Genome-Wide Identification and Expression Analysis of LBD Transcription Factor Genes in Passion Fruit (Passiflora edulis)
Abstract
:1. Introduction
2. Results
2.1. Whole-Genome Characterization of PeLBD Genes in Passion Fruit
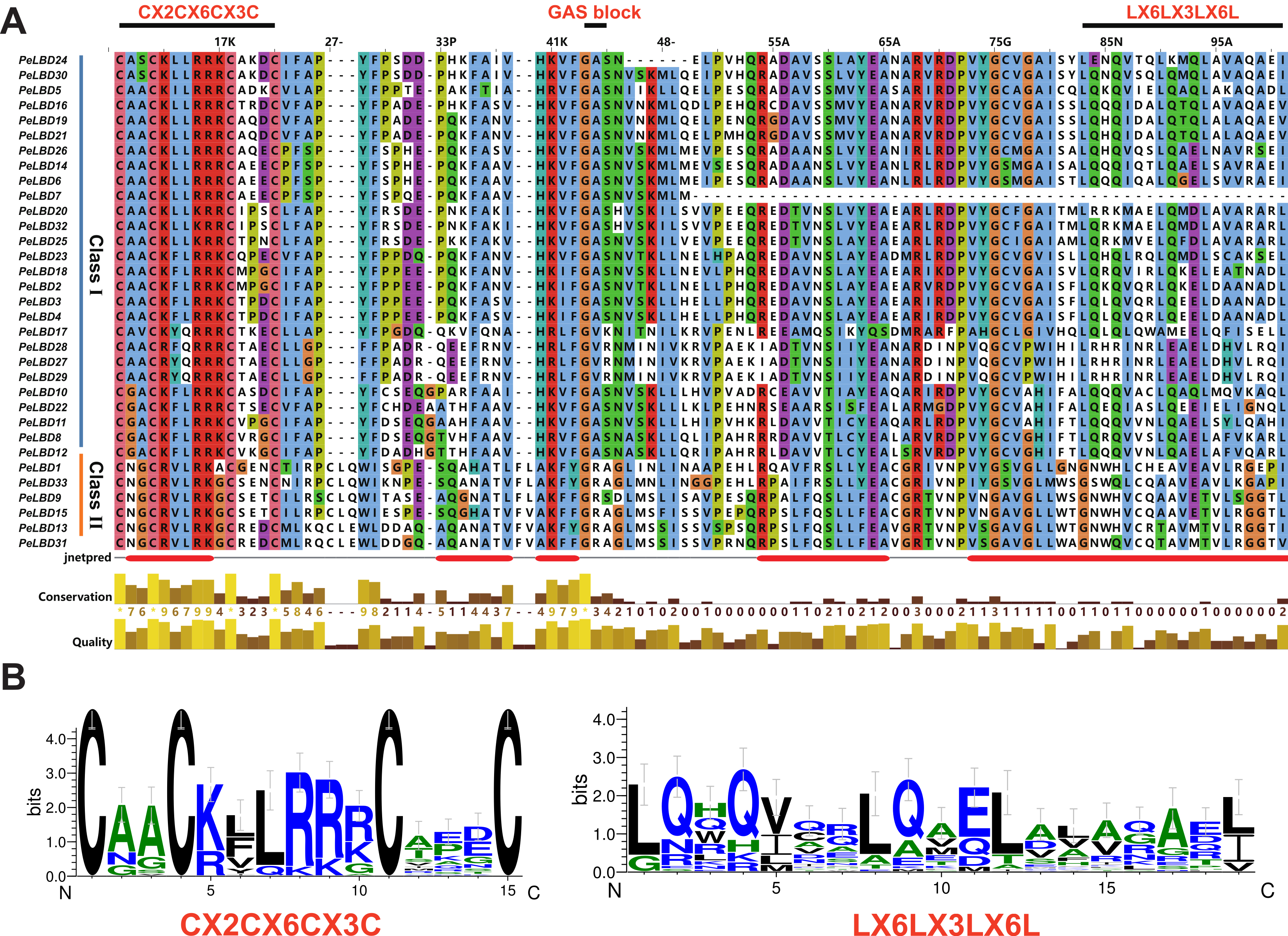
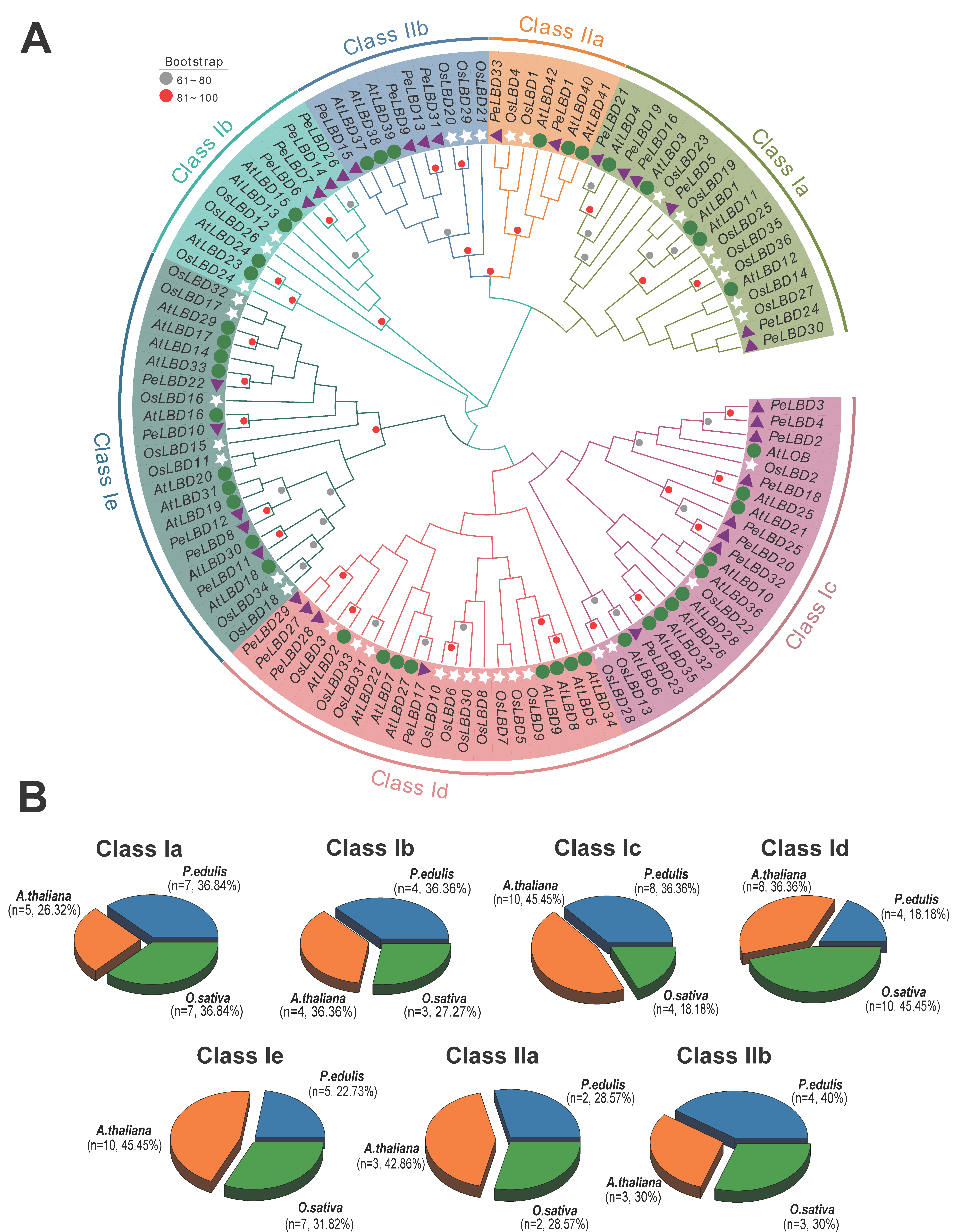
2.2. Multiple Sequence Alignment and Phylogenetic Analysis of PeLBD Genes
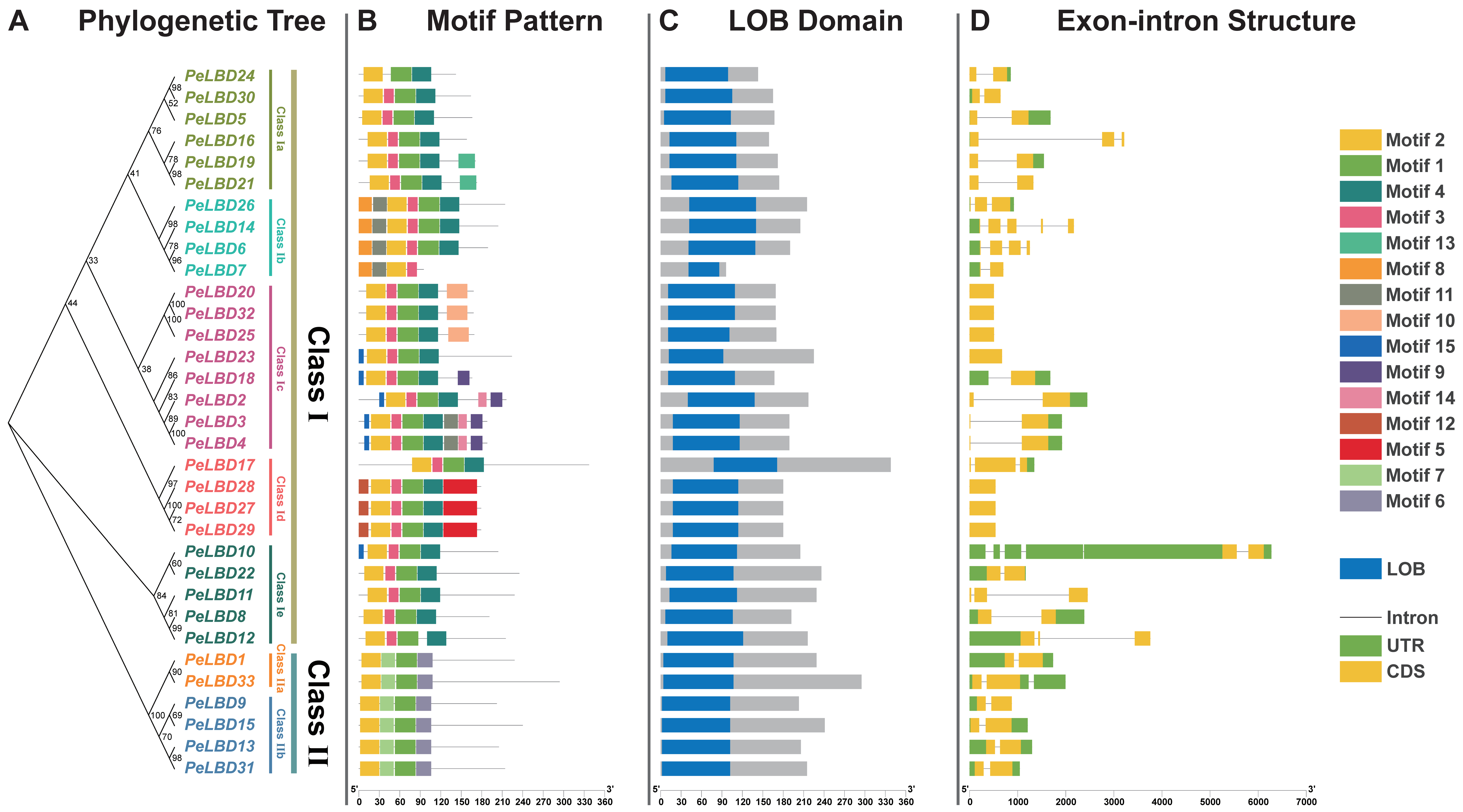
2.3. Gene Structure and Conserved Motif Analysis of PeLBD Genes
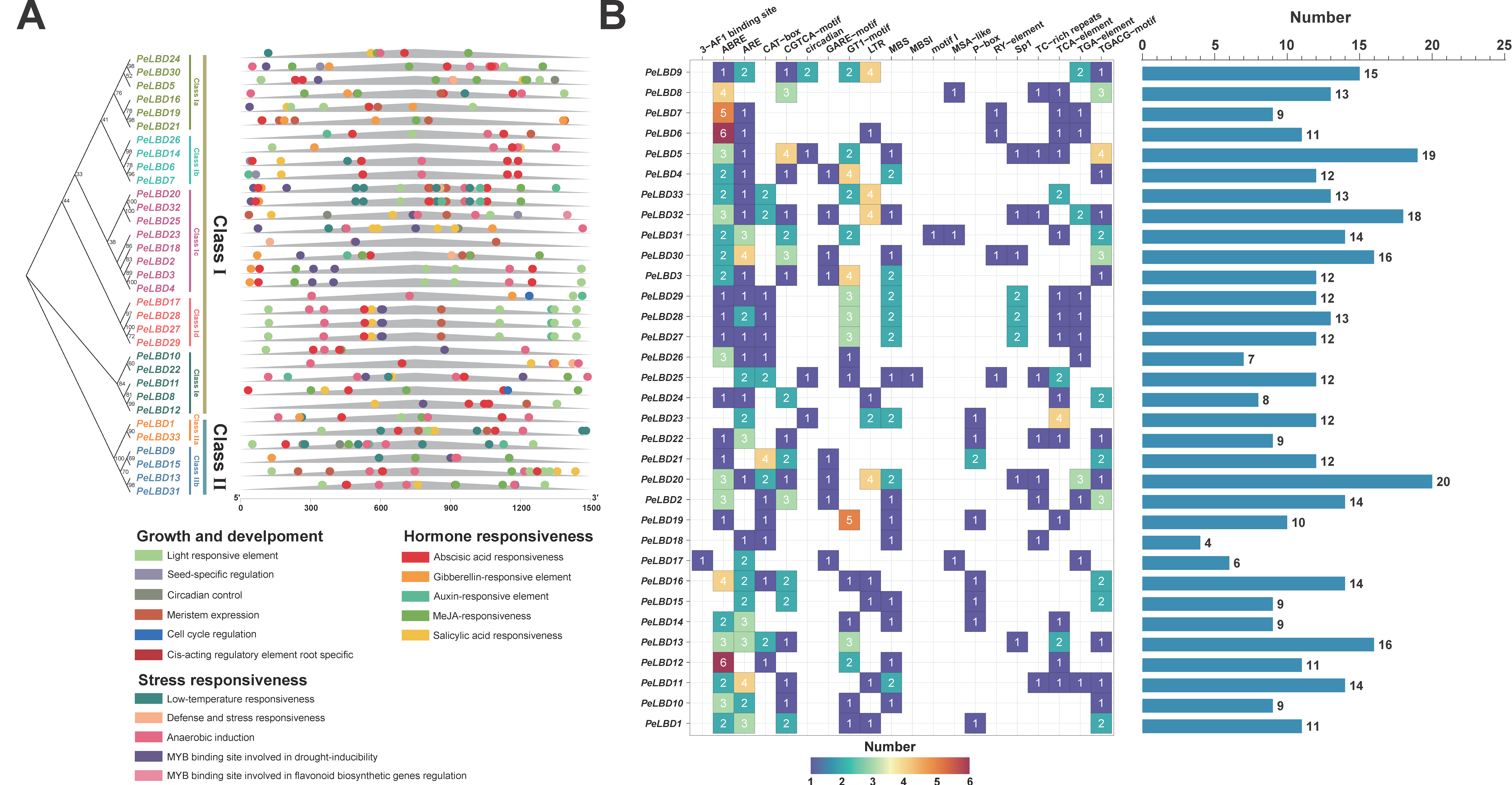
2.4. Analysis of Cis-Acting Elements in PeLBD Promoters
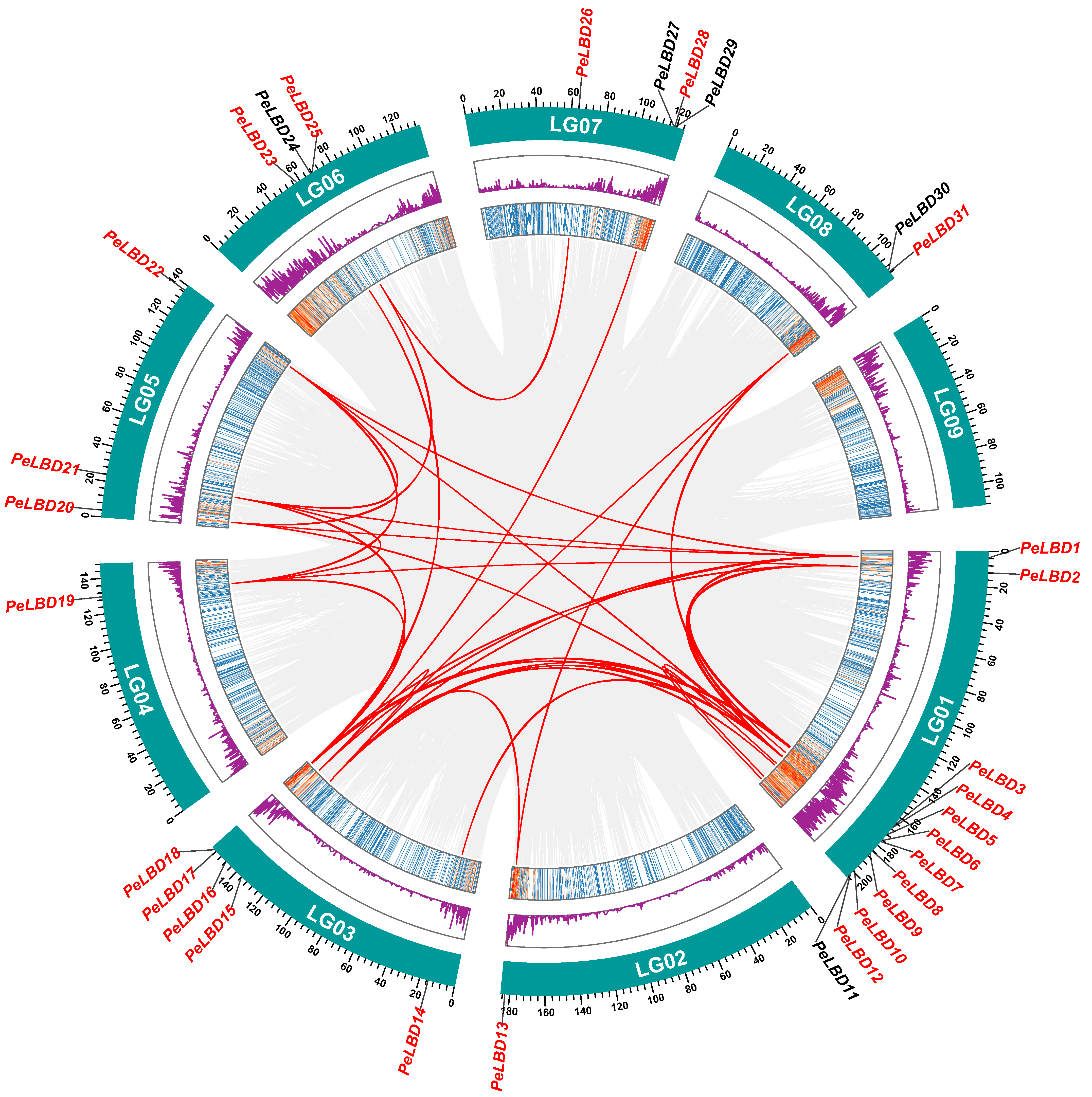
2.5. Chromosomal Location, Collinearity and Evolution Analysis of PeLBD Genes
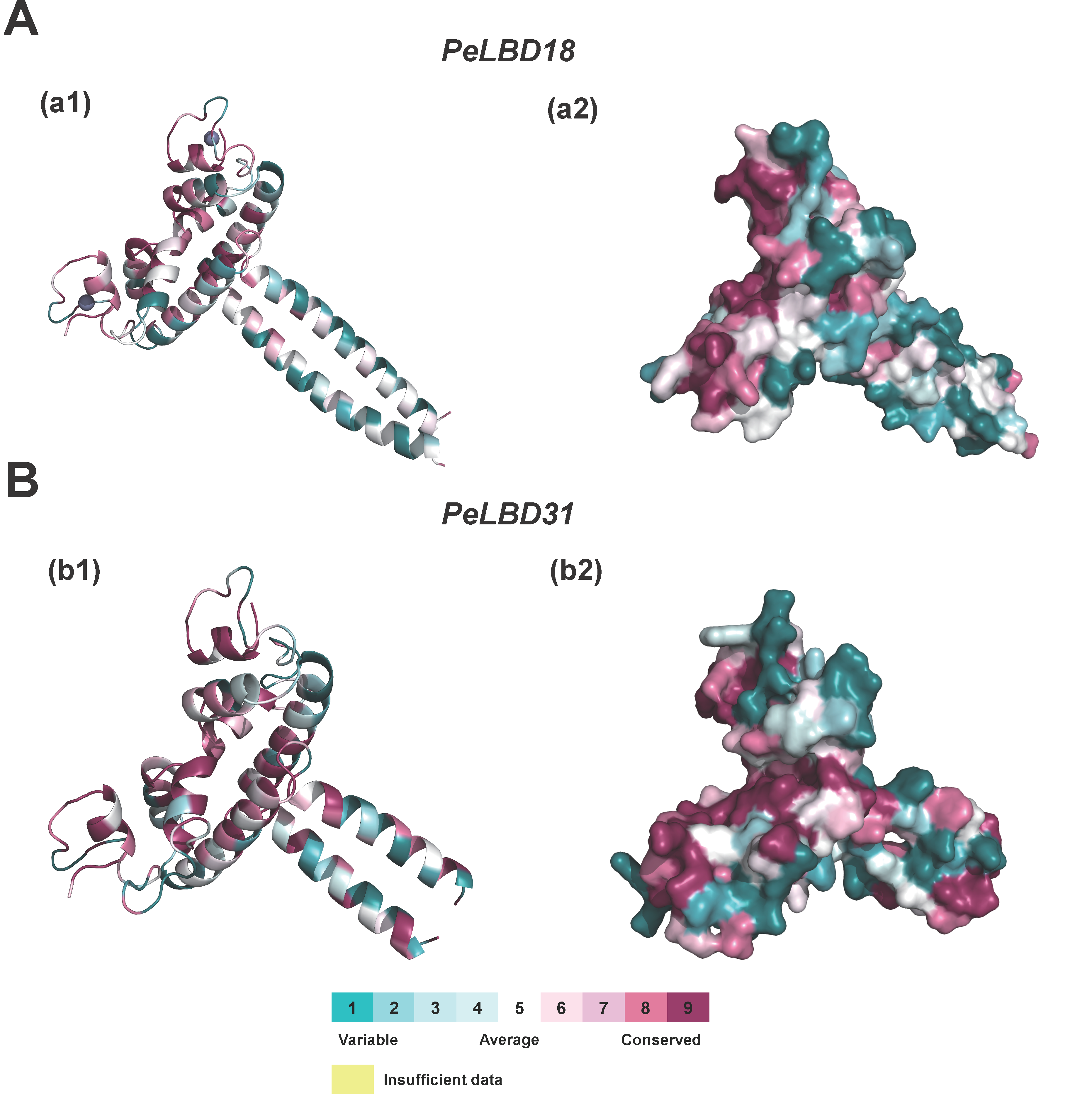
2.6. Homology Modeling of PeLBD Tertiary Structures
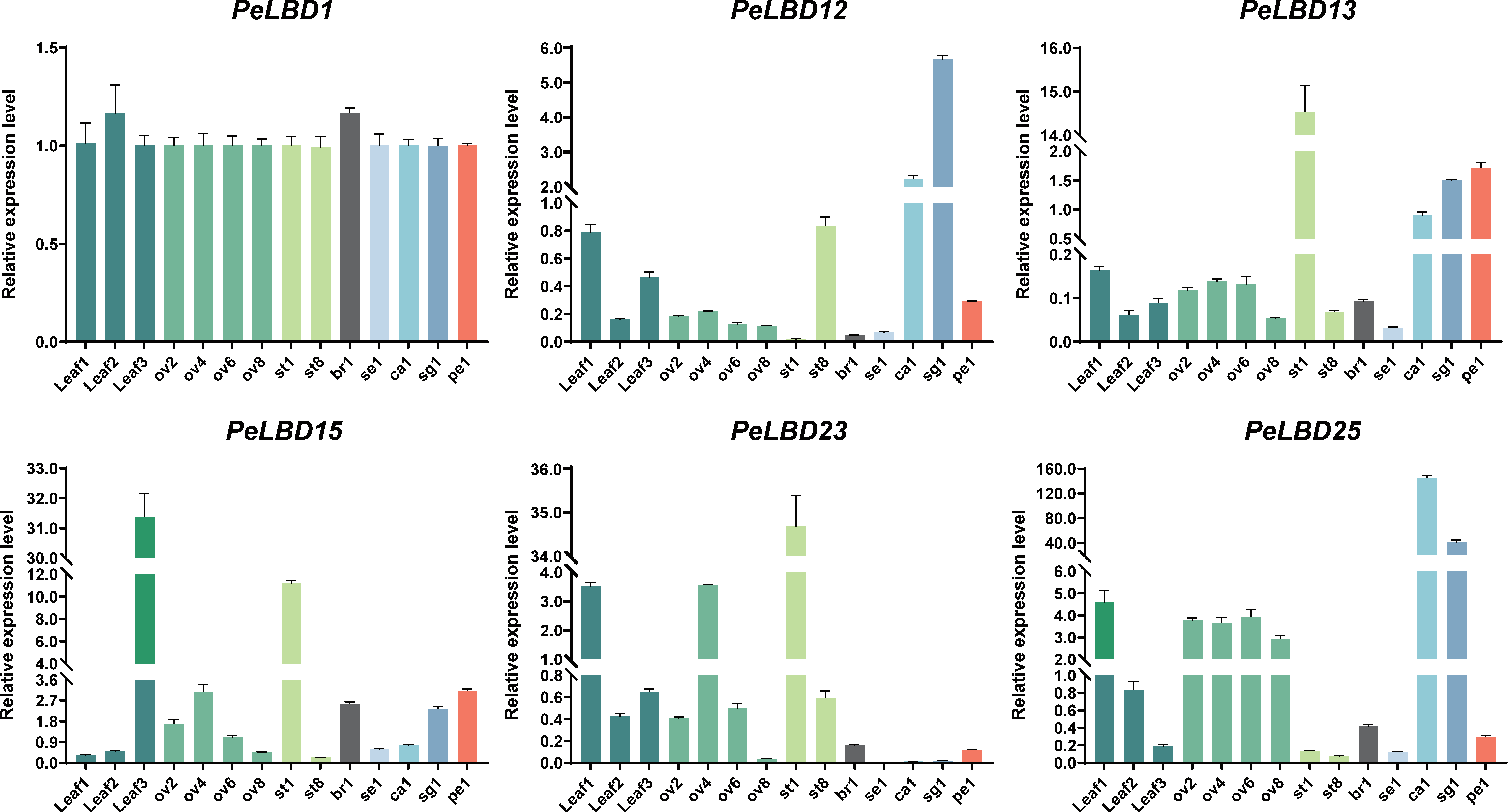
2.7. Expression Patterns of PeLBDs in Different Tissues of Passion Fruit
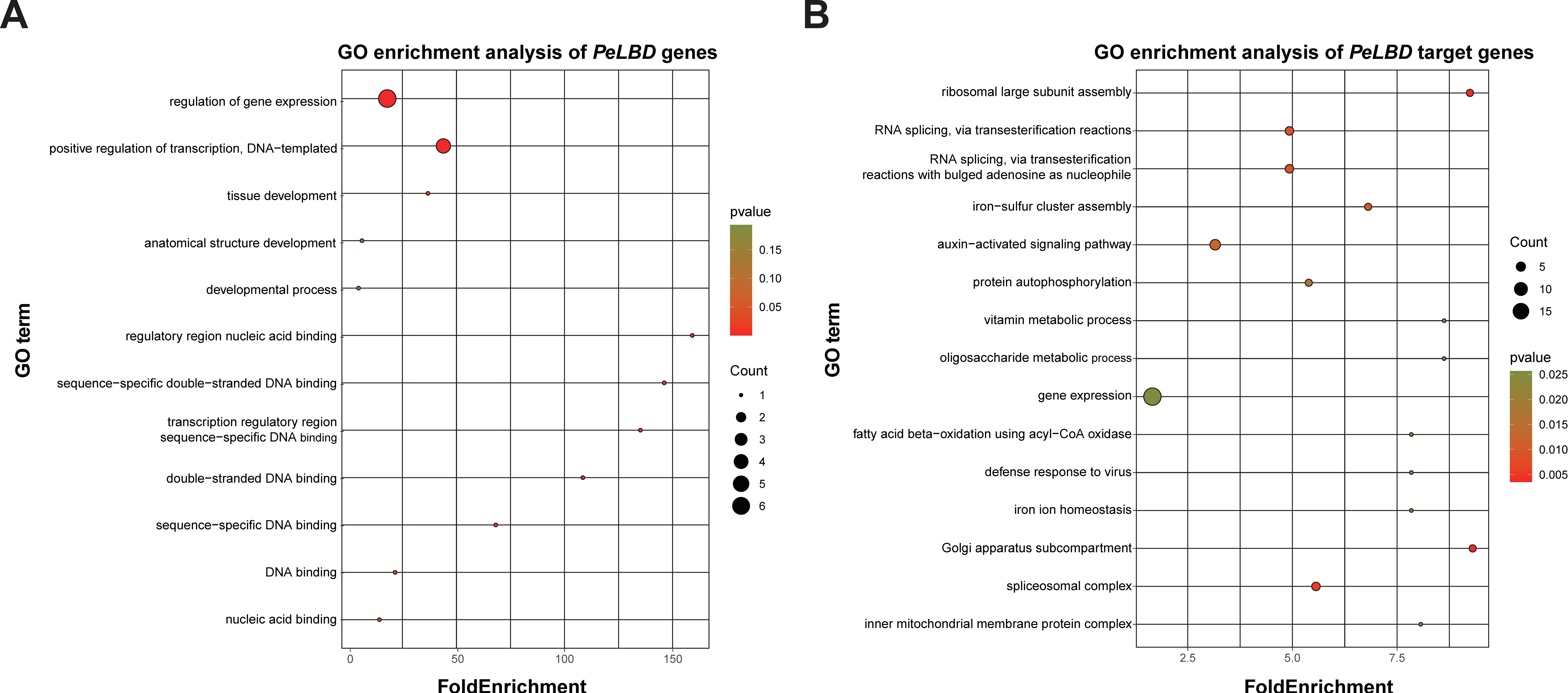
2.8. Identification and Annotation of PeLBD Target Genes
3. Discussion
4. Materials and Methods
4.1. Identification and Sequence Analysis of LBD Proteins from Passion Fruit
4.2. Multiple Sequence Alignment and Phylogenetic Tree Construction
4.3. Gene Structure, Conserved Motif and Cis-Regulatory Elements Analysis
4.4. Chromosomal Distribution and Gene Duplication
4.5. Three-Dimensional (3D) Structural Modeling of LBD Family Proteins
4.6. Identification and Annotation of LBD Target Genes
4.7. PeLBDs Transcriptome Analysis Based on RNA-Seq Data
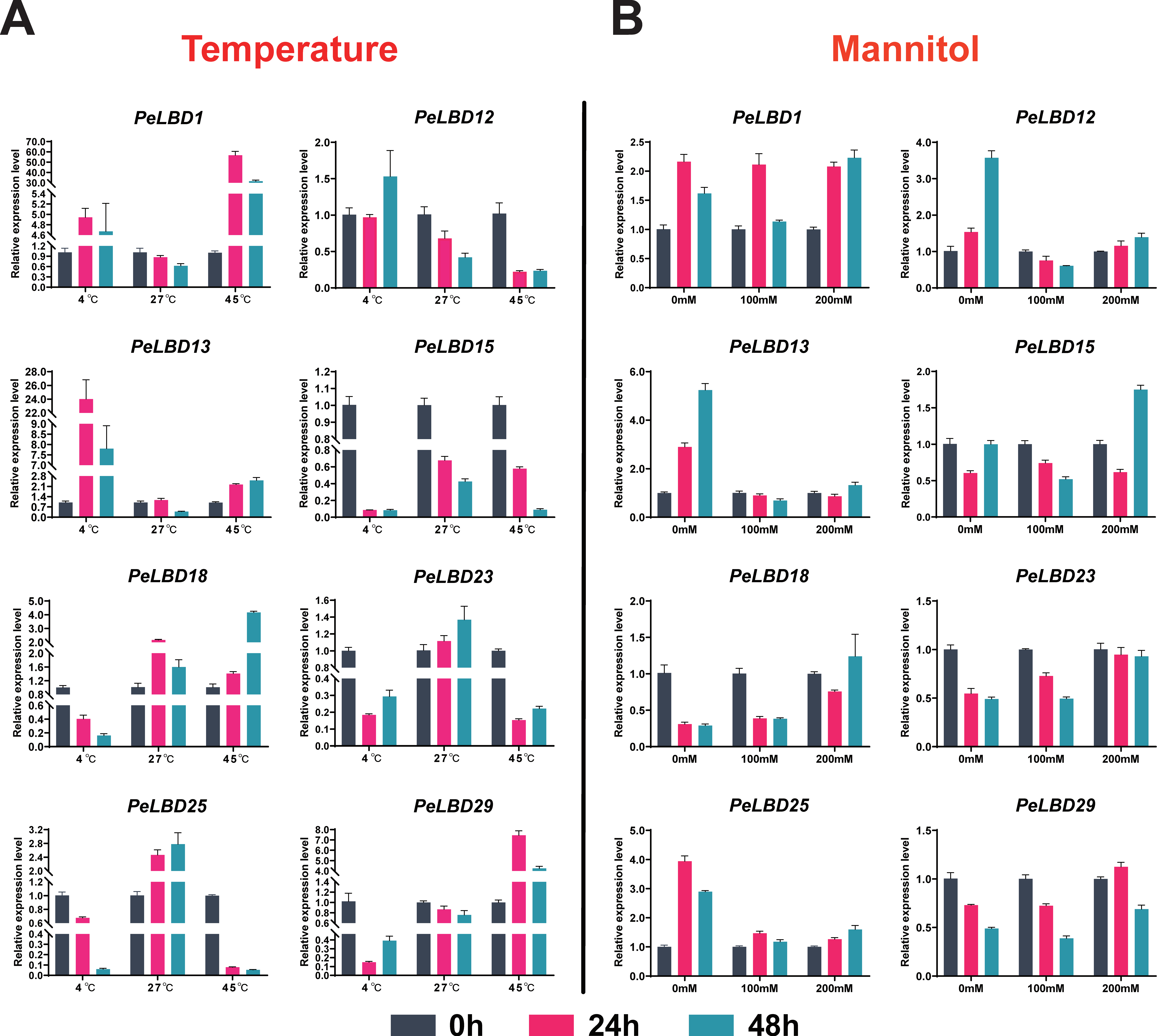
4.8. Plant Material, Stress Treatment, RNA Extraction, and qRT–PCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lopez-Vargas, J.H.; Fernandez-Lopez, J.; Perez-Alvarez, J.A.; Viuda-Martos, M. Chemical, physico-chemical, technological, antibacterial and antioxidant properties of dietary fiber powder obtained from yellow passion fruit (Passiflora edulis var. flavicarpa) co-products. Food Res. Int. 2013, 51, 756–763. [Google Scholar] [CrossRef]
- Antognoni, F.; Zheng, S.P.; Pagnucco, C.; Baraldi, R.; Poli, F.; Biondi, S. Induction of flavonoid production by UV-B radiation in Passiflora quadrangularis callus cultures. Fitoterapia 2007, 78, 345–352. [Google Scholar] [CrossRef] [PubMed]
- Rudnicki, M.; Silveira, M.; Pereira, T.; Oliveira, M.; Reginatto, F.; Dal-Pizzol, F.; Moreira, J. Protective effects of Passiflora alata extract pretreatment on carbon tetrachloride induced oxidative damage in rats. Food Chem. Toxicol. 2007, 45, 656–661. [Google Scholar] [CrossRef] [PubMed]
