Applications of Nanozymology in the Detection and Identification of Viral, Bacterial and Fungal Pathogens
Abstract
:1. Introduction
2. The Pathogen Challenge
3. Mechanistic Considerations
4. Recent Approaches of Pathogen Detection
5. Nanozymology
6. Indirect Nanozyme-Based Fungal Detection
7. Direct Nanozyme-Based Cancer Cell Detection
8. Direct Nanozyme-Based Parasite Ova Detection
9. Bacterial Detection and Identification
10. Pathogen Biosensors
11. Microfluidic Biosensors
12. The Paper Dipstick or Spot Test Methods
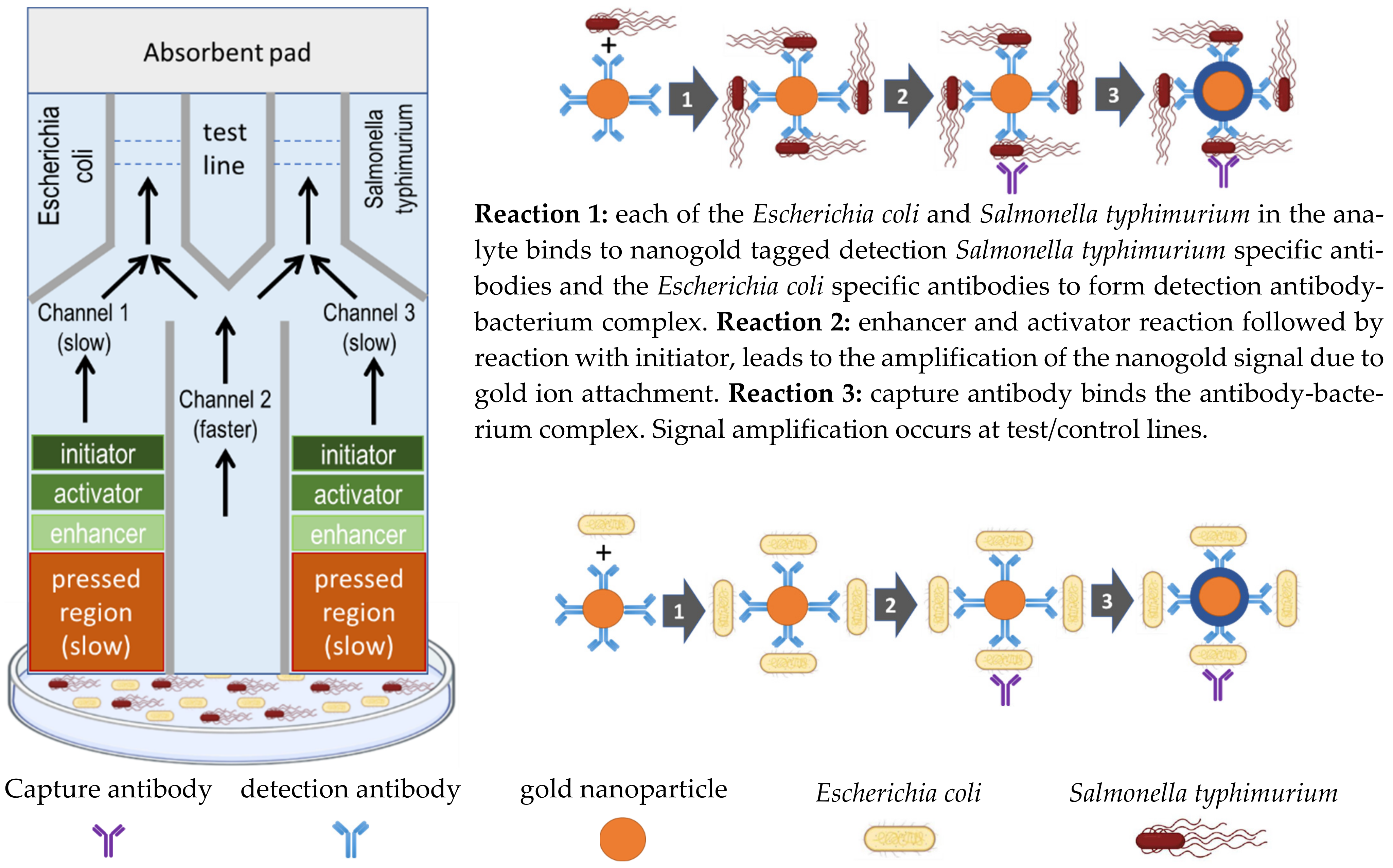
13. Multiple Flow Rate Multiple Channel Colorimetric Paper Strip
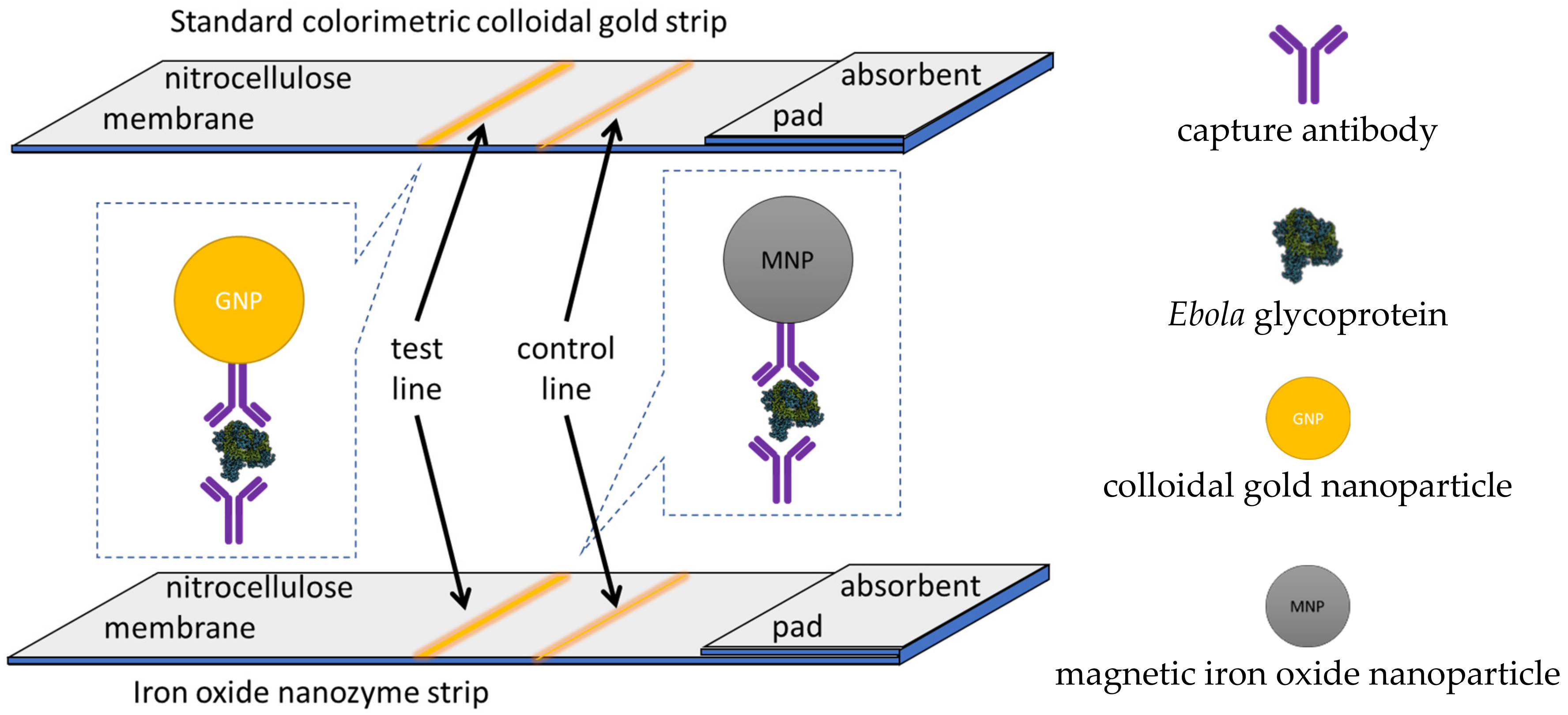
14. Chemiluminescence Nanozyme-Linked Immunoassay Paper Strip
15. Future Implications with Therapeutics and Applications
16. Conclusions
Funding
Conflicts of Interest
References
- McNamara, K.; Tofail, S.A.M. Nanoparticles in biomedical applications. Adv. Phys. X 2016, 2, 54–88. [Google Scholar] [CrossRef]
- Bharathala, S.; Sharma, P. Biomedical applications of nanoparticles. In Nanotechnology in Modern Animal Biotechnology; Maurya, P.K., Singh, S., Eds.; Elsevier: Amsterdam, The Netherlands, 2019; Volume 8, pp. 113–132. ISBN 9780128188231. [Google Scholar] [CrossRef]
- Ramos, A.P.; Cruz, M.; Tovani, C.B.; Ciancaglini, P. Biomedical applications of nanotechnology. Biophys. Rev. 2017, 9, 79–89. [Google Scholar] [CrossRef]
- Savaliya, R.; Shah, D.; Singh, R.; Kumar, A.; Shanker, R.; Dhawan, A.; Singh, S. Nanotechnology in disease diagnostic techniques. Curr. Drug Metab. 2015, 16, 645–661. [Google Scholar] [CrossRef]
- Gomez-Marquez, J.; Hamad-Schifferli, K. Local development of nanotechnology-based diagnostics. Nat. Nanotechnol. 2021, 16, 484–486. [Google Scholar] [CrossRef]
- Murthy, S.K. Nanoparticles in modern medicine: State of the art and future challenges. Int. J. Nanomed. 2007, 2, 129–141. [Google Scholar]
- Rezaei, R.; Safaei, M.; Mozaffari, H.R.; Moradpoor, H.; Karami, S.; Golshah, A.; Salimi, B.; Karami, H. The role of nanomaterials in the treatment of diseases and their effects on the immune system. Open Access Maced. J. Med. Sci. 2019, 7, 1884–1890. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zolnik, B.S.; Gonzaález-Fernaández, A.; Sadrieh, N.; Dobrovolskaia, M.A. Nanoparticles and the immune system. Endocrinology 2010, 151, 458–465. [Google Scholar] [CrossRef] [PubMed]
- Nogrady, B. How nanotechnology can flick the immunity switch. Nano immuno-engineering shows promise against autoimmune conditions, cancer and allergies. Nature 2021, 595, S18–S19. [Google Scholar] [CrossRef]
- Sirsat, S.A.; Neal, J.A. Titanium dioxide nanoparticles as an environmental sanitizing agent. J. Microb. Biochem. Technol. 2015, 7, 61–63. [Google Scholar] [CrossRef] [Green Version]
- Talebian, S.; Wallace, G.G.; Schroeder, A.; Stellacci, F.; Conde, J. Nanotechnology-based disinfectants and sensors for SARS-CoV-2. Nat. Nanotechnol. 2020, 15, 618–621. [Google Scholar] [CrossRef] [PubMed]
- Muhammad, M.; Dong-Jin, L.; Hansoo, P.; Dokyun, N. Nanotechnology for diagnosis and treatment of infectious diseases. J. Nanosci. Nanotechnol. 2014, 14, 7374–7387. [Google Scholar] [CrossRef]
- Kirtane, A.R.; Verma, M.; Karandikar, P.; Furin, J.; Langer, R.; Traverso, G. Nanotechnology approaches for global infectious diseases. Nat. Nanotechnol. 2021, 16, 369–384. [Google Scholar] [CrossRef]
- Kang, J.; Tahir, A.; Wang, H.; Chang, J. Applications of nanotechnology in virus detection, tracking, and infection mechanisms. WIREs Nanomed. Nanobiotechnol. 2021, 13, e1700. [Google Scholar] [CrossRef]
- Tharayil, A.; Rajakumari, R.; Kumar, A.; Choudhary, M.D.; Palit, P.; Thomas, S. New insights into application of nanoparticles in the diagnosis and screening of novel coronavirus (SARS-CoV-2). Emergent Mater. 2021, 4, 101–117. [Google Scholar] [CrossRef]
