An Effect of Culture Media on Epithelial Differentiation Markers in Breast Cancer Cell Lines MCF7, MDA-MB-436 and SkBr3
Abstract
1. Introduction
2. Material and Methods
2.1. Breast Cancer Cell Lines
2.2. Basal Growth Media and Supplements
2.3. Cell Culturing and Growth and Viability Analysis
2.4. Reverse-Transcription and qReal-Time PCR
2.5. Statistical Analysis
3. Results
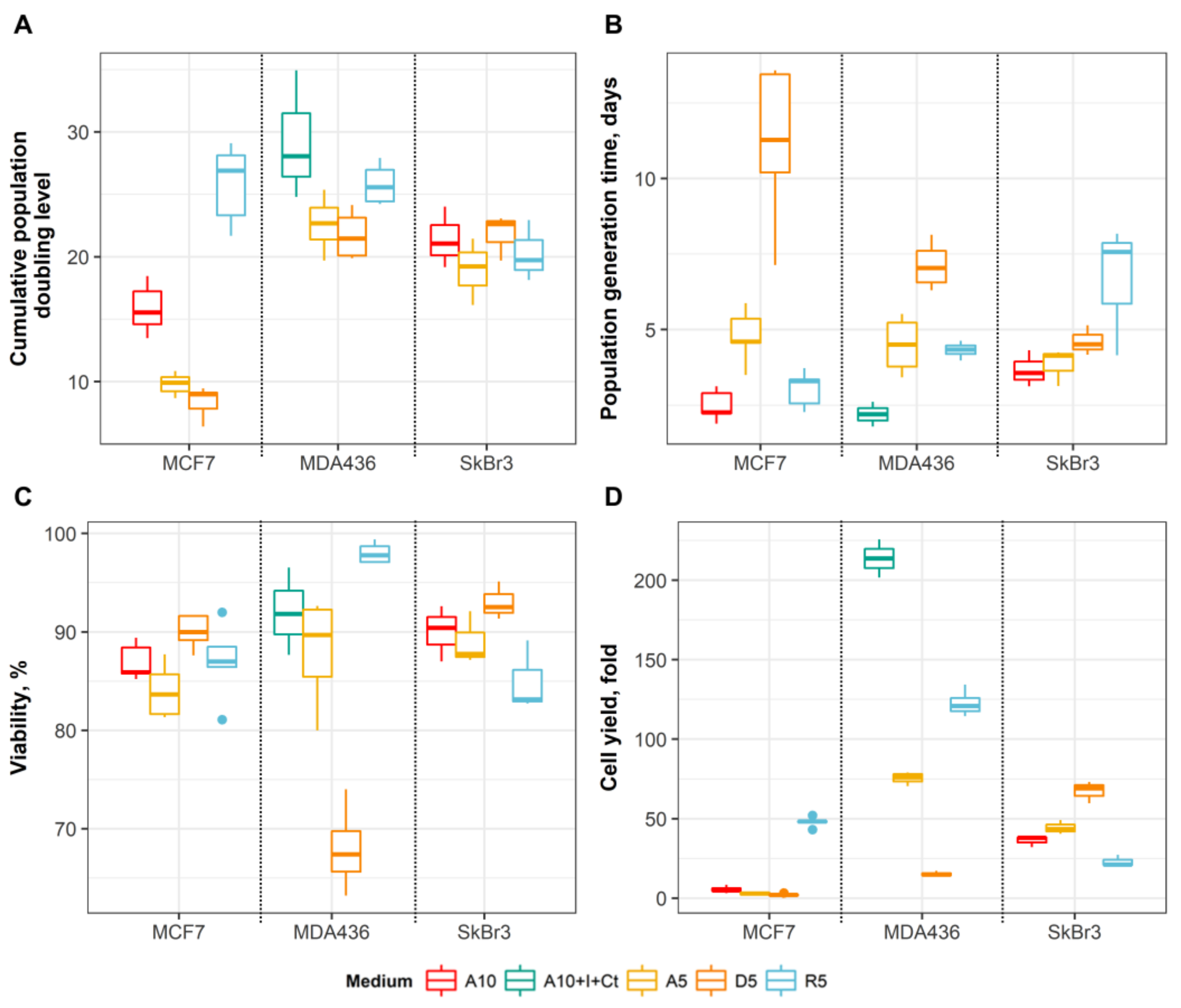
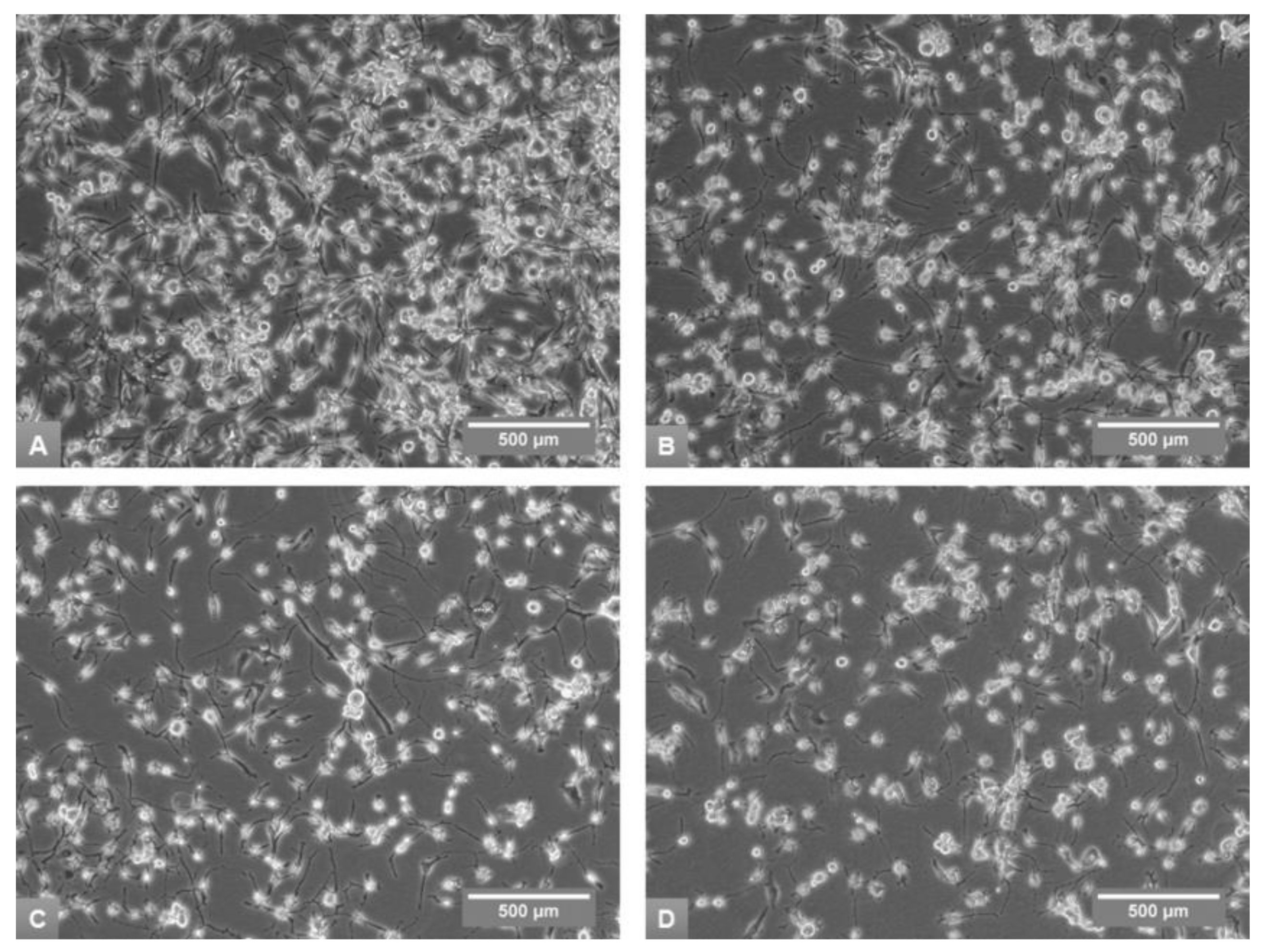
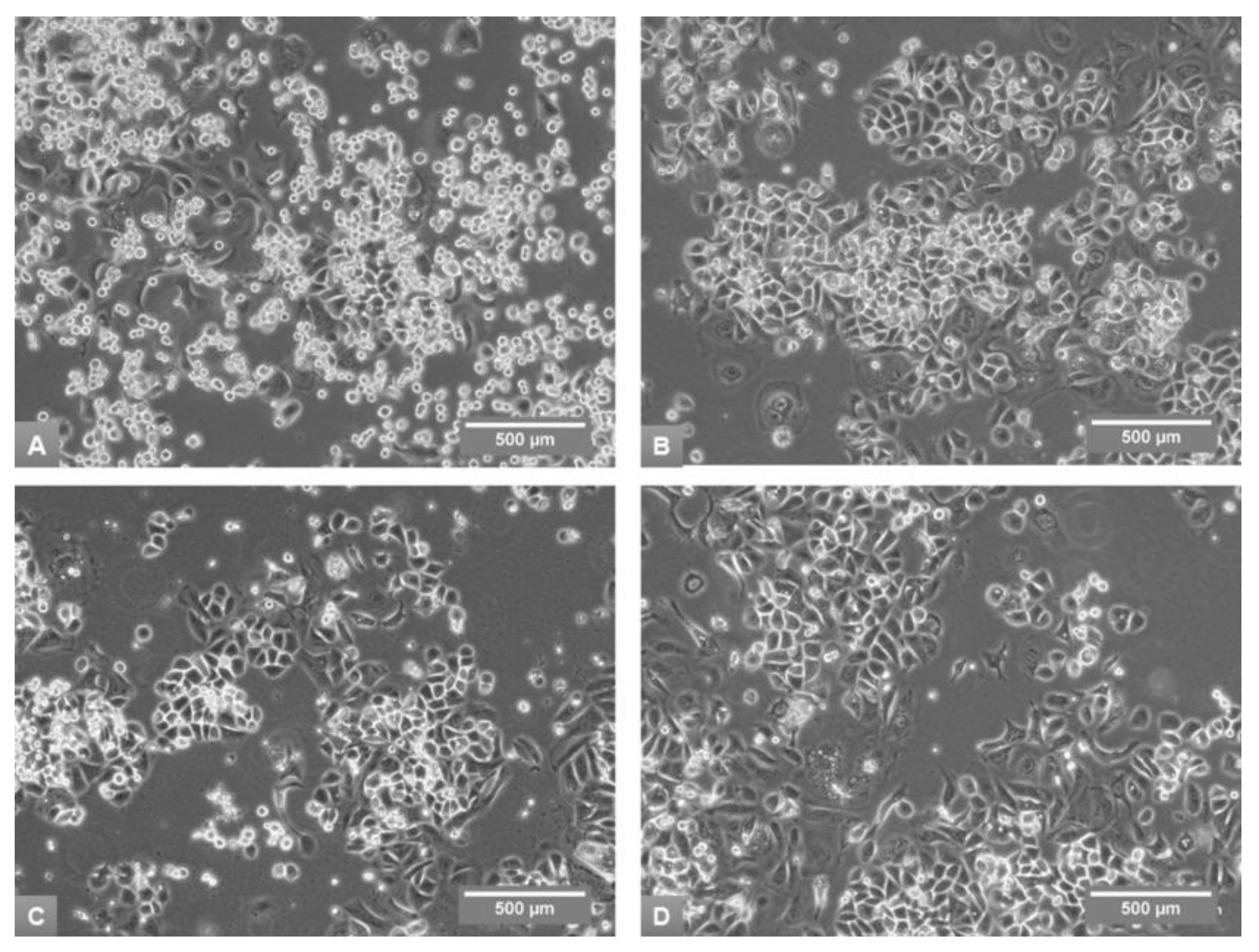
3.1. Effects of Media on Cell Growth and Morphology
3.2. Substitution of Serum with Hormones/Growth Factors Shifts MCF7, SkBr3 and MDA-MB-436 Cells Towards Less Differentiated Phenotype
3.3. An Effect of Basal Media on Epithelial Differentiation/Breast Cancer Marker Expression Depends on Initial Differentiation State and Genomic Context of the Cancer Cells
3.4. Transcriptional Profiles
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Vargo-Gogola, T.; Rosen, J.M. Modelling breast cancer: One size does not fit all. Nat. Rev. Cancer 2007, 7, 659–672. [Google Scholar] [CrossRef] [PubMed]
- Holliday, D.L.; Speirs, V. Choosing the right cell line for breast cancer research. Breast Cancer Res. 2011, 13, 215. [Google Scholar] [CrossRef] [PubMed]
- Eccles, S.A.; Aboagye, E.O.; Ali, S.; Anderson, A.S.; Armes, J.; Berditchevski, F.; Blaydes, J.P.; Brennan, K.; Brown, N.J.; Bryant, H.E.; et al. Critical research gaps and translational priorities for the successful prevention and treatment of breast cancer. Breast Cancer Res. 2013, 15, R92. [Google Scholar] [CrossRef] [PubMed]
- Lacroix, M.; Leclercq, G. Relevance of breast cancer cell lines as models for breast tumors: An update. Breast Cancer Res. Treat. 2004, 83, 249–289. [Google Scholar] [CrossRef] [PubMed]
- Vinci, M.; Gowan, S.; Boxall, F.; Patterson, L.; Zimmermann, M.; Court, W.; Lomas, C.; Mendiola, M.; Hardisson, D.; Eccles, S.A. Advances in establishment and analysis of three-dimensional tumor spheroid-based functional assays for target validation and drug evaluation. BMC Biol. 2012, 10, 29. [Google Scholar] [CrossRef] [PubMed]