- Foudah, A.I.; Alam, P.; Kamal, Y.T.; Alqasoumi, S.I.; Alqarni, M.H.; Ross, S.A.; Yusufoglu, H.S. Development and validation of a high-performance thin-layer chromatographic method for the quantitative analysis of vitexin in Passiflora foetida herbal formulations. Saudi Pharm. J. 2019, 27, 1157–1163. [Google Scholar] [CrossRef]
- Gupta, R.K.; Kumar, D.; Chaudhary, A.K.; Maithani, M.; Singh, R. Antidiabetic activity of Passiflora incarnata Linn. in streptozotocin-induced diabetes in mice. J. Ethnopharmacol. 2012, 139, 801–806. [Google Scholar] [CrossRef]
- Faleiro, F.G.; Junqueira, N.T.V.; Junghans, T.G.; Jesus, O.N.D.; Miranda, D.; Otoni, W.C. Advances in passion fruit (Passiflora spp.) propagation. Rev. Bras. De Frutic. 2019, 41, e-155. [Google Scholar] [CrossRef]
- Shuai, B.; Reynaga-Pena, C.G.; Springer, P.S. The LATERAL ORGAN BOUNDARIES gene defines a novel, plant-specific gene family. Plant Physiol. 2002, 129, 747–761. [Google Scholar] [CrossRef] [Green Version]
- Iwakawa, H.; Ueno, Y.; Semiarti, E.; Onouchi, H.; Kojima, S.; Tsukaya, H.; Hasebe, M.; Soma, T.; Ikezaki, M.; Machida, C. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol. 2002, 43, 467–478. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.J.; Wang, S.F.; Yu, X.B.; Yu, J.; He, X.W.; Zhang, S.L.; Shou, H.X.; Wu, P. ARL1, a LOB-domain protein required for adventitious root formation in rice. Plant J. 2005, 43, 47–56. [Google Scholar] [CrossRef]
- Xu, B.; Li, Z.; Zhu, Y.; Wang, H.; Ma, H.; Dong, A.; Huang, H. Arabidopsis genes AS1, AS2, and JAG negatively regulate boundary-specifying genes to promote sepal and petal development. Plant Physiol. 2008, 146, 566–575. [Google Scholar] [CrossRef] [Green Version]
- Xu, C.; Luo, F.; Hochholdinger, F. LOB domain proteins: Beyond lateral organ boundaries. Trends Plant Sci. 2016, 21, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Naito, T.; Yamashino, T.; Kiba, T.; Koizumi, N.; Kojima, M.; Sakakibara, H.; Mizuno, T. A link between cytokinin and ASL9 (ASYMMETRIC LEAVES 2 LIKE 9) that belongs to the AS2/LOB (LATERAL ORGAN BOUNDARIES) family genes in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 2007, 71, 1269–1278. [Google Scholar] [CrossRef] [PubMed]
- Rubin, G.; Tohge, T.; Matsuda, F.; Saito, K.; Scheible, W.R. Members of the LBD Family of Transcription Factors Repress Anthocyanin Synthesis and Affect Additional Nitrogen Responses in Arabidopsis. Plant Cell 2009, 21, 3567–3584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okushima, Y.; Fukaki, H.; Onoda, M.; Theologis, A.; Tasaka, M. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 2007, 19, 118–130. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.W.; Kim, M.J.; Kim, N.Y.; Lee, S.H.; Kim, J. LBD18 acts as a transcriptional activator that directly binds to the EXPANSIN14 promoter in promoting lateral root emergence of Arabidopsis. Plant J. 2013, 73, 212–224. [Google Scholar] [CrossRef] [PubMed]
