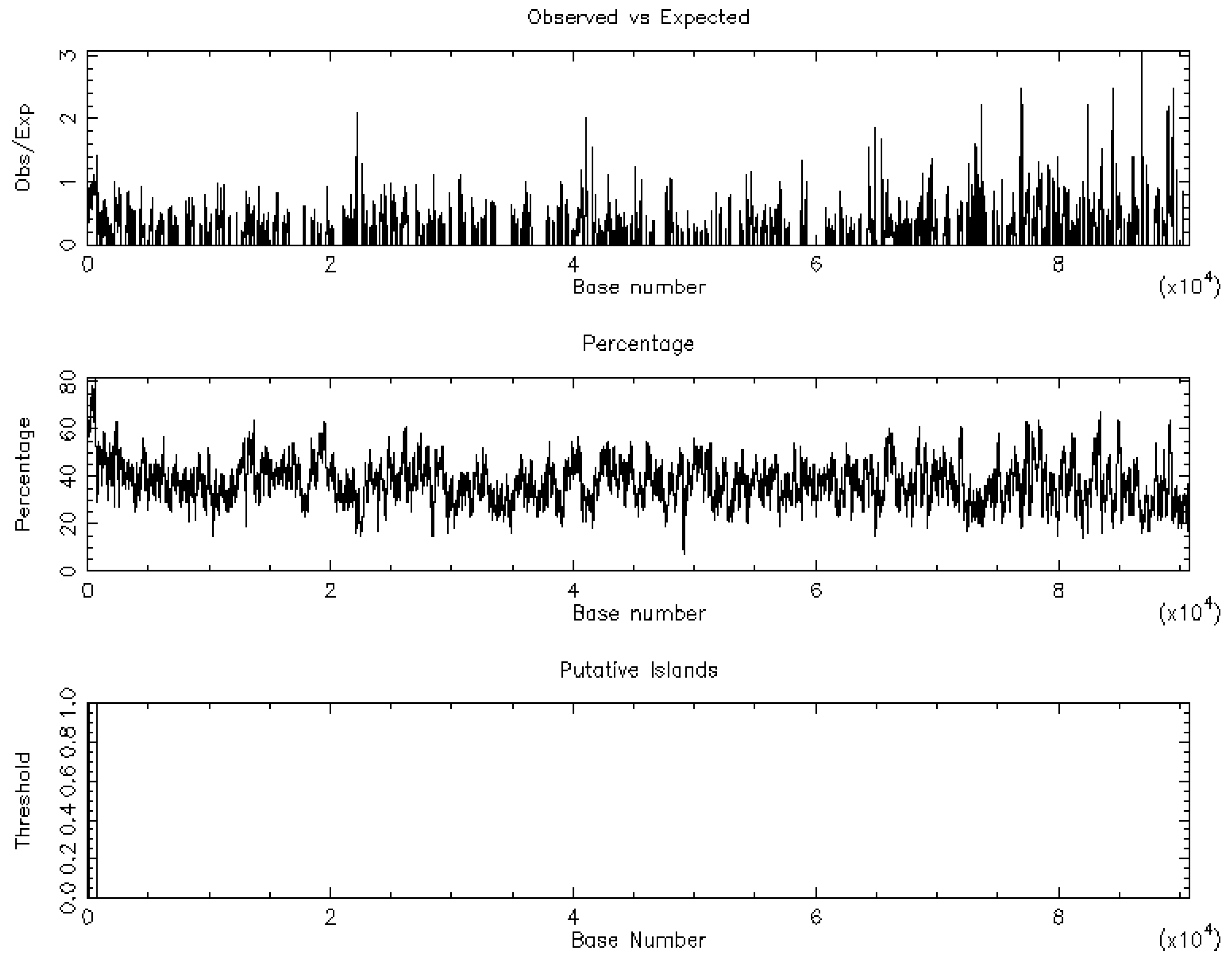
3.1. CLDN16 Gene
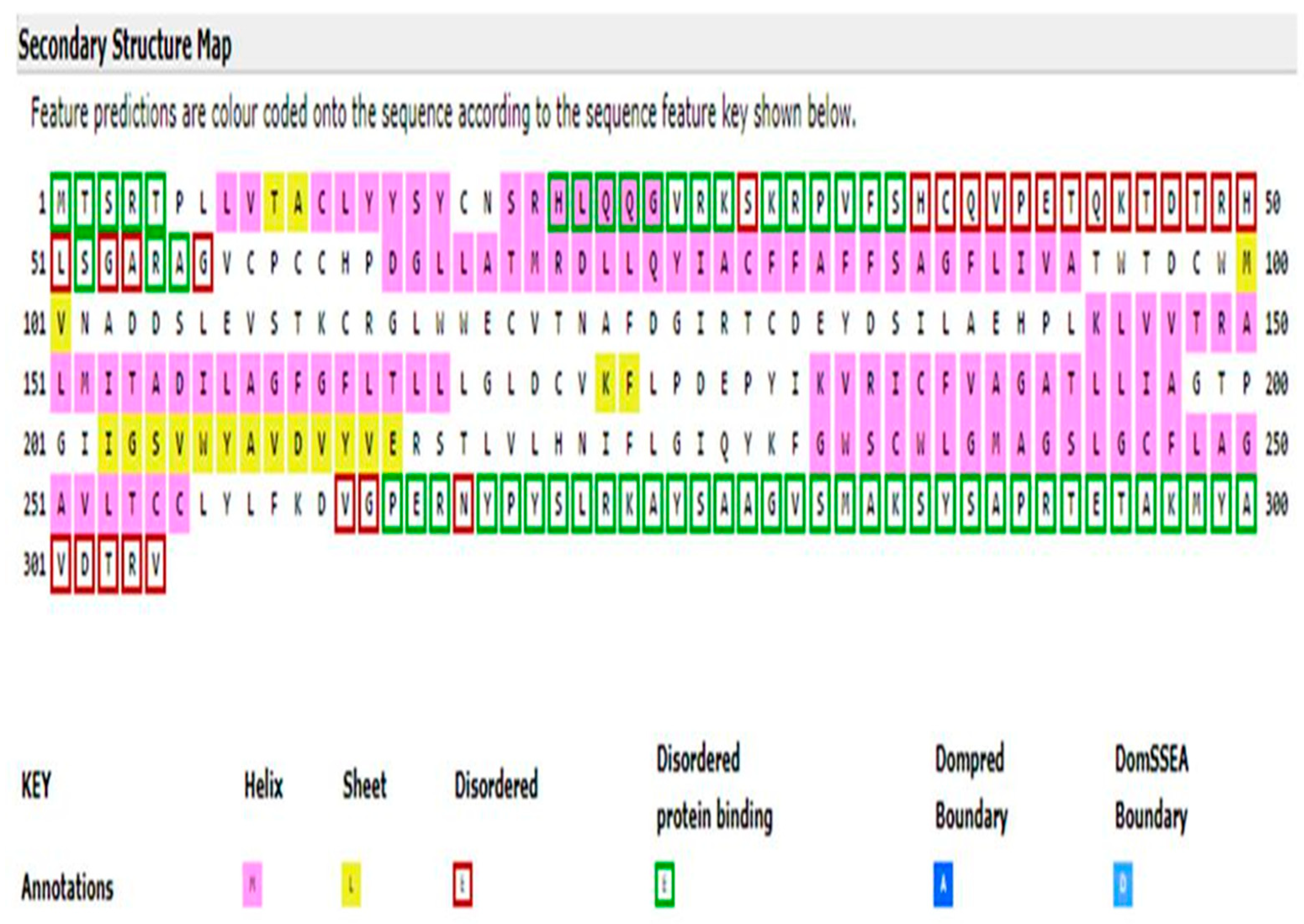
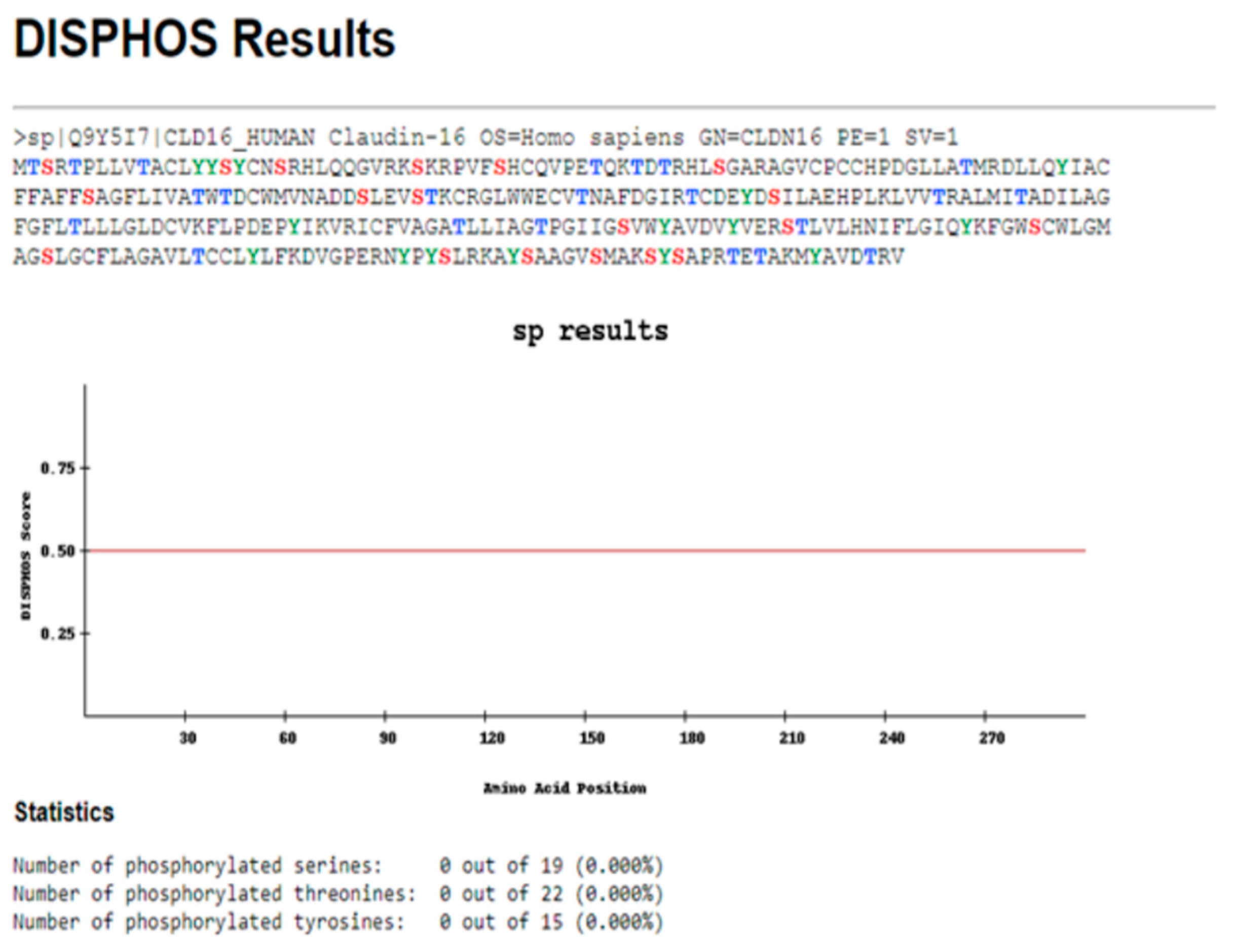
3.2. CLDN16 Protein
3.3. miRNA Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Viering, D.H.H.M.; de Baaij, J.H.F.; Walsh, S.B.; Kleta, R.; Bockenhauer, D. Genetic causes of hypomagnesemia, a clinical overview. Pediatric Nephrol. 2017, 32, 1123–1135. [Google Scholar] [CrossRef]
- Arteaga, M.E.; Hunziker, W.; Teo, A.S.M.; Hillmer, A.M.; Mutchinick, O.M. Familiar hypomagnesemia with hypercalciuria and nephrocalcinosis: Variable phenotypic expression in three affected sisters from Mexican ancestry. Ren. Fail. 2015, 37, 180–183. [Google Scholar] [CrossRef] [PubMed]
- Hanssen, O.; Castermans, E.; Bovy, C.; Weekers, L.; Erpicum, P.; Dubois, B.; Bours, V.; Krzesinski, J.-M.; Jouret, F. Two novel mutations of the CLDN16 gene cause familiar hypomagnesaemia with hypercalciuria and nephrocalcinosis. Clin. Kidney J. 2014, 7, 282–285. [Google Scholar] [CrossRef] [PubMed]
- Claverie-Martin, F. Familiar hypomagnesaemia with hypercalciuria and nephrocalcinosis: Clinical and molecular characteristics. Clin. Kidney J. 2015, 8, 656–664. [Google Scholar] [CrossRef] [PubMed]
- Kausalya, P.J.; Amasheh, S.; Günzel, D.; Wurps, H.; Müller, D.; Fromm, M.; Hunziker, W. Disease-associated mutations affect intracellular traffic and paracellular Mg2+ transport function of Claudin-16. J. Clin. Investig. 2006, 116, 878–891. [Google Scholar] [CrossRef] [PubMed]
- Ikari, A.; Tonegawa, C.; Sanada, A.; Kimura, T.; Sakai, H.; Hayashi, H.; Hasegawa, H.; Yamaguchi, M.; Yamazaki, Y.; Endo, S.; et al. Tight junctional localization of claudin-16 is regulated by syntaxin 8 in renal tubular epithelial cells. J. Biol. Chem. 2014, 289, 13112–13123. [Google Scholar] [CrossRef] [PubMed]