- Prat, A.; Karginova, O.; Parker, J.S.; Fan, C.; He, X.; Bixby, L.; Harrell, J.C.; Roman, E.; Adamo, B.; Troester, M.; et al. Characterization of cell lines derived from breast cancers and normal mammary tissues for the study of the intrinsic molecular subtypes. Breast Cancer Res. Treat. 2013, 142, 237–255. [Google Scholar] [CrossRef] [PubMed]
- Neve, R.M.; Chin, K.; Fridlyand, J.; Yeh, J.; Baehner, F.L.; Fevr, T.; Clark, L.; Bayani, N.; Coppe, J.P.; Tong, F.; et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 2006, 10, 515–527. [Google Scholar] [CrossRef] [PubMed]
- Lim, E.; Vaillant, F.; Wu, D.; Forrest, N.C.; Pal, B.; Hart, A.H.; Asselin-Labat, M.L.; Gyorki, D.E.; Ward, T.; Partanen, A.; et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat. Med. 2009, 15, 907–913. [Google Scholar] [CrossRef] [PubMed]
- Shoemaker, R.H. The NCI60 human tumor cell line anticancer drug screen. Nat. Rev. Cancer 2006, 6, 813–823. [Google Scholar] [CrossRef] [PubMed]
- Monks, A.; Scudiero, D.; Skehan, P.; Shoemaker, R.; Paull, K.; Vistica, D.; Hose, C.; Langley, J.; Cronise, P.; Vaigro-Wolff, A.; et al. Feasibility of a high-flux anticancer drug screen using a diverse panel of cultured human tumor cell lines. J. Natl. Cancer Inst. 1991, 83, 757–766. [Google Scholar] [CrossRef] [PubMed]
- Prat, A.; Perou, C.M. Deconstructing the molecular portraits of breast cancer. Mol. Oncol. 2011, 5, 5–23. [Google Scholar] [CrossRef] [PubMed]
- Sorlie, T.; Perou, C.M.; Tibshirani, R.; Aas, T.; Geisler, S.; Johnsen, H.; Hastie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 2001, 98, 10869–10874. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Fan, C.; Oh, D.S.; Marron, J.S.; He, X.; Qaqish, B.F.; Livasy, C.; Carey, L.A.; Reynolds, E.; Dressler, L.; et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genom. 2006, 7, 96. [Google Scholar]
- Prat, A.; Parker, J.S.; Karginova, O.; Fan, C.; Livasy, C.; Herschkowitz, J.I.; He, X.; Perou, C.M. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010, 12, R68. [Google Scholar] [CrossRef] [PubMed]
- Mackay, A.; Tamber, N.; Fenwick, K.; Iravani, M.; Grigoriadis, A.; Dexter, T.; Lord, C.J.; Reis-Filho, J.S.; Ashworth, A. A high-resolution integrated analysis of genetic and expression profiles of breast cancer cell lines. Breast Cancer Res. Treat. 2009, 118, 481–498. [Google Scholar] [CrossRef] [PubMed]