- Salieb-Beugelaar, G.B.; Hunziker, P.R. Towards nano-diagnostics for bacterial infections. Eur. J. Nanomed. 2015, 7, 37–50. [Google Scholar] [CrossRef]
- Karaman, D.Ş.; Pamukçu, A.; Karakaplan, M.B.; Kocaoglu, O.; Rosenholm, J.M. Recent advances in the use of mesoporous silica nanoparticles for the diagnosis of bacterial infections. Int. J. Nanomed. 2021, 16, 6575–6591. [Google Scholar] [CrossRef]
- Sojinrin, T.; Conde, J.; Liu, K.; Curtin, J.; Byrne, H.J.; Cui, D.; Tian, F. Plasmonic gold nanoparticles for detection of fungi and human cutaneous fungal infections. Anal. Bioanal. Chem. 2017, 409, 4647–4658. [Google Scholar] [CrossRef] [Green Version]
- Abd-Elsalam, K. Special issue: Fungal nanotechnology. J. Fungi 2021, 7, 583. [Google Scholar] [CrossRef]
- Teles, F.S.R.R.; Da Luz, M.M.; Vieira, M.R.; Fonseca, L.J.P. Nanotechnology for the diagnosis of parasitic infections. In Nanotechnology in Dermatology; Nasir, A., Friedman, A., Wang, S., Eds.; Springer: New York, NY, USA, 2012; pp. 209–219. ISBN 978-1-4614-5033-7. [Google Scholar] [CrossRef]
- Hikal, W.M.; Bratovcic, A.; Baeshen, R.S.; Tkachenko, K.G.; Ahl, H.A.H.S.-A. Nanobiotechnology for the detection and control of waterborne parasites. Open J. Ecol. 2021, 11, 203–223. [Google Scholar] [CrossRef]
- Collinge, J. Prion diseases of humans and animals: Their causes and molecular basis. Annu. Rev. Neurosci. 2001, 24, 519–550. [Google Scholar] [CrossRef] [Green Version]
- Myers, R.; Cembran, A.; Fernandez-Funez, P. Insight from animals resistant to prion diseases: Deciphering the genotype–morphotype–phenotype code for the prion protein. Front. Cell. Neurosci. 2020, 14, 254. [Google Scholar] [CrossRef] [PubMed]
- Herrero-Uribe, L. Viruses, definitions and reality. Rev. Biol. Trop. Int. J. Trop. Biol. Conserv. 2011, 59, 993–998. [Google Scholar] [CrossRef] [Green Version]
- Woolhouse, M.; Scott, F.; Hudson, Z.; Howey, R.; Chase-Topping, M. Human viruses: Discovery and emergence. Philos. Trans. R. Soc. B Biol. Sci. 2012, 367, 2864–2871. [Google Scholar] [CrossRef] [Green Version]
- Hart, C.A.; Kariuki, S. Antimicrobial resistance in developing countries. BMJ 1998, 317, 647–650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boutayeb, A. The impact of infectious diseases on the development of Africa. In Handbook of Disease Burdens and Quality of Life Measures; Preedy, V.R., Watson, R.R., Eds.; Springer: New York, NY, USA, 2010; Chapter 66; pp. 1171–1188. [Google Scholar] [CrossRef]
- Dong, M.; Wang, C.; Li, H.; Yan, Y.; Ma, X.; Li, H.; Li, X.; Wang, H.; Zhang, Y.; Qi, W.; et al. Aerobic Vaginitis Diagnosis Criteria Combining Gram Stain with Clinical Features: An Establishment and Prospective Validation Study. Diagnostics 2022, 12, 185. [Google Scholar] [CrossRef] [PubMed]
- Duquenoy, A.; Bellais, S.; Gasc, C.; Schwintner, C.; Dore, J.; Thomas, V. Assessment of gram- and viability-staining methods for quantifying bacterial community dynamics using flow cytometry. Front. Microbiol. 2020, 11, 1469. [Google Scholar] [CrossRef] [PubMed]
- Odds, F.C.; Gow, N.A.; Brown, A.J. Fungal virulence studies come of age. Genome Biol. 2001, 2, 1009. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Rubio, R.; De Oliveira, H.C.; Rivera, J.; Trevijano-Contador, N. The fungal cell wall: Candida, Cryptococcus, and Aspergillus species. Front. Microbiol. 2020, 10, 2993. [Google Scholar] [CrossRef]
- Adekiya, T.A.; Kondiah, P.P.D.; Choonara, Y.E.; Kumar, P.; Pillay, V. A review of nanotechnology for targeted anti-schistosomal therapy. Front. Bioeng. Biotechnol. 2020, 8, 32. [Google Scholar] [CrossRef]
- Yan, X.; Gao, L. Nanozymology: An overview. In Nanozymology: Connecting Biology and Nanotechnology; Yan, X., Ed.; Springer: Singapore, 2020. [Google Scholar] [CrossRef]
- Liang, M.; Yan, X. Nanozymes: From new concepts, mechanisms, and standards to applications. Acc. Chem. Res. 2019, 52, 2190–2200. [Google Scholar] [CrossRef]