- Rice, P.; Longden, I.; Bleasby, A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000, 16, 276–277. [Google Scholar] [CrossRef]
- Takai, D.; Jones, P.A. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc. Natl. Acad. Sci. USA 2002, 99, 3740–3745. [Google Scholar] [CrossRef]
- Solovyev, V.V.; Shahmuradov, I.A.; Salamov, A.A. Identification of promoter regions and regulatory sites. Methods Mol. Biol. 2010, 674, 57–83. [Google Scholar]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T.; et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010, 38, 214–220. [Google Scholar] [CrossRef]
- Rappaport, N.; Fishilevich, S.; Nudel, R.; Twik, M.; Belinky, F.; Plaschkes, I.; Stein, T.I.; Cohen, D.; Oz-Levi, D.; Safran, M.; et al. Rational confederation of genes and diseases: NGS interpretation via GeneCards, MalaCards and VarElect. Biomed. Eng. Online 2017, 16, 72. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Bardes, E.E.; Aronow, B.J.; Jegga, A.G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009, 37, W305–W311. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server; Walker, J.M., Ed.; The Proteomics Protocols Handbook Humana Press: New York, NY, USA, 2005; pp. 571–607. [Google Scholar]
- Nielsen, H. Predicting Secretory Proteins with SignalP. Methods Mol. Biol. 2017, 1611, 59–73. [Google Scholar] [PubMed]
- Käll, L.; Krogh, A.; Sonnhammer, E.L. Advantages of combined transmembrane topology and signal peptide prediction—The Phobius web server. Nucleic Acids Res. 2007, 35, 429–432. [Google Scholar] [CrossRef] [PubMed]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, 585–587. [Google Scholar] [CrossRef]
- Claros, M.G.; Vincens, P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 1996, 241, 779–786. [Google Scholar] [CrossRef] [PubMed]