- Rast, M.I.; Simon, R. Arabidopsis JAGGED LATERAL ORGANS acts with ASYMMETRIC LEAVES2 to coordinate KNOX and PIN expression in shoot and root meristems. Plant Cell 2012, 24, 2917–2933. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.J.; Kim, M.; Lee, M.R.; Park, S.K.; Kim, J. LATERAL ORGAN BOUNDARIES DOMAIN (LBD) 10 interacts with SIDECAR POLLEN/LBD 27 to control pollen development in A rabidopsis. Plant J. 2015, 81, 794–809. [Google Scholar] [CrossRef]
- Goh, T.; Toyokura, K.; Yamaguchi, N.; Okamoto, Y.; Uehara, T.; Kaneko, S.; Takebayashi, Y.; Kasahara, H.; Ikeyama, Y.; Okushima, Y.; et al. Lateral root initiation requires the sequential induction of transcription factors LBD16 and PUCHI in Arabidopsis thaliana. New Phytol. 2019, 224, 749–760. [Google Scholar] [CrossRef]
- Fan, M.; Xu, C.; Xu, K.; Hu, Y. LATERAL ORGAN BOUNDARIES DOMAIN transcription factors direct callus formation in Arabidopsis regeneration. Cell Res. 2012, 22, 1169–1180. [Google Scholar] [CrossRef] [Green Version]
- Majer, C.; Hochholdinger, F. Defining the boundaries: Structure and function of LOB domain proteins. Trends Plant Sci. 2011, 16, 47–52. [Google Scholar] [CrossRef]
- Kong, Y.M.; Xu, P.; Jing, X.Y.; Chen, L.X.; Li, L.G.; Li, X. Decipher the ancestry of the plant-specific LBD gene family. BMC Genom. 2017, 18, 951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chanderbali, A.S.; He, F.M.; Soltis, P.S.; Soltis, D.E. Out of the Water: Origin and Diversification of the LBD Gene Family. Mol. Biol. Evol. 2015, 32, 1996–2000. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.-H.; Liu, X.; An, J.-P.; Hao, Y.-J.; Wang, X.-F.; You, C.-X. Cloning and elucidation of the functional role of apple MdLBD13 in anthocyanin biosynthesis and nitrate assimilation. Plant Cell Tissue Organ Cult. 2017, 130, 47–59. [Google Scholar] [CrossRef]
- Yang, Y.; Yu, X.B.; Wu, P. Comparison and evolution analysis of two rice subspecies LATERAL ORGAN BOUNDARIES domain gene family and their evolutionary characterization from Arabidopsis. Mol. Phylogenet. Evol. 2006, 39, 248–262. [Google Scholar] [CrossRef]
- Wang, X.F.; Liu, X.; Ling, S.U.; Sun, Y.J.; Zhang, S.Z.; Hao, Y.J.; You, C.X. Identification, Evolution and Expression Analysis of the LBD Gene Family in Tomato. Sci. Agric. Sin. 2013, 46, 2501–2513. [Google Scholar]
- Cao, H.; Liu, C.Y.; Liu, C.X.; Zhao, Y.L.; Xu, R.R. Genomewide analysis of the lateral organ boundaries domain gene family in Vitis vinifera. J. Genet. 2016, 95, 515–526. [Google Scholar] [CrossRef] [PubMed]