- Dong, H.; Fan, Y.; Zhang, W.; Gu, N.; Zhang, Y. Catalytic mechanisms of nanozymes and their applications in biomedicine. Bioconjug. Chem. 2019, 30, 1273–1296. [Google Scholar] [CrossRef]
- Lockwood, D.J. Nanozymology-Connecting Biology and Nanotechnology, Nanostructure Science and Technology Series; Springer Nature: Singapore, 2020; ISBN 978-981-15-1490-6. [Google Scholar] [CrossRef]
- Liu, Q.; Zhang, A.; Wang, R.; Zhang, Q.; Cui, D. A review on metal- and metal oxide-based nanozymes: Properties, mechanisms, and applications. Nano-Micro Lett. 2021, 13, 154. [Google Scholar] [CrossRef]
- Jutla, A.; Whitcombe, E.; Hasan, N.; Haley, B.; Akanda, A.; Huq, A.; Alam, M.; Sack, R.B.; Colwell, R. Environmental factors influencing epidemic cholera. Am. J. Trop. Med. Hyg. 2013, 89, 597–607. [Google Scholar] [CrossRef]
- Chopra, M.; Wilkinson, D.; Stirling, S. Epidemic shigella dysentery in children in northern KwaZulu-Natal. S. Afr. Med. J. 1997, 87, 48–51. [Google Scholar]
- Azuma, K.; Yanagi, U.; Kagi, N.; Kim, H.; Ogata, M.; Hayashi, M. Environmental factors involved in SARS-CoV-2 transmission: Effect and role of indoor environmental quality in the strategy for COVID-19 infection control. Environ. Health Prev. Med. 2020, 25, 66. [Google Scholar] [CrossRef]
- Eslami, H.; Jalili, M. The role of environmental factors to transmission of SARS-CoV-2 (COVID-19). AMB Express 2020, 10, 92. [Google Scholar] [CrossRef]
- Glogowsky, U.; Schächtele, S. How effective are social distancing policies? Evidence on the fight against COVID-19. PLoS ONE 2021, 16, e0257363. [Google Scholar] [CrossRef]
- Howard, J.; Huang, A.; Li, Z.; Tufekci, Z.; Zdimal, V.; Van der Westhuizen, H.-M.; von Delft, A.; Price, A.; Fridman, L.; Tang, L.-H.; et al. An evidence review of face masks against COVID-19. Proc. Natl. Acad. Sci. USA 2021, 118, e2014564118. [Google Scholar] [CrossRef]
- Sharafi, S.M.; Ebrahimpour, K.; Nafez, A. Environmental disinfection against COVID-19 in different areas of health care facilities: A review. Rev. Environ. Health 2020, 36, 193–198. [Google Scholar] [CrossRef]
- Holmes, E.C.; Goldstein, S.A.; Rasmussen, A.L.; Robertson, D.L.; Crits-Christoph, A.; Wertheim, J.O.; Anthony, S.J.; Barclay, W.S.; Boni, M.F.; Doherty, P.C.; et al. The origins of SARS-CoV-2: A critical review. Cell 2021, 184, 4848–4856. [Google Scholar] [CrossRef]
- Konda, M.; Dodda, B.; Konala, V.M.; Naramala, S.; Adapa, S. Potential zoonotic origins of SARS-CoV-2 and insights for preventing future pandemics through one health approach. Cureus 2020, 12, e8932. [Google Scholar] [CrossRef] [PubMed]
- Barreiro, C.; Albano, H.; Silva, J.; Teixeira, P. Role of flies as vectors of foodborne pathogens in rural areas. ISRN Microbiol. 2013, 2013, 718780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dahmana, H.; Granjon, L.; Diagne, C.; Davoust, B.; Fenollar, F.; Mediannikov, O. Rodents as hosts of pathogens and related zoonotic disease risk. Pathogens 2020, 9, 202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boiocchi, F.; Davies, M.P.; Hilton, A.C. An examination of flying insects in seven hospitals in the United Kingdom and carriage of bacteria by true flies (Diptera: Calliphoridae, Dolichopodidae, Fanniidae, Muscidae, Phoridae, Psychodidae, Sphaeroceridae). J. Med. Entomol. 2019, 56, 1684–1697. [Google Scholar] [CrossRef]
- Tijjani, M.; Majid, R.A.; Abdullahi, S.A.; Unyah, N.Z. Detection of rodent-borne parasitic pathogens of wild rats in Serdang, Selangor, Malaysia: A potential threat to human health. Int. J. Parasitol. Parasites Wildl. 2020, 11, 174–182. [Google Scholar] [CrossRef]
- Clack, L.; Scotoni, M.; Wolfensberger, A.; Sax, H. “First-person view” of pathogen transmission and hand hygiene–Use of a new head-mounted video capture and coding tool. Antimicrob. Resist. Infect. Control 2017, 6, 108. [Google Scholar] [CrossRef] [Green Version]