- Fukasawa, Y.; Tsuji, J.; Fu, S.C.; Tomii, K.; Horton, P.; Imai, K. Mitofates: Improved Prediction of Mitochondrial Targeting Sequences and Their Cleavage Sites. Mol. Cell. Proteom. 2015, 14, 1113–1126. [Google Scholar] [CrossRef]
- Buchan, D.W.; Minneci, F.; Nugent, T.C.; Bryson, K.; Jones, D.T. Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res. 2013, 41, 349–357. [Google Scholar] [CrossRef]
- Jamroz, M.; Niemyska, W.; Rawdon, E.J.; Stasiak, A.; Millett, K.C.; Sułkowski, P.; Sulkowska, J.I. KnotProt: A database of proteins with knots and slipknots. Nucleic Acids Res. 2015, 43, 306–314. [Google Scholar] [CrossRef]
- Iakoucheva, L.M.; Radivojac, P.; Brown, C.J.; O’Connor, T.R.; Sikes, J.G.; Obradovic, Z.; Dunker, A.K. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004, 32, 1037–1049. [Google Scholar] [CrossRef]
- Dweep, H.; Gretz, N. miRWalk 2.0: A comprehensive atlas of microRNA-target interactions. Nat. Methods 2015, 12, 697. [Google Scholar] [CrossRef] [PubMed]
- Rabouille, C. Pathways of Unconventional Protein Secretion. Trends Cell Biol. 2017, 27, 230–240. [Google Scholar] [CrossRef]
- Hung, M.C.; Link, W. Protein localization in disease and therapy. J. Cell Sci. 2011, 124, 3381–3392. [Google Scholar] [CrossRef] [PubMed]
- Marunaka, K.; Furukawa, C.; Fujii, N.; Kimura, T.; Furuta, T.; Matsunaga, T.; Endo, S.; Hasegawa, H.; Anzai, N.; Yamazaki, Y.; et al. The RING finger-and PDZ domain-containing protein PDZRN3 controls localization of the Mg2+ regulator claudin-16 in renal tube epithelial cells. J. Biol. Chem. 2017, 292, 13034–13044. [Google Scholar] [CrossRef]
- Ferecatu, I.; Le Floch, N.; Bergeaud, M.; Rodríguez-Enfedaque, A.; Rincheval, V.; Oliver, L.; Vallette, F.M.; Mignotte, B.; Vayssière, J.-L. Evidence for a mitochondrial localization of the retinoblastoma protein. BMC Cell Biol. 2009, 10, 50. [Google Scholar] [CrossRef] [PubMed]