- Zhongfan, Z.; Yali, Z.; Can, H.; Xiongze, D.; Feng, L.; Zuhua, Y. Genome-wide identification and expressing analysis of LBD transcription factors in pepper. Acta Hortic. Sin. 2016, 43, 12. [Google Scholar]
- Lu, Q.; Shao, F.; Qiu, D. Genome-Wide Analysis of Gene Family of Lateral Organ Boundaries Domain in Populus trichocarpa. Genom. Appl. Biol. 2018, 37, 313–325. [Google Scholar]
- Zhao, J.M.; Zhai, Z.W.; Li, Y.A.; Geng, S.F.; Song, G.Y.; Guan, J.T.; Jia, M.L.; Wang, F.L.; Sun, G.L.; Feng, N.; et al. Genome-Wide Identification and Expression Profiling of the TCP Family Genes in Spike and Grain Development of Wheat (Triticum aestivum L.). Front. Plant Sci. 2018, 9, 1282. [Google Scholar] [CrossRef] [Green Version]
- Huang, B.; Huang, Z.; Ma, R.; Ramakrishnan, M.; Chen, J.; Zhang, Z.; Yrjälä, K. Genome-wide identification and expression analysis of LBD transcription factor genes in Moso bamboo (Phyllostachys edulis). BMC Plant Biol. 2021, 21, 296. [Google Scholar] [CrossRef]
- Lynch, M.; Conery, J.S. The Evolutionary Fate and Consequences of Duplicate Genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, J.; Xie, Q.; Li, C.; Dong, Y.; Zhu, S.; Chen, J. Comprehensive characterization and gene expression patterns of LBD gene family in Gossypium. Planta 2020, 251, 81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jianxiang, L.; Jinzhang, W.; Yao, Z.; Shijiang, C. Genomic identification and evolutionary analysis of the boundary domain of lateral organs in Myrica rubra. J. For. Environ. 2021, 41, 172–179. [Google Scholar] [CrossRef]
- Scorza, L.C.T.; Hernandes-Lopes, J.; Melo-de-Pinna, G.F.A.; Dornelas, M.C. Expression patterns of Passiflora edulis APETALA1/FRUITFULL homologues shed light onto tendril and corona identities. Evodevo 2017, 8, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.M.; Zhang, S.Z.; Zheng, C.C. Genomewide analysis of LATERAL ORGAN BOUNDARIES Domain gene family in Zea mays. J. Genet. 2014, 93, 79–91. [Google Scholar] [CrossRef] [PubMed]
- Song, B.; Tang, Z.; Li, X.; Li, J.; Zhang, M.; Zhao, K.; Liu, H.; Zhang, S.; Wu, J. Mining and evolution analysis of lateral organ boundaries domain (LBD) genes in Chinese white pear (Pyrus bretschneideri). BMC Genom. 2020, 21, 644. [Google Scholar] [CrossRef] [PubMed]
- Kaul, S.; Koo, H.L.; Jenkins, J.; Rizzo, M.; Rooney, T.; Tallon, L.J.; Feldblyum, T.; Nierman, W.; Benito, M.-I.; Lin, X. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 2000, 408, 796–815. [Google Scholar]
- Yu, J.; Hu, S.; Wang, J.; Wong, G.K.-S.; Li, S.; Liu, B.; Deng, Y.; Dai, L.; Zhou, Y.; Zhang, X. A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 2002, 296, 79–92. [Google Scholar] [CrossRef]