- Kao, J.; Salari, K.; Bocanegra, M.; Choi, Y.-L.; Girard, L.; Gandhi, J.; Kwei, K.A.; Hernandez-Boussard, T.; Wang, P.; Gazdar, A.F.; et al. Molecular profiling of breast cancer cell lines defines relevant tumor models and provides a resource for cancer gene discovery. PLoS ONE 2009, 4, e6146. [Google Scholar] [CrossRef] [PubMed]
- Ross, D.T.; Perou, C.M. A comparison of gene expression signatures from breast tumors and breast tissue derived cell lines. Dis. Mark. 2001, 17, 99–109. [Google Scholar] [CrossRef]
- Perou, C.M.; Sorlie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular portraits of human breast tumors. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef] [PubMed]
- Parker, J.S.; Mullins, M.; Cheang, M.C.; Leung, S.; Voduc, D.; Vickery, T.; Davies, S.; Fauron, C.; He, X.; Hu, Z.; et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 2009, 27, 1160–1167. [Google Scholar] [CrossRef] [PubMed]
- Van Keymeulen, A.; Rocha, A.S.; Ousset, M.; Beck, B.; Bouvencourt, G.; Rock, J.; Sharma, N.; Dekoninck, S.; Blanpain, C. Distinct stem cells contribute to mammary gland development and maintenance. Nature 2011, 479, 189–193. [Google Scholar] [CrossRef] [PubMed]
- Shehata, M.; Teschendorff, A.; Sharp, G.; Novcic, N.; Russell, I.A.; Avril, S.; Prater, M.; Eirew, P.; Caldas, C.; Watson, C.J.; et al. Phenotypic and functional characterisation of the luminal cell hierarchy of the mammary gland. Breast Cancer Res. 2012, 14, R134. [Google Scholar] [CrossRef] [PubMed]
- Granit, R.Z.; Slyper, M.; Ben-Porath, I. Axes of Differentiation in Breast Cancer: Untangling Stemness, Lineage Identity, and the Epithelial to Mesenchymal Transition; Wiley Interdisciplinary Reviews. Systems Biology and Medicine; John Wiley & Sons, Inc.: New York, NY, USA, 2014; Volume 6, pp. 93–106. [Google Scholar]
- Chou, J.; Provot, S.; Werb, Z. GATA3 in development and cancer differentiation: Cells GATA have it! J. Cell. Physiol. 2010, 222, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Kouros-Mehr, H.; Kim, J.W.; Bechis, S.K.; Werb, Z. GATA-3 and the regulation of the mammary luminal cell fate. Curr. Opin. Cell Biol. 2008, 20, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Douglas, N.C.; Papaioannou, V.E. The T-box transcription factors TBX2 and TBX3 in mammary gland development and breast cancer. J. Mammary Gland Biol. Neoplasia 2013, 18, 143–147. [Google Scholar] [CrossRef] [PubMed]