- Schaeffer, C.; Creatore, A.; Rampoldi, L. Protein trafficking defects in inherited kidney diseses. Nephrol. Dial. Transplant. 2014, 29, 33–44. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Scholl, U.; Hebeisen, S.; Janssen, A.G.; Müller-Newen, G.; Alekov, A.; Fahlke, C. Barttin modulates trafficking and function of CIC-K channels. Proc. Natl. Acad. Sci. USA 2006, 103, 11411–11416. [Google Scholar] [CrossRef] [PubMed]
- Wojciechowski, D.; Thiemann, S.; Schaal, C.; Rahtz, A.; de la Roche, J.; Begemann, B.; Becher, T.; Fischer, M. Activation of renal CIC-K chloride channels depends on an intact N terminus of their accessory subunit barttin. J. Biol. Chem. 2018, 293, 8626–8637. [Google Scholar] [CrossRef]
- Hou, J. Lecture: New light on the role of claudins in the kidney. Organogenesis 2012, 8, 1–9. [Google Scholar] [CrossRef]
- Himmerkus, N.; Shan, Q.; Goerke, B.; Hou, J.; Goodenough, D.A.; Bleich, M. Salt and acid-base metabolism in claudin-16 knockdown mice: Impact for the pathophysiology of FHHNC patients. Am. J. Physiol. Ren. Physiol. 2008, 295, F1641–F1647. [Google Scholar] [CrossRef]
- Gong, Y.; Renigunta, V.; Himmerkus, N.; Zhang, J.; Renigunta, A.; Bleich, M.; Hou, J. Claudin-14 regulates renal Ca++ transport in response to CaSR signalling via a novel microRNA pathway. EMBO J. 2012, 31, 1999–2012. [Google Scholar] [CrossRef]
- Breiderhoff, T.; Himmerkus, N.; Drewell, H.; Plain, A.; Günzel, D.; Mutig, K.; Willnow, T.E.; Müller, D.; Bleich, M. Deletion of claudin-10 rescues claudin-16-deficient mice from hypomagnesemia and hypercalciuria. Kidney Int. 2018, 93, 580–588. [Google Scholar] [CrossRef]
- Chen, G.; Wang, Y.; Wang, L.; Xu, W. Identifying prognostic biomarkers based on aberrant DNA methylation in kidney renal clear cell carcinoma. Oncotarget 2017, 8, 5268–5280. [Google Scholar] [CrossRef]