- Xia, Z.; Huang, D.; Zhang, S.; Wang, W.; Ma, F.; Wu, B.; Xu, Y.; Xu, B.; Chen, D.; Zou, M. Chromosome-scale genome assembly provides insights into the evolution and flavor synthesis of passion fruit (Passiflora edulis Sims). Hortic. Res. 2021, 8, 14. [Google Scholar] [CrossRef]
- Matsumura, Y.; Iwakawa, H.; Machida, Y.; Machida, C. Characterization of genes in the ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARIES (AS2/LOB) family in Arabidopsis thaliana, and functional and molecular comparisons between AS2 and other family members. Plant J. 2009, 58, 525–537. [Google Scholar] [CrossRef] [Green Version]
- Staiger, D.; Brown, J.W.S. Alternative Splicing at the Intersection of Biological Timing, Development, and Stress Responses. Plant Cell 2013, 25, 3640–3656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Butler, J.E.F.; Kadonaga, J.T. The RNA polymerase II core promoter: A key component in the regulation of gene expression. Genes Dev. 2002, 16, 2583–2592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeon, B.W.; Kim, J. Role of LBD14 during ABA-mediated control of root system architecture in Arabidopsis. Plant Signal. Behav. 2018, 13, e1507405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Zhang, W.; Cheng, Y.; Feng, L. Genome-Wide Identification of LATERAL ORGAN BOUNDARIES DOMAIN (LBD) Transcription Factors and Screening of Salt Stress Candidates of Rosa rugosa Thunb. Biology 2021, 10, 992. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; He, Y.; He, W.; Su, H.; Wang, Y.; Hong, G.; Xu, P. Structural and functional insights into the LBD family involved in abiotic stress and flavonoid synthases in Camellia sinensis. Sci. Rep. 2019, 9, 15651. [Google Scholar] [CrossRef]
- Liu, H.; Cao, M.; Chen, X.; Ye, M.; Zhao, P.; Nan, Y.; Li, W.; Zhang, C.; Kong, L.; Kong, N.; et al. Genome-Wide Analysis of the Lateral Organ Boundaries Domain (LBD) Gene Family in Solanum tuberosum. Int. J. Mol. Sci. 2019, 20, 5360. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Hu, P.; Tao, Y.; Song, P.; Gao, H.; Guan, Y. Genome-wide identification and characterization of the Lateral Organ Boundaries Domain (LBD) gene family in polyploid wheat and related species. PeerJ 2021, 9, e11811. [Google Scholar] [CrossRef]
- Cucinotta, M.; Colombo, L.; Roig-Villanova, I. Ovule development, a new model for lateral organ formation. Front. Plant Sci. 2014, 5, 117. [Google Scholar] [CrossRef] [Green Version]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef] [Green Version]