- Bouras, T.; Pal, B.; Vaillant, F.; Harburg, G.; Asselin-Labat, M.L.; Oakes, S.R.; Lindeman, G.J.; Visvader, J.E. Notch signaling regulates mammary stem cell function and luminal cell-fate commitment. Cell Stem Cell 2008, 3, 429–441. [Google Scholar] [CrossRef] [PubMed]
- Visvader, J.E.; Stingl, J. Mammary stem cells and the differentiation hierarchy: Current status and perspectives. Genes Dev. 2014, 28, 1143–1158. [Google Scholar] [CrossRef] [PubMed]
- Heiser, L.M.; Sadanandam, A.; Kuo, W.L.; Benz, S.C.; Goldstein, T.C.; Ng, S.; Gibb, W.J.; Wang, N.J.; Ziyad, S.; Tong, F.; et al. Subtype and pathway specific responses to anticancer compounds in breast cancer. Proc. Natl. Acad. Sci. USA 2012, 109, 2724–2729. [Google Scholar] [CrossRef] [PubMed]
- Niepel, M.; Hafner, M.; Pace, E.A.; Chung, M.; Chai, D.H.; Zhou, L.; Schoeberl, B.; Sorger, P.K. Profiles of basal and stimulated receptor signaling networks predict drug response in breast cancer lines. Sci Signal. 2013, 6, ra84. [Google Scholar] [CrossRef]
- Liu, X.; Ory, V.; Chapman, S.; Yuan, H.; Albanese, C.; Kallakury, B.; Timofeeva, O.A.; Nealon, C.; Dakic, A.; Simic, V.; et al. ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. Am. J. Pathol. 2012, 180, 599–607. [Google Scholar] [CrossRef] [PubMed]
- Ethier, S.P.; Mahacek, M.L.; Gullick, W.J.; Frank, T.S.; Weber, B.L. Differential isolation of normal luminal mammary epithelial cells and breast cancer cells from primary and metastatic sites using selective media. Cancer Res. 1993, 53, 627–635. [Google Scholar] [PubMed]
- Ethier, S.P.; Ammerman, C.A.; Dziubinski, M.L. Isolation and Culture of Human Breast Cancer Cells from Primary Tumors and Metastases. In Methods in Mammary Gland Biology and Breast Cancer Research; Ip, M.M., Asch, B.B., Eds.; Springer: Boston, MA, USA, 2000; pp. 195–208. [Google Scholar]
- Wallden, B.; Storhoff, J.; Nielsen, T.; Dowidar, N.; Schaper, C.; Ferree, S.; Liu, S.; Leung, S.; Geiss, G.; Snider, J.; et al. Development and verification of the PAM50-based Prosigna breast cancer gene signature assay. BMC Med. Genom. 2015, 8, 54. [Google Scholar] [CrossRef] [PubMed]