- Hou, J.; Zhao, D. MicroRNA Regulation in Renal Pathophysiology. Int. J. Mol. Sci. 2013, 14, 13078–13092. [Google Scholar] [CrossRef]
- Mladinov, D.; Liu, Y.; Mattson, D.L.; Liang, M. MicroRNAs contribute to the maintenance of cell-type-specific physiological characteristics: MiR-192 targets Na(+)/K(+)-ATPase β1. Nucleic Acids Res. 2013, 41, 1273–1283. [Google Scholar] [CrossRef]
- Vall-Palomar, M.; Arévalo, J.; Ariceta, G.; Meseguer, A. Establishment of urinary exosome-like vesicles isolation protocol for FHHNC patients and evaluation of different exosomal RNA extraction methods. J. Transl. Med. 2018, 16, 278. [Google Scholar] [CrossRef]
- Deaton, A.M.; Bird, A. CpG islands and the regulation of transcription. Genes Dev. 2011, 25, 1010–1022. [Google Scholar] [CrossRef]
- Beck, S.; Rhee, C.; Song, J.; Lee, B.K.; LeBlanc, L.; Cannon, L.; Kim, J. Implications of CpG islands on chromosomal architectures and modes of global gene regulation. Nucleic Acids Res. 2018, 46, 4382–4391. [Google Scholar] [CrossRef]
- Kim, T.K.; Shiekhattar, R. Architectural and Functional Commonalities between Enhancers and Promoters. Cell 2015, 162, 948–959. [Google Scholar] [CrossRef]
- Anish, R.; Hossain, M.B.; Jacobson, R.H.; Takada, S. Characterization of transcription from TATA-less promoters: Identification of a new core promoter element XCPE2 and analysis of factor requirements. PLoS ONE 2009, 4, e5103. [Google Scholar] [CrossRef]
- Mehmood, M.A.; Sehar, U.; Ahmad, N. Use of Bioinformatics Tools in Different Spheres of Life Sciences. J. Data Min. Genom. Proteom. 2014, 5, 2. [Google Scholar]

| Promoter Position | 49220 LDF 1: +9.953 |
| Promoter Position | 559 LDF: +6.606 TATA box at 527 +3.708 GATTTAAA; Enchancer at: 622 Score: +10.194 |
| Promoter Position | 58406 LDF: +5.233 TATA box at 58379 +4.957 TATATAGA |