- Pittet, D.; Allegranzi, B.; Sax, H.; Dharan, S.; Pessoa-Silva, C.L.; Donaldson, L.; Boyce, J.M. Evidence-based model for hand transmission during patient care and the role of improved practices. Lancet Infect. Dis. 2006, 6, 641–652. [Google Scholar] [CrossRef]
- Bintsis, T. Foodborne pathogens. AIMS Microbiol. 2017, 3, 529–563. [Google Scholar] [CrossRef]
- Stein, R.A.; Chirilã, M. Routes of transmission in the food chain. In Foodborne Diseases; Academic Press: Cambridge, MA, USA, 2017; pp. 65–103. [Google Scholar] [CrossRef]
- Reen, D.J. Enzyme-Linked Immunosorbent Assay (ELISA). In Basic Protein and Peptide Protocols. Methods in Molecular Biology™; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 1994; Volume 32. [Google Scholar] [CrossRef]
- Chen, Y.J.; Chen, M.; Hsieh, Y.C.; Su, Y.C.; Wang, C.H.; Cheng, C.M.; Kao, A.P.; Wang, K.H.; Cheng, J.J.; Chuang, K.H. Development of a highly sensitive enzyme-linked immunosorbent assay (ELISA) through use of poly-protein G-expressing cell-based microplates. Sci. Rep. 2018, 8, 17868. [Google Scholar] [CrossRef] [Green Version]
- Koczula, K.M.; Gallotta, A. Lateral flow assays. Essays Biochem. 2016, 60, 111–120. [Google Scholar] [CrossRef]
- Han, J.; Zhang, L.; Hu, L.; Xing, K.; Lu, X.; Huang, Y.; Zhang, J.; Lai, W.; Chen, T. Nanozyme-based lateral flow assay for the sensitive detection of Escherichia coli O157:H7 in milk. J. Dairy Sci. 2018, 101, 5770–5779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, H.; Cai, S.; Wang, C.; Chen, Y.; Yang, R. Recent progress of nanozymes in the detection of pathogenic microorganisms. ChemBioChem 2020, 21, 2572–2584. [Google Scholar] [CrossRef]
- Shin, H.Y.; Park, T.J.; Kim, M.I. Recent research trends and future prospects in nanozymes. J. Nanomater. 2015, 2015, 756278. [Google Scholar] [CrossRef] [Green Version]
- Mao, M.X.; Zheng, R.; Peng, C.F.; Wei, X.L. DNA–Gold Nanozyme-modified paper device for enhanced colorimetric detection of mercury ions. Biosensors 2020, 10, 211. [Google Scholar] [CrossRef]
- Chen, W.; Fang, X.; Ye, X.; Wang, X.; Kong, J. Colorimetric DNA assay by exploiting the DNA-controlled peroxidase mimicking activity of mesoporous silica loaded with platinum nanoparticles. Mikrochim. Acta 2018, 185, 544. [Google Scholar] [CrossRef]
- Ali, J.; Elahi, S.; Ali, A.; Waseem, H.; Abid, R.; Mohamed, M. Unveiling the potential role of nanozymes in combating the COVID-19 outbreak. Nanomaterials 2021, 11, 1328. [Google Scholar] [CrossRef]
- Oh, S.; Kim, J.; Tran, V.T.; Lee, D.K.; Ahmed, S.R.; Hong, J.C.; Lee, J.; Park, E.Y.; Lee, J. Magnetic nanozyme-linked immunosorbent assay for ultrasensitive influenza A virus detection. ACS Appl. Mater. Interfaces 2018, 10, 12534–12543. [Google Scholar] [CrossRef]
- Kim, J.; Tran, V.T.; Oh, S.; Jang, M.; Lee, D.K.; Hong, J.C.; Park, T.J.; Kim, H.-J.; Lee, J. Clinical trial: Magnetoplasmonic ELISA for urine-based active tuberculosis detection and anti-tuberculosis therapy monitoring. ACS Cent. Sci. 2021, 7, 1898–1907. [Google Scholar] [CrossRef]
- Liu, P.; Wang, Y.; Han, L.; Cai, Y.; Ren, H.; Ma, T.; Li, X.; Petrenko, V.A.; Liu, A. Colorimetric assay of bacterial pathogens based on Co3O4 magnetic nanozymes conjugated with specific fusion phage proteins and magnetophoretic chromatography. ACS Appl. Mater. Interfaces 2020, 12, 9090–9097. [Google Scholar] [CrossRef]
- Alahi, E.E.; Mukhopadhyay, S.C. Detection methodologies for pathogen and toxins: A review. Sensors 2017, 17, 1885. [Google Scholar] [CrossRef] [Green Version]
- Law, J.W.-F.; Ab Mutalib, N.-S.; Chan, K.-G.; Lee, L.-H. Rapid methods for the detection of foodborne bacterial pathogens: Principles, applications, advantages and limitations. Front. Microbiol. 2015, 5, 770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kadri, K. Polymerase Chain Reaction (PCR): Principle and applications. In Synthetic Biology-New Interdisciplinary Science; Nagpal, M.L., Boldura, O.M., Baltă, C., Enany, S., Eds.; IntechOpen: London, UK, 2019. [Google Scholar] [CrossRef] [Green Version]