- Planas-Silva, M.D.; Waltz, P.K. Estrogen promotes reversible epithelial-to-mesenchymal-like transition and collective motility in MCF-7 breast cancer cells. J. Steroid Biochem. Mole. Biol. 2007, 104, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Li, J.; Wang, L.; Bian, C.; Wang, Q.; Liao, L.; Dou, X.; Bian, X.; Zhao, R.C. Overexpression of DeltaNp63alpha induces a stem cell phenotype in MCF7 breast carcinoma cell line through the Notch pathway. Cancer Sci. 2010, 101, 2417–2424. [Google Scholar] [CrossRef] [PubMed]
- Al-Hussaini, H.; Subramanyam, D.; Reedijk, M.; Sridhar, S.S. Notch signaling pathway as a therapeutic target in breast cancer. Mol. Cancer Ther. 2011, 10, 9–15. [Google Scholar] [CrossRef] [PubMed]
- D’Angelo, R.C.; Ouzounova, M.; Davis, A.; Choi, D.; Tchuenkam, S.M.; Kim, G.; Luther, T.; Quraishi, A.A.; Senbabaoglu, Y.; Conley, S.J.; et al. Notch reporter activity in breast cancer cell lines identifies a subset of cells with stem cell activity. Molecular Cancer. Therapeutics 2015, 14, 779–787. [Google Scholar]
- Mohammed, H.; Russell, I.A.; Stark, R.; Rueda, O.M.; Hickey, T.E.; Tarulli, G.A.; Serandour, A.A.; Birrell, S.N.; Bruna, A.; Saadi, A.; et al. Progesterone receptor modulates ERalpha action in breast cancer. Nature 2015, 523, 313–317. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.Y.; Bay, B.H.; Aw, S.E.; Lin, V.C. A novel antiestrogenic mechanism in progesterone receptor-transfected breast cancer cells. J. Biol. Chem. 2005, 280, 17480–17487. [Google Scholar] [CrossRef] [PubMed]
- Fan, W.; Huang, X.; Chen, C.; Gray, J.; Huang, T. TBX3 and its isoform TBX3+2a are functionally distinctive in inhibition of senescence and are overexpressed in a subset of breast cancer cell lines. Cancer Res. 2004, 64, 5132–5139. [Google Scholar] [CrossRef] [PubMed]
- Fillmore, C.M.; Gupta, P.B.; Rudnick, J.A.; Caballero, S.; Keller, P.J.; Lander, E.S.; Kuperwasser, C. Estrogen expands breast cancer stem-like cells through paracrine FGF/Tbx3 signaling. Proc. Natl. Acad. Sci. USA 2010, 107, 21737–21742. [Google Scholar] [CrossRef] [PubMed]