| Promoter Position | 40172 LDF: +3.884 TATA box at 40145 +8.509 TATAAAAC |
| Promoter Position | 29232 LDF: +2.816 TATA box at 29200 +3.764 CATATAGA |
| Promoter Position | 27968 LDF: +2.513 TATA box at 27936 +7.471 TATATAAG |
| Promoter Position | 906 LDF: +1.626 TATA box at 865 +5.343 TATTAAAA; Enchancer at: 622 Score: +10.194 |
| Promoter Position | 41678 LDF: +1.619 TATA box at 41643 +4.703 TATATATA; Enchancer at: 41701 Score: +10.937 |
| Promoter Position | 3335 LDF: +1.403 TATA box at 3304 +5.535 TATATAAT |
| Promoter Position | 48910 LDF: +1.227 TATA box at 48877 +6.554 AATATAAA |
| Promoter Position | 44869 LDF: +0.663 TATA box at 44839 +4.130 TATAACAG |
| Promoter Position | 65626 LDF: +0.437 TATA box at 65594 +5.926 TATATAAA |
| Promoter Position | 69400 LDF: +0.423 TATA box at 69370 +7.797 TATAAAAG |
| Promoter Position | 32529 LDF: +0.116 TATA box at 32500 +8.328 TATAAAAA |
| Promoter Position | 51931 LDF: +0.008 TATA box at 51900 +3.876 TATTAAAA |
| Promoter Position | 4793 LDF: −0.042 TATA box at 4762 +3.800 CATAAAAC |
| Promoter Position | 81748 LDF: −0.140 TATA box at 81718 +4.492 CTTAAAAA |
| Promoter Position | 27538 LDF: −0.398 TATA box at 27508 +5.750 TATAAATT |
| Promoter Position | 50088 LDF: −0.458 TATA box at 50057 +5.632 CATATAAG |
| Promoter Position | 67124 LDF: −0.489 TATA box at 67094 +3.638 TTTAAATC |
| Promoter Position | 36521 LDF: −0.500 TATA box at 36492 +5.469 AATAAAAA |
| Promoter Position | 52521 LDF: −0.572 TATA box at 52491 +4.197 CATAAAAG |
| Promoter Position | 34329 LDF: −0.821 TATA box at 34301 +4.292 AATAAAAA |
| Promoter Position | 68333 LDF: −0.879 TATA box at 68303 +4.256 TTTAAAGG |
| Promoter Position | 85754 LDF: −0.905 TATA box at 85723 +5.966 TATTTAAA |
| Gene name | Gene Description |
|---|---|
| CLDN19 | Claudin 19 |