- Coudray-Meunier, C.; Fraisse, A.; Martin-Latil, S.; Delannoy, S.; Fach, P.; Perelle, S. A novel high-throughput method for molecular detection of human pathogenic viruses using a nanofluidic real-time PCR system. PLoS ONE 2016, 11, e0147832. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Cao, Y.; Wang, T.; Dong, Q.; Li, J.; Niu, C. Detection of 12 common food-borne bacterial pathogens by TaqMan real-time PCR using a single set of reaction conditions. Front. Microbiol. 2019, 10, 222. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, Y. PCR in diagnosis of infection: Detection of bacteria in cerebrospinal fluids. Clin. Vaccine Immunol. 2002, 9, 508–514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Järvinen, A.-K.; Laakso, S.; Piiparinen, P.; Aittakorpi, A.; Lindfors, M.; Huopaniemi, L.; Piiparinen, H.; Mäki, M. Rapid identification of bacterial pathogens using a PCR- and microarray-based assay. BMC Microbiol. 2009, 9, 161. [Google Scholar] [CrossRef] [Green Version]
- Mancini, V.; Murolo, S.; Romanazzi, G. Diagnostic methods for detecting fungal pathogens on vegetable seeds. Plant Pathol. 2016, 65, 691–703. [Google Scholar] [CrossRef]
- Hariharan, G.; Prasannath, K. Recent advances in molecular diagnostics of fungal plant pathogens: A mini review. Front. Cell. Infect. Microbiol. 2021, 10, 600234. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, R.T.; Ning, H.L.; Li, W.X.; Bai, Y.L.; Li, Y.G. Evaluation and management of fungal-infected carrot seeds. Sci. Rep. 2020, 10, 8. [Google Scholar] [CrossRef]
- Wagner, K.; Springer, B.; Pires, V.P.; Keller, P.M. Molecular detection of fungal pathogens in clinical specimens by 18S rDNA high-throughput screening in comparison to ITS PCR and culture. Sci. Rep. 2018, 8, 6964. [Google Scholar] [CrossRef]
- Weerathunge, P.; Ramanathan, R.; Torok, V.A.; Hodgson, K.; Xu, Y.; Goodacre, R.; Behera, B.K.; Bansal, V. Ultrasensitive colorimetric detection of murine norovirus using NanoZyme aptasensor. Anal. Chem. 2019, 91, 3270–3276. [Google Scholar] [CrossRef]
- Giamberardino, A.; Labib, M.; Hassan, E.M.; Tetro, J.A.; Springthorpe, S.; Sattar, S.A.; Berezovski, M.V.; DeRosa, M.C. Ultrasensitive norovirus detection using DNA aptasensor technology. PLoS ONE 2013, 8, e79087. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Lai, B.S.; Juhas, M. Recent advances in aptamer discovery and applications. Molecules 2019, 24, 941. [Google Scholar] [CrossRef] [Green Version]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef]
- Ning, Y.; Hu, J.; Lu, F. Aptamers used for biosensors and targeted therapy. Biomed. Pharmacother. 2020, 132, 110902. [Google Scholar] [CrossRef]
- Sharma, T.K.; Ramanathan, R.; Weerathunge, P.; Mohammadtaheri, M.; Daima, H.K.; Shukla, R.; Bansal, V. Aptamer-mediated ‘turn-off/turn-on’ nanozyme activity of gold nanoparticles for kanamycin detection. Chem. Commun. 2014, 50, 15856–15859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chatterjee, B.; Das, S.J.; Anand, A.; Sharma, T.K. Nanozymes and aptamer-based biosensing. Mater. Sci. Energy Technol. 2019, 3, 127–135. [Google Scholar] [CrossRef]
- Savas, S.; Altintas, Z. Graphene quantum dots as nanozymes for electrochemical sensing of Yersinia enterocolitica in milk and human serum. Materials 2019, 12, 2189. [Google Scholar] [CrossRef] [Green Version]
- Qingzhi, W.; Zou, S.; Wang, Q.; Chen, L.; Yan, X.; Gao, L. Catalytic defense against fungal pathogens using nanozymes. Nanotechnol. Rev. 2021, 10, 1277–1292. [Google Scholar] [CrossRef]
- Sun, S.; Zhao, R.; Feng, S.; Xie, Y. Colorimetric zearalenone assay based on the use of an aptamer and of gold nanoparticles with peroxidase-like activity. Mikrochim. Acta 2018, 185, 535. [Google Scholar] [CrossRef]