- Letunic, I.; Doerks, T.; Bork, P. SMART 7: Recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012, 40, D302–D305. [Google Scholar] [CrossRef]
- Chou, K.C.; Shen, H.B. Cell-PLoc 2.0: An improved package of web-servers for predicting subcellular localization of proteins in various organisms. Nat. Sci. 2010, 2, 1090–1103. [Google Scholar] [CrossRef] [Green Version]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Hochstrasser, D.F. Protein Identification and Analysis Tools in the ExPASy Server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar] [PubMed]
- Jin, J.P.; Tian, F.; Yang, D.C.; Meng, Y.Q.; Kong, L.; Luo, J.C.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.C.; Soc, I.C. MUSCLE: Multiple sequence alignment with improved accuracy and speed. In Proceedings of the IEEE Computational Systems Bioinformatics Conference (CSB 2004), Stanford, CA, USA, 16–19 August 2004; pp. 728–729. [Google Scholar]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11 Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Procter, J.; Martin, D.A.; Barton, G.J. Jalview: Visualization and Analysis of Molecular Sequences, Alignments, and Structures. BMC Bioinform. 2005, 6, P28. [Google Scholar] [CrossRef] [Green Version]
- Troshin, P.V.; Procter, J.B.; Sherstnev, A.; Barton, D.L.; Madeira, F.; Barton, G.J. JABAWS 2.2 distributed web services for Bioinformatics: Protein disorder, conservation and RNA secondary structure. Bioinformatics 2018, 34, 1939–1940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator—eScholarship. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [Green Version]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.Y.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.P.; Tang, H.B.; DeBarry, J.D.; Tan, X.; Li, J.P.; Wang, X.Y.; Lee, T.H.; Jin, H.Z.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, Z.L.; Cavalcanti, A.; Chen, F.C.; Bouman, P.; Li, W.H. Extent of gene duplication in the genomes of Drosophila, nematode, and yeast. Mol. Biol. Evol. 2002, 19, 256–262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, D.P.; Wan, H.L.; Zhang, S.; Yu, J. gamma-MYN: A new algorithm for estimating Ka and Ks with consideration of variable substitution rates. Biol. Direct 2009, 4, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashkenazy, H.; Abadi, S.; Martz, E.; Chay, O.; Mayrose, I.; Pupko, T.; Ben-Tal, N. ConSurf 2016: An improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016, 44, W344–W350. [Google Scholar] [CrossRef] [Green Version]
- Delano, W.L. The PyMol Molecular Graphics System. Proteins Struct. Funct. Bioinform. 2002, 30, 442–454. [Google Scholar]