- Pendse, S.N.; Maertens, A.; Rosenberg, M.; Roy, D.; Fasani, R.A.; Vantangoli, M.M.; Madnick, S.J.; Boekelheide, K.; Fornace, A.J.; Odwin, S.A.; et al. Information-dependent enrichment analysis reveals time-dependent transcriptional regulation of the estrogen pathway of toxicity. Arch. Toxicol. 2017, 91, 1749–1762. [Google Scholar] [CrossRef] [PubMed]
- Stender, J.D.; Frasor, J.; Komm, B.; Chang, K.C.N.; Kraus, W.L.; Katzenellenbogen, B.S. Estrogen-Regulated Gene Networks in Human Breast Cancer Cells: Involvement of E2F1 in the Regulation of Cell Proliferation. Mol. Endocrinol. 2007, 21, 2112–2123. [Google Scholar] [CrossRef] [PubMed]
- Frasor, J.; Danes, J.M.; Komm, B.; Chang, K.C.N.; Lyttle, C.R.; Katzenellenbogen, B.S. Profiling of Estrogen Up- and Down-Regulated Gene Expression in Human Breast Cancer Cells: Insights into Gene Networks and Pathways Underlying Estrogenic Control of Proliferation and Cell Phenotype. Endocrinology 2003, 144, 4562–4574. [Google Scholar] [CrossRef] [PubMed]
- Nagai, M.A.; Brentani, M.M. Gene Expression Profiles in Breast Cancer to Identify Estrogen Receptor Target Genes. Recent Adv. Med. Chem. 2015, 1, 381–403. [Google Scholar]
- Yamaga, R.; Ikeda, K.; Horie-Inoue, K.; Ouchi, Y.; Suzuki, Y.; Inoue, S. RNA sequencing of MCF-7 breast cancer cells identifies novel estrogen-responsive genes with functional estrogen receptor-binding sites in the vicinity of their transcription start sites. Hormones Cancer 2013, 4, 222–232. [Google Scholar] [CrossRef] [PubMed]
- Omoto, Y.; Hayashi, S. A study of estrogen signaling using DNA microarray in human breast cancer. Breast Cancer 2002, 9, 308–311. [Google Scholar] [CrossRef] [PubMed]


| Cell Line | Source * | Tumor Type * | ER * | PR * | HER2 * | Cluster/Subtype ** | Complete Growth Medium According to ATCC * |
|---|---|---|---|---|---|---|---|
| MCF7 | PE | IDC | + | + | 0 | luminal | Eagle’s Minimum Essential Medium + 10% FBS + 0.01 mg/mL hr insulin |
| SkBr3 | PE | AC | 0 | 0 | + | luminal/HER2+ | McCoy’s 5a +10% FBS |
| MDA-MB-436 | PE | IDC | 0 | 0 | 0 | basal B/claudin-low | Leibovitz’s L-15 + 10% FBS + 10 μg/mL insulin + 16 μg/mL glutathione |
| Medium | Basis | FBS (%) | Supplements * |
|---|---|---|---|
| A10 | DMEM/F12 (1:1) | 10 | P/S |
| A5 | DMEM/F12 (1:1) | 5 | P/S, I, HC, EGF, E2, HEPES, T, T3, Se, Ct |