- Xu, Z.; Long, L.-L.; Chen, Y.-Q.; Chen, M.-L.; Cheng, Y.-H. A nanozyme-linked immunosorbent assay based on metal–organic frameworks (MOFs) for sensitive detection of aflatoxin B1. Food Chem. 2020, 338, 128039. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.-L.; Chen, J.-H.; Ding, L.; Xu, Z.; Wen, L.; Wang, L.-B.; Cheng, Y.-H. Study of the detection of bisphenol A based on a nano-sized metal–organic framework crystal and an aptamer. Anal. Methods 2017, 9, 906–909. [Google Scholar] [CrossRef]
- Poste, G.; Fidler, I.J. The pathogenesis of cancer metastasis. Nature 1980, 283, 139–146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fidler, I.J. The pathogenesis of cancer metastasis: The ‘seed and soil’ hypothesis revisited. Nat. Cancer 2003, 3, 453–458. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Liu, Y.; Zhu, H.; Zhang, L.; Liao, X. Nanozymes in tumor theranostics. Front. Oncol. 2021, 11, 666017. [Google Scholar] [CrossRef]
- Zhang, L.-N.; Deng, H.-H.; Lin, F.-L.; Xu, X.-W.; Weng, S.-H.; Liu, A.-L.; Lin, X.-H.; Xia, X.-H.; Chen, W. In situ growth of porous platinum nanoparticles on graphene oxide for colorimetric detection of cancer cells. Anal. Chem. 2014, 86, 2711–2718. [Google Scholar] [CrossRef] [PubMed]
- Fernández, M.; Javaid, F.; Chudasama, V. Advances in targeting the folate receptor in the treatment/imaging of cancers. Chem. Sci. 2018, 9, 790–810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, L.; Qi, J.; Qian, K.; Oderinde, O.; Liu, Q.; Yao, C.; Song, W.; Wang, Y. Copper (II) oxide nanozyme based electrochemical cytosensor for high sensitive detection of circulating tumor cells in breast cancer. J. Electroanal. Chem. 2018, 812, 1–9. [Google Scholar] [CrossRef]
- Ravindran, V.B.; Soni, S.K.; Ball, A.S. A review on the current knowledge and prospects for the development of improved detection methods for soil-transmitted helminth ova for the safe reuse of wastewater and mitigation of public health risks. Water 2019, 11, 1212. [Google Scholar] [CrossRef] [Green Version]
- Ravindran, V.B.; Khallaf, B.; Surapaneni, A.; Crosbie, N.D.; Soni, S.K.; Ball, A.S. Detection of helminth ova in wastewater using recombinase polymerase amplification coupled to lateral flow strips. Water 2020, 12, 691. [Google Scholar] [CrossRef] [Green Version]
- Diouani, M.F.; Ouerghi, O.; Belgacem, K.; Sayhi, M.; Ionescu, R.; Laouini, D. Casein-conjugated gold nanoparticles for amperometric detection of leishmania infantum. Biosensors 2019, 9, 68. [Google Scholar] [CrossRef] [Green Version]
- Gomaa, M.M. Early diagnosis of experimental Trichinella spiralis infection by nano-based enzyme-linked immunosorbent assay (nano-based ELISA). Exp. Parasitol. 2020, 212, 107867. [Google Scholar] [CrossRef]
- Wang, K.-Y.; Bu, S.-J.; Ju, C.-J.; Li, C.-T.; Li, Z.-Y.; Han, Y.; Ma, C.-Y.; Wang, C.-Y.; Hao, Z.; Liu, W.-S.; et al. Hemin-incorporated nanoflowers as enzyme mimics for colorimetric detection of foodborne pathogenic bacteria. Bioorgan. Med. Chem. Lett. 2018, 28, 3802–3807. [Google Scholar] [CrossRef]
- Nguyen, Q.H.; Kim, M.I. Nanomaterial-mediated paper-based biosensors for colorimetric pathogen detection. TrAC Trends Anal. Chem. 2020, 132, 116038. [Google Scholar] [CrossRef]
- Kuswandi, B.; Ensafi, A.A. Perspective—Paper-based biosensors: Trending topic in clinical diagnostics developments and commercialization. J. Electrochem. Soc. 2019, 167, 037509. [Google Scholar] [CrossRef]
- Antiochia, R. Paper-based biosensors: Frontiers in point-of-care detection of COVID-19 disease. Biosensors 2021, 11, 110. [Google Scholar] [CrossRef]
- Cheng, N.; Song, Y.; Zeinhom, M.M.A.; Chang, Y.-C.; Sheng, L.; Li, H.; Du, D.; Li, L.; Zhu, M.-J.; Luo, Y.; et al. Nanozyme-mediated dual immunoassay integrated with smartphone for use in simultaneous detection of pathogens. ACS Appl. Mater. Interfaces 2017, 9, 40671–40680. [Google Scholar] [CrossRef]