- Khan, A.; Fornes, O.; Stigliani, A.; Gheorghe, M.; Castro-Mondragon, J.A.; van der Lee, R.; Bessy, A.; Cheneby, J.; Kulkarni, S.R.; Tan, G.; et al. JASPAR 2018: Update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 2018, 46, D260–D266. [Google Scholar] [CrossRef] [Green Version]
- Chen, P.; Li, Y.; Zhao, L.; Hou, Z.; Yan, M.; Hu, B.; Liu, Y.; Azam, S.M.; Zhang, Z.; Rahman, Z.U. Genome-wide identification and expression profiling of ATP-binding cassette (ABC) transporter gene family in pineapple (Ananas comosus (L.) Merr.) reveal the role of AcABCG38 in pollen development. Front. Plant Sci. 2017, 8, 2150. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Wu, Y.; Tian, Q.; Huang, W.; Liu, J.; Xia, X.; Yang, X.; Mou, H. Identification and evaluation of reference genes for quantitative real-time PCR analysis in Passiflora edulis under stem rot condition. Mol. Biol. Rep. 2020, 47, 2951–2962. [Google Scholar] [CrossRef]


| Gene Name | Gene ID | Chromosome | Size (aa) | MW (Da) | pI | A.I. | Stability | GRAVY | Predicted Location |
|---|---|---|---|---|---|---|---|---|---|
| PeLBD1 | P_edulia010000163.g | LG01 | 228 | 24,788.28 | 6.7 | 92.37 | U | −0.208 | Nucleus |
| PeLBD2 | P_edulia010000500.g | LG01 | 216 | 23,829.2 | 6.3 | 71.44 | U | −0.26 | Nucleus |
| PeLBD3 | P_edulia010001973.g | LG01 | 188 | 20,672.23 | 5.64 | 68.56 | U | −0.353 | Nucleus |
| PeLBD4 | P_edulia010002080.g | LG01 | 188 | 20,702.3 | 5.64 | 70.64 | U | −0.324 | Nucleus |
| PeLBD5 | P_edulia010002578.g | LG01 | 166 | 18,452.21 | 5.36 | 85.3 | U | −0.087 | Nucleus |
| PeLBD6 | P_edulia010002985.g | LG01 | 189 | 20,945.79 | 9.08 | 72.91 | U | −0.441 | Nucleus |
| PeLBD7 | P_edulia010003009.g | LG01 | 95 | 10,772.56 | 9.64 | 57.58 | U | −0.58 | Nucleus |
| PeLBD8 | P_edulia010003878.g | LG01 | 191 | 20,985.21 | 9.28 | 86.28 | U | −0.121 | Nucleus |
| PeLBD9 | P_edulia010004119.g | LG01 | 202 | 21,696.82 | 9.37 | 83.56 | U | −0.045 | Nucleus |
| PeLBD10 | P_edulia010005184.g | LG01 | 204 | 22,205.36 | 8.37 | 82.35 | U | −0.107 | Nucleus |
| PeLBD11 | P_edulia010005391.g | LG01 | 228 | 24,231.57 | 7.63 | 77.94 | U | −0.132 | Nucleus |
| PeLBD12 | P_edulia010005394.g | LG01 | 215 | 23,314.77 | 8.9 | 85.77 | U | −0.17 | Nucleus |
| PeLBD13 | P_edulia020007264.g | LG02 | 205 | 22,251.98 | 6.95 | 62.78 | U | −0.412 | Nucleus |
| PeLBD14 | P_edulia030008046.g | LG03 | 204 | 23,008.4 | 9.04 | 72.79 | U | −0.45 | Nucleus |
| PeLBD15 | P_edulia030008474.g | LG03 | 240 | 26,282.99 | 8.05 | 76.75 | U | −0.275 | Nucleus |
| PeLBD16 | P_edulia030008742.g | LG03 | 158 | 17,413.83 | 7.64 | 72.91 | U | −0.333 | Nucleus |
| PeLBD17 | P_edulia030009145.g | LG03 | 337 | 38,858.07 | 8.74 | 71.78 | U | −0.612 | Nucleus |