| D5 | DMEM/F12 (1:3) | 5 | P/S, I, HC, EGF, E2, HEPES, T, T3, Se, Ct |
| R5 | RPMI1640 | 5 | P/S, I, HC, EGF, E2, T, T3, Se, Ct |
| Gene Class ** | Gene | MCF7 | SkBr3 | MDA-MB-436 |
|---|---|---|---|---|
| MLR | AR | ↓↓ | ↑↑ | ↓↓ |
| ELF5 | NS | ↑↑ | NS | |
| ESR1 | ↓ | NS | NS | |
| FOXA1 | ↓ | NS | ↑ | |
| GATA3 | NS | ↓↓ | ↑↑ | |
| HES1 | NS | ↑ | NS | |
| PGR | ↑↑ | NS | NS | |
| TBX3 | ↓↓ | NS | ↑ | |
| LPR | EZH2 | ↓ | NS | NS |
| FOXM1 | NS | ↓↓ | NS | |
| FOXQ1 | NS | NS | NS | |
| SOX9 | NS | ↑↑ | ↓↓ | |
| VGLL1 | ↓↓ | ↓ | NS | |
| BR | HEY1 | ↑↑ | ↑↑↑ | NS |
| ID3 | NS | NS | NS | |
| IFI16 | ↑↑ | ↑↑↑ | ↓ | |
| SNAI2 | ↑ | NS | NS | |
| TBX2 | NS | ↑↑ | ↑↑↑ | |
| TP63 | ↑↑↑ | NS | NS | |
| LM | EPCAM | ↓↓ | NS | NS |
| ESR2 | NS | NS | NS | |
| KRT18 | NS | ↓ | NS | |
| KRT19 | NS | NS | ↓ | |
| KRT7 | NS | ↓ | ↑↑ | |
| KRT8 | NS | NS | ↓ | |
| MUC1 | NS | NS | ↓↓↓ | |
| NAT1 | NS | NS | NS | |
| SLC39A6 | ↓ | NS | ↓ | |
| TMEM45B | NS | ↑ | ↑↑ | |
| XBP1 | NS | NS | ↓ | |
| BM | EGFR | NS | ↓ | ↑ |
| ERBB2 | ↑↑ | NS | NS | |
| FOXC1 | NS | ↑↑ | ↑↑ | |
| ITGB3 | NS | ↑↑↑ | NS | |
| KRT16 | ↑↑↑ | ↑↑ | ↓↓↓ | |
| KRT17 | NS | ↑ | ↓↓↓ | |
| KRT6A | ↑↑↑ | ↑↑↑ | ↑↑ | |
| KRTAP5-6 | NS | ↑ | NS | |
| MIA | ↑↑↑ | NS | NS | |
| PROCR | ↑ | NS | ↑ |
| Gene Class ** | Gene | MCF7 | SkBr3 | MDA-MB-436 | |||
|---|---|---|---|---|---|---|---|
| D5 | R5 | D5 | R5 | D5 | R5 | ||
| MLR | AR | NS | ↑ | NS | ↑ | NS | NS |
| ELF5 | NS | NS | NS | ↑ | NS | ↓↓ | |
| ESR1 | ↑ | ↑↑ | NS | ↓ | NS | NS | |
| FOXA1 | NS | ↑ | ↑ | ↑ | NS | ↓ | |
| GATA3 | ↓↓ | ↑ | NS | ↑↑↑ | NS | NS | |
| HES1 | ↓ | NS | ↑↑↑ | NS | NS | ↓ | |
| PGR | ↑ | ↑↑ | NS | NS | NS | NS | |
| TBX3 | NS | NS | NS | ↑ | NS | ↓ | |
| LPR | EZH2 | ↑ | NS | ↑ | ↑ | NS | ↓ |
| FOXM1 | NS | NS | NS | ↓↓ | NS | ↓ | |
| FOXQ1 | NS | NS | ↑↑↑ | NS | NS | ↓↓↓ | |
| SOX9 | NS | ↓↓ | ↓↓ | ↑↑ | NS | NS | |
| VGLL1 | NS | NS | NS | ↑ | NS | NS | |
| BR | HEY1 | NS | ↓↓↓ | ↑↑ | ↑↑ | NS | ↓ |
| ID3 | NS | NS | ↑↑↑ | NS | NS | NS | |
| IFI16 | NS | NS | ↓ | NS | NS | NS | |
| SNAI2 | ↓ | ↓↓↓ | ↑↑↑ | NS | NS | NS | |
| TBX2 | NS | NS | NS | ↑↑ | NS | NS | |
| TP63 | ↓↓↓ | ↓↓↓ | NS | NS | NS | NS | |
| LM | EPCAM | NS | ↑ | NS | ↓ | NS | ↓↓ |