| CLDN14 | Claudin 14 |
| CLDN10 | Claudin 10 |
| CLDN6 | Claudin 6 |
| TJP1 | Tight Junction Protein 1 |
| TJP3 | Tight Junction Protein 3 |
| NPHS1 | NPHS1, Nephrin |
| PATJ | PATJ, Crumbs Cell Polarity Complex Component |
| MYO3B | Myosin IIIB |
| C14orf105 | Coiled-Coil Domain Containing 198 |
| CLCNKA | Chloride Voltage-Gated Channel Ka |
| CLCNKB | Chloride Voltage-Gated Channel Kb |
| CTXN3 | Cortexin 3 |
| FCAMR | Fc Fragment Of IgA And IgM Receptor |
| CDH16 | Cadherin 16 |
| LYG1 | Lysozyme G1 |
| FBXO40 | F-Box Protein 40 |
| KCNJ1 | Potassium Voltage-Gated Channel Subfamily J Member 1 |
| TMEM178B | Transmembrane Protein 178B |
| ADGRF3 | Adhesion G Protein-Coupled Receptor F3 |
| ID | Name | Source | p Value | FDR B&H | FDR B&Y | Bonferroni | Genes from Input | Genes in Annotation | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | C1846352 | Increased urinary chloride | DisGeNET Curated | 7.888 × 10−9 | 1.037 × 10−6 | 6.376 × 10−6 | 2.074 × 10−6 | 3 | 5 |
| 2 | C1846351 | Increased urinary potassium | DisGeNET Curated | 7.888 × 10−9 | 1.037 × 10−6 | 6.376 × 10−6 | 2.074 × 10−6 | 3 | 5 |
| 3 | C0085680 | Hypochloremia (disorder) | DisGeNET Curated | 1.577 × 10−8 | 1.037 × 10−6 | 6.376 × 10−6 | 4.146 × 10−6 | 3 | 6 |
| 4 | C0595901 | Serum chloride level decreased (finding) | DisGeNET Curated | 1.577 × 10−8 | 1.037 × 10−6 | 6.376 × 10−6 | 4.146 × 10−6 | 3 | 6 |
| 5 | C1865279 | Fetal polyuria | DisGeNET Curated | 4.409 × 10−8 | 2.319 × 10−6 | 1.427 × 10−5 | 1.160 × 10−5 | 3 | 8 |
| 6 | C1846347 | Renal salt wasting | DisGeNET Curated | 6.386 × 10−7 | 2.799 × 10−5 | 1.722 × 10−4 | 1.680 × 10−4 | 3 | 18 |
| 7 | cv:C2751312 | Bartter syndrome, type 4b | Clinical Variations | 9.148 × 10−7 | 3.007 × 10−5 | 1.850 × 10−4 | 2.406 × 10−4 | 2 | 2 |