| PeLBD18 | P_edulia030009282.g | LG03 | 166 | 18,330.5 | 5.93 | 62.95 | U | −0.499 | Nucleus |
| PeLBD19 | P_edulia040010647.g | LG04 | 171 | 18,559.21 | 8.24 | 71.35 | U | −0.258 | Nucleus |
| PeLBD20 | P_edulia050011298.g | LG05 | 168 | 18,645.42 | 7.52 | 74.94 | U | −0.181 | Nucleus |
| PeLBD21 | P_edulia050011880.g | LG05 | 173 | 18,601.25 | 8.59 | 73.93 | U | −0.205 | Nucleus |
| PeLBD22 | P_edulia050012869.g | LG05 | 235 | 25,742.65 | 4.5 | 75.23 | U | −0.161 | Nucleus |
| PeLBD23 | P_edulia060015822.g | LG06 | 224 | 24,306.31 | 8.25 | 71.61 | U | −0.401 | Nucleus |
| PeLBD24 | P_edulia060015864.g | LG06 | 142 | 15,627 | 7.61 | 82.46 | U | −0.062 | Nucleus |
| PeLBD25 | P_edulia060015871.g | LG06 | 169 | 18,867.63 | 7.52 | 72.78 | U | −0.227 | Nucleus |
| PeLBD26 | P_edulia070017232.g | LG07 | 214 | 23,195.4 | 8.83 | 72.57 | U | −0.3 | Nucleus |
| PeLBD27 | P_edulia070018122.g | LG07 | 179 | 20,573.48 | 6.22 | 88.27 | U | −0.419 | Nucleus |
| PeLBD28 | P_edulia070018138.g | LG07 | 179 | 20,557.48 | 6.22 | 88.27 | U | −0.396 | Nucleus |
| PeLBD29 | P_edulia070018148.g | LG07 | 179 | 20,573.48 | 6.22 | 88.27 | U | −0.419 | Nucleus |
| PeLBD30 | P_edulia080020093.g | LG08 | 164 | 18,188.81 | 6.8 | 87.44 | U | −0.17 | Nucleus |
| PeLBD31 | P_edulia080020165.g | LG08 | 214 | 23,141.16 | 8.62 | 66.07 | U | −0.379 | Nucleus |
| PeLBD32 | P_eduliaContig20022702.g | Contig2 | 168 | 18,617.36 | 6.71 | 74.94 | U | −0.175 | Nucleus |
| PeLBD33 | P_eduliaContig60022431.g | Contig6 | 294 | 31,614.88 | 8.75 | 71.67 | U | −0.402 | Nucleus |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, J.; Hou, Z.; Liao, J.; Qin, Y.; Wang, L.; Wang, X.; Su, W.; Cai, Z.; Fang, Y.; Aslam, M.; et al. Genome-Wide Identification and Expression Analysis of LBD Transcription Factor Genes in Passion Fruit (Passiflora edulis). Int. J. Mol. Sci. 2022, 23, 4700. https://doi.org/10.3390/ijms23094700
Liang J, Hou Z, Liao J, Qin Y, Wang L, Wang X, Su W, Cai Z, Fang Y, Aslam M, et al. Genome-Wide Identification and Expression Analysis of LBD Transcription Factor Genes in Passion Fruit (Passiflora edulis). International Journal of Molecular Sciences. 2022; 23(9):4700. https://doi.org/10.3390/ijms23094700
Chicago/Turabian StyleLiang, Jianxiang, Zhimin Hou, Jingyi Liao, Yuan Qin, Lulu Wang, Xiaomei Wang, Weiqiang Su, Zhaoyan Cai, Yunying Fang, Mohammad Aslam, and et al. 2022. "Genome-Wide Identification and Expression Analysis of LBD Transcription Factor Genes in Passion Fruit (Passiflora edulis)" International Journal of Molecular Sciences 23, no. 9: 4700. https://doi.org/10.3390/ijms23094700