- Urusov, A.E.; Zherdev, A.V.; Dzantiev, B.B. Towards lateral flow quantitative assays: Detection approaches. Biosensors 2019, 9, 89. [Google Scholar] [CrossRef] [Green Version]
- Nasseri, B.; Soleimani, N.; Rabiee, N.; Kalbasi, A.; Karimi, M.; Hamblin, M.R. Point-of-care microfluidic devices for pathogen detection. Biosens. Bioelectron. 2018, 117, 112–128. [Google Scholar] [CrossRef]
- Jiang, X.; Jing, W.; Zheng, L.; Zhao, W.; Sui, G. Rapid diagnosis by microfluidic techniques, proof and concepts. In Rapid Diagnostic Tests and Technologies; Saxena, S.K., Ed.; IntechOpen: London, UK, 2016. [Google Scholar] [CrossRef] [Green Version]
- Adegoke, O.; Zolotovskaya, S.; Abdolvand, A.; Nic Daeid, N. Rapid and highly selective colorimetric detection of nitrite based on the catalytic-enhanced reaction of mimetic Au nanoparticle-CeO2 nanoparticle-graphene oxide hybrid nanozyme. Talanta 2020, 224, 121875. [Google Scholar] [CrossRef]
- Wang, M.; Liu, P.; Zhu, H.; Liu, B.; Niu, X. Ratiometric colorimetric detection of nitrite realized by stringing nanozyme catalysis and diazotization together. Biosensors 2021, 11, 280. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Tang, F.; Wang, L.; Tang, M.; Jiang, Y.-Z.; Liu, C. Nanozyme sensor based-on platinum-decorated polymer nanosphere for rapid and sensitive detection of Salmonella typhimurium with the naked eye. Sens. Actuators B Chem. 2021, 346, 130560. [Google Scholar] [CrossRef]
- Alizadeh, N.; Salimi, A. Multienzymes activity of metals and metal oxide nanomaterials: Applications from biotechnology to medicine and environmental engineering. J. Nanobiotechnol. 2021, 19, 26. [Google Scholar] [CrossRef] [PubMed]
- Fu, G.; Sanjay, S.T.; Zhou, W.; Brekken, R.A.; Kirken, R.A.; Li, X. Exploration of nanoparticle-mediated photothermal effect of TMB-H2O2 colorimetric system and its application in a visual quantitative photothermal immunoassay. Anal. Chem. 2018, 90, 5930–5937. [Google Scholar] [CrossRef]
- Tarn, M.; Pamme, N. Microfluidics, reference module in chemistry. In Molecular Sciences and Chemical Engineering; Elsevier: Amsterdam, The Netherlands, 2014. [Google Scholar] [CrossRef]
- Garrison, J.; Li, Z.; Palanisamy, B.; Wang, L.; Seker, E. An electrically-controlled programmable microfluidic concentration waveform generator. J. Biol. Eng. 2018, 12, 31. [Google Scholar] [CrossRef]
- Rao, M.V.; Shah, J.J.; Geist, J.; Gaitan, M. Microwave dielectric heating of fluids in microfluidic devices. In The Development and Application of Microwave Heating; Cao, W., Ed.; IntechOpen: London, UK, 2012. [Google Scholar] [CrossRef] [Green Version]
- Phurimsak, C.; Yildirim, E.; Tarn, M.D.; Trietsch, S.J.; Hankemeier, T.; Pamme, N.; Vulto, P. Phaseguide assisted liquid lamination for magnetic particle-based assays. Lab Chip 2014, 14, 2334–2343. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez-Ruiz, I.; Ackermann, T.N.; Muñoz-Berbel, X.; Llobera, A. Photonic lab-on-a-chip: Integration of optical spectroscopy in microfluidic systems. Anal. Chem. 2016, 88, 6630–6637. [Google Scholar] [CrossRef] [Green Version]
- Hsieh, Y.-L.; Ho, T.-Y.; Chakrabarty, K. A reagent-saving mixing algorithm for preparing multiple-target biochemical samples using digital microfluidics. IEEE Trans. Comput. Des. Integr. Circuits Syst. 2012, 31, 1656–1669. [Google Scholar] [CrossRef]
- Gu, W.; Zhu, X.; Futai, N.; Cho, B.S.; Takayama, S. Computerized microfluidic cell culture using elastomeric channels and Braille displays. Proc. Natl. Acad. Sci. USA 2004, 101, 15861–15866. [Google Scholar] [CrossRef] [Green Version]
- Davis, A.N.; Samlali, K.; Kapadia, J.B.; Perreault, J.; Shih, S.C.C.; Kharma, N. Digital microfluidics chips for the execution and real-time monitoring of multiple ribozymatic cleavage reactions. ACS Omega 2021, 6, 22514–22524. [Google Scholar] [CrossRef]