| 8 | OMIN:613090 | Bartter syndrome, type 4b | OMIM | 9.148 × 10−7 | 3.007 × 10−5 | 1.850 × 10−4 | 2.406 × 10−4 | 2 | 2 |
| 9 | C0740896 | Hypokalemic hypochloremic metabolic alkalosis | DisGeNET Curated | 2.740 × 10−6 | 6.226 × 10−5 | 3.830 × 10−4 | 7.207 × 10−4 | 2 | 3 |
| 10 | OMIN:602522 | Bartter syndrome, type 4a | OMIM | 2.740 × 10−6 | 6.226 × 10−5 | 3.830 × 10−4 | 7.207 × 10−4 | 2 | 3 |
| miRNA | Seed Length | p-Value |
|---|---|---|
| hsa-miR-6076 | 11 | 0.0005 |
| hsa-miR-6878-3p | 10 | 0.0020 |
| hsa-miR-328-5p | 10 | 0.0020 |
| hsa-miR-559 | 10 | 0.0020 |
| hsa-miR-1256 | 10 | 0.0020 |
| hsa-miR-6859-5p | 10 | 0.0020 |
| hsa-miR-95-5p | 10 | 0.0020 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rouka, E.; Liakopoulos, V.; Gourgoulianis, K.I.; Hatzoglou, C.; Zarogiannis, S.G. In-Depth Bioinformatic Study of the CLDN16 Gene and Protein: Prediction of Subcellular Localization to Mitochondria. Medicina 2019, 55, 409. https://doi.org/10.3390/medicina55080409
Rouka E, Liakopoulos V, Gourgoulianis KI, Hatzoglou C, Zarogiannis SG. In-Depth Bioinformatic Study of the CLDN16 Gene and Protein: Prediction of Subcellular Localization to Mitochondria. Medicina. 2019; 55(8):409. https://doi.org/10.3390/medicina55080409
Chicago/Turabian StyleRouka, Erasmia, Vassilios Liakopoulos, Konstantinos I. Gourgoulianis, Chrissi Hatzoglou, and Sotirios G. Zarogiannis. 2019. "In-Depth Bioinformatic Study of the CLDN16 Gene and Protein: Prediction of Subcellular Localization to Mitochondria" Medicina 55, no. 8: 409. https://doi.org/10.3390/medicina55080409
APA StyleRouka, E., Liakopoulos, V., Gourgoulianis, K. I., Hatzoglou, C., & Zarogiannis, S. G. (2019). In-Depth Bioinformatic Study of the CLDN16 Gene and Protein: Prediction of Subcellular Localization to Mitochondria. Medicina, 55(8), 409. https://doi.org/10.3390/medicina55080409