| ESR2 | NS | NS | ↓↓↓ | NS | NS | NS | |
| KRT18 | NS | ↑↑ | ↑↑ | NS | NS | ↓ | |
| KRT19 | NS | ↑↑ | NS | NS | NS | ↓ | |
| KRT7 | NS | ↓↓↓ | ↑↑↑ | NS | NS | ↑↑ | |
| KRT8 | NS | NS | NS | NS | NS | ↓ | |
| MUC1 | NS | NS | ↑↑ | NS | NS | ↑ | |
| NAT1 | NS | ↑ | NS | NS | NS | ↓ | |
| SLC39A6 | NS | ↑↑↑ | NS | NS | NS | ↓ | |
| TMEM45B | NS | NS | ↑ | ↓↓ | NS | ↓↓↓ | |
| XBP1 | NS | ↑ | ↑ | NS | NS | NS | |
| BM | EGFR | NS | ↓↓ | ↓ | ↓ | NS | ↓ |
| ERBB2 | NS | NS | ↑ | ↓↓ | NS | NS | |
| FOXC1 | NS | NS | NS | ↓ | NS | ↓ | |
| ITGB3 | ↓ | ↓↓↓ | NS | NS | NS | NS | |
| KRT16 | NS | ↓↓↓ | ↑ | ↓↓ | ↑↑ | NS | |
| KRT17 | NS | NS | NS | ↓↓↓ | NS | ↓ | |
| KRT6A | NS | ↓↓↓ | ↑↑ | ↓↓ | ↑↑↑ | ↑↑↑ | |
| KRTAP5-6 | NS | NS | NS | ↓↓↓ | NS | NS | |
| MIA | ↓ | NS | NS | NS | NS | ↓ | |
| PROCR | ↓↓ | NS | NS | NS | NS | ↓ | |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pirsko, V.; Cakstina, I.; Priedite, M.; Dortane, R.; Feldmane, L.; Nakazawa-Miklasevica, M.; Daneberga, Z.; Gardovskis, J.; Miklasevics, E. An Effect of Culture Media on Epithelial Differentiation Markers in Breast Cancer Cell Lines MCF7, MDA-MB-436 and SkBr3. Medicina 2018, 54, 11. https://doi.org/10.3390/medicina54020011
Pirsko V, Cakstina I, Priedite M, Dortane R, Feldmane L, Nakazawa-Miklasevica M, Daneberga Z, Gardovskis J, Miklasevics E. An Effect of Culture Media on Epithelial Differentiation Markers in Breast Cancer Cell Lines MCF7, MDA-MB-436 and SkBr3. Medicina. 2018; 54(2):11. https://doi.org/10.3390/medicina54020011
Chicago/Turabian StylePirsko, Valdis, Inese Cakstina, Marta Priedite, Rasma Dortane, Linda Feldmane, Miki Nakazawa-Miklasevica, Zanda Daneberga, Janis Gardovskis, and Edvins Miklasevics. 2018. "An Effect of Culture Media on Epithelial Differentiation Markers in Breast Cancer Cell Lines MCF7, MDA-MB-436 and SkBr3" Medicina 54, no. 2: 11. https://doi.org/10.3390/medicina54020011
APA StylePirsko, V., Cakstina, I., Priedite, M., Dortane, R., Feldmane, L., Nakazawa-Miklasevica, M., Daneberga, Z., Gardovskis, J., & Miklasevics, E. (2018). An Effect of Culture Media on Epithelial Differentiation Markers in Breast Cancer Cell Lines MCF7, MDA-MB-436 and SkBr3. Medicina, 54(2), 11. https://doi.org/10.3390/medicina54020011