- Xue, L.; Jin, N.; Guo, R.; Wang, S.; Qi, W.; Liu, Y.; Li, Y.; Lin, J. Microfluidic colorimetric biosensors based on MnO2 nanozymes and convergence–divergence spiral micromixers for rapid and sensitive detection of salmonella. ACS Sens. 2021, 6, 2883–2892. [Google Scholar] [CrossRef]
- Hao, L.; Xue, L.; Huang, F.; Cai, G.; Qi, W.; Zhang, M.; Han, Q.; Wang, Z.; Lin, J. A microfluidic biosensor based on magnetic nanoparticle separation, quantum dots labeling and MnO2 nanoflower amplification for rapid and sensitive detection of salmonella typhimurium. Micromachines 2020, 11, 281. [Google Scholar] [CrossRef] [Green Version]
- Duan, D.; Fan, K.; Zhang, D.; Tan, S.; Liang, M.; Liu, Y.; Zhang, J.; Zhang, P.; Liu, W.; Qiu, X.; et al. Nanozyme-strip for rapid local diagnosis of Ebola. Biosens. Bioelectron. 2015, 74, 134–141. [Google Scholar] [CrossRef] [PubMed]
- Sheng, X.; Song, J.; Zhan, W. Development of a colloidal gold immunochromatographic test strip for detection of lymphocystis disease virus in fish. J. Appl. Microbiol. 2012, 113, 737–744. [Google Scholar] [CrossRef]
- Park, J.; Shin, J.H.; Park, J.-K. Pressed paper-based dipstick for detection of foodborne pathogens with multistep reactions. Anal. Chem. 2016, 88, 3781–3788. [Google Scholar] [CrossRef]
- Liu, D.; Ju, C.; Han, C.; Shi, R.; Chen, X.; Duan, D.; Yan, J.; Yan, X. Nanozyme chemiluminescence paper test for rapid and sensitive detection of SARS-CoV-2 antigen. Biosens. Bioelectron. 2020, 173, 112817. [Google Scholar] [CrossRef]
- Kumawat, M.; Umapathi, A.; Lichtfouse, E.; Daima, H.K. Nanozymes to fight the COVID-19 and future pandemics. Environ. Chem. Lett. 2021, 19, 3951–3957. [Google Scholar] [CrossRef]
- Wang, Q.; Jiang, J.; Gao, L. Nanozyme-based medicine for enzymatic therapy: Progress and challenges. Biomed. Mater. 2021, 16, 042002. [Google Scholar] [CrossRef]
- Wang, H.; Cui, Z.; Wang, X.; Sun, S.; Zhang, D.; Fu, C. Therapeutic applications of nanozymes in chronic inflammatory diseases. BioMed Res. Int. 2021, 2021, 9980127. [Google Scholar] [CrossRef]
- Das, B.; Franco, J.L.; Logan, N.; Balasubramanian, P.; Kim, M.I.; Cao, C. Nanozymes in point-of-care diagnosis: An emerging futuristic approach for biosensing. Nano-Micro Lett. 2021, 13, 193. [Google Scholar] [CrossRef]
- Fang, R.; Liu, J. Cleaving DNA by nanozymes. J. Mater. Chem. B 2020, 8, 7135–7142. [Google Scholar] [CrossRef] [PubMed]
- Petree, J.R.; Yehl, K.; Galior, K.; Glazier, R.; Deal, B.; Salaita, K. Site-selective RNA splicing nanozyme: DNAzyme and RtcB conjugates on a gold nanoparticle. ACS Chem. Biol. 2017, 13, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Ali, Z.; Zou, J.; Jin, G.; Zhu, J.; Yang, J.; Dai, J. Detection methods for Pseudomonas aeruginosa: History and future perspective. RSC Adv. 2017, 7, 51789–51800. [Google Scholar] [CrossRef] [Green Version]
- Zhu, H.; Liu, P.; Xu, L.; Li, X.; Hu, P.; Liu, B.; Pan, J.; Yang, F.; Niu, X. Nanozyme-participated biosensing of pesticides and cholinesterases: A critical review. Biosensors 2021, 11, 382. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.; Goegebuer, T.; De Leener, K.; Maes, P.; Matthijnssens, J.; Podder, G.; Azim, T.; Van Ranst, M. Chromatography paper strip method for collection, transportation, and storage of rotavirus RNA in stool samples. J. Clin. Microbiol. 2004, 42, 1605–1608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez, A.W.; Phillips, S.T.; Whitesides, G.M.; Carrilho, E. Diagnostics for the developing world: Microfluidic paper-based analytical devices. Anal. Chem. 2009, 82, 3–10. [Google Scholar] [CrossRef]
- Li, J.; Zhu, Y.; Wu, X.; Hoffmann, M.R. Rapid detection methods for bacterial pathogens in ambient waters at the point of sample collection: A brief review. Clin. Infect. Dis. 2020, 71, S84–S90. [Google Scholar] [CrossRef]
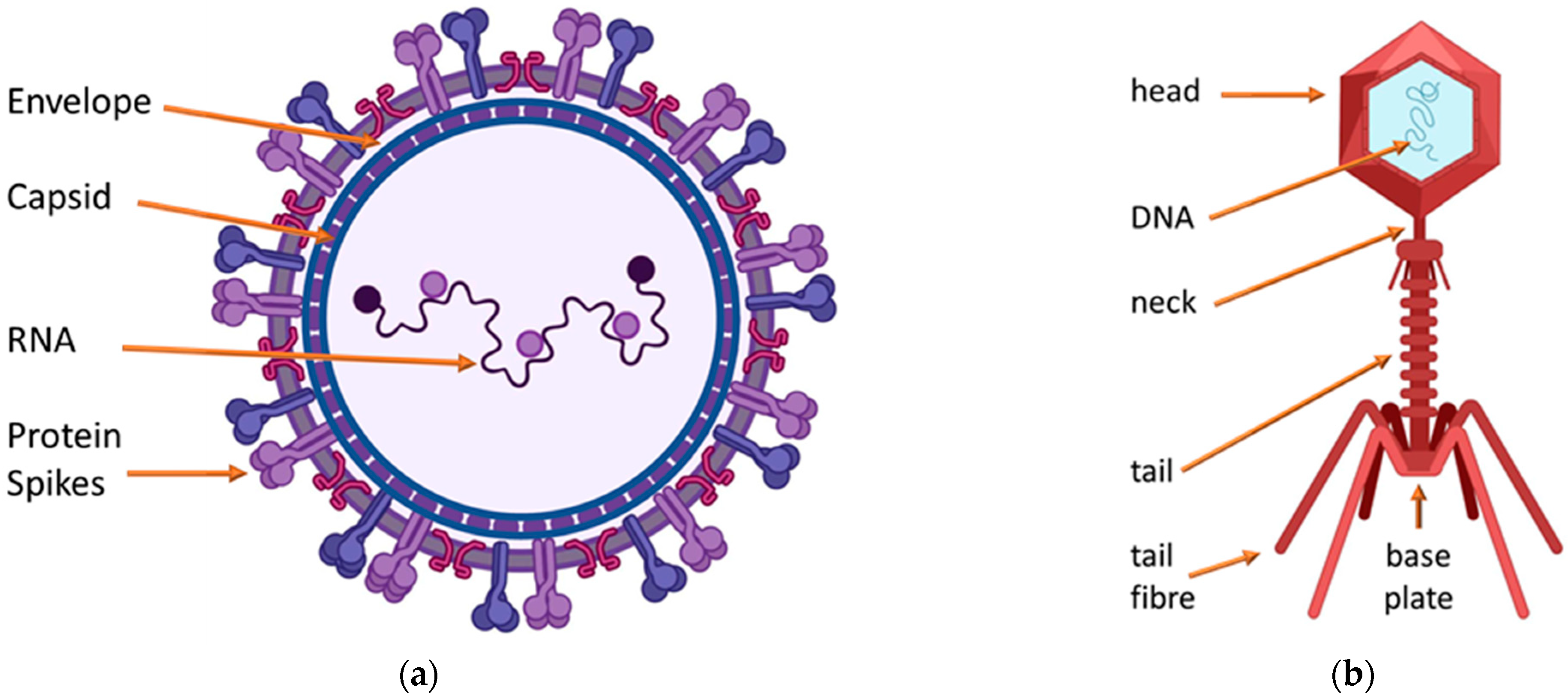
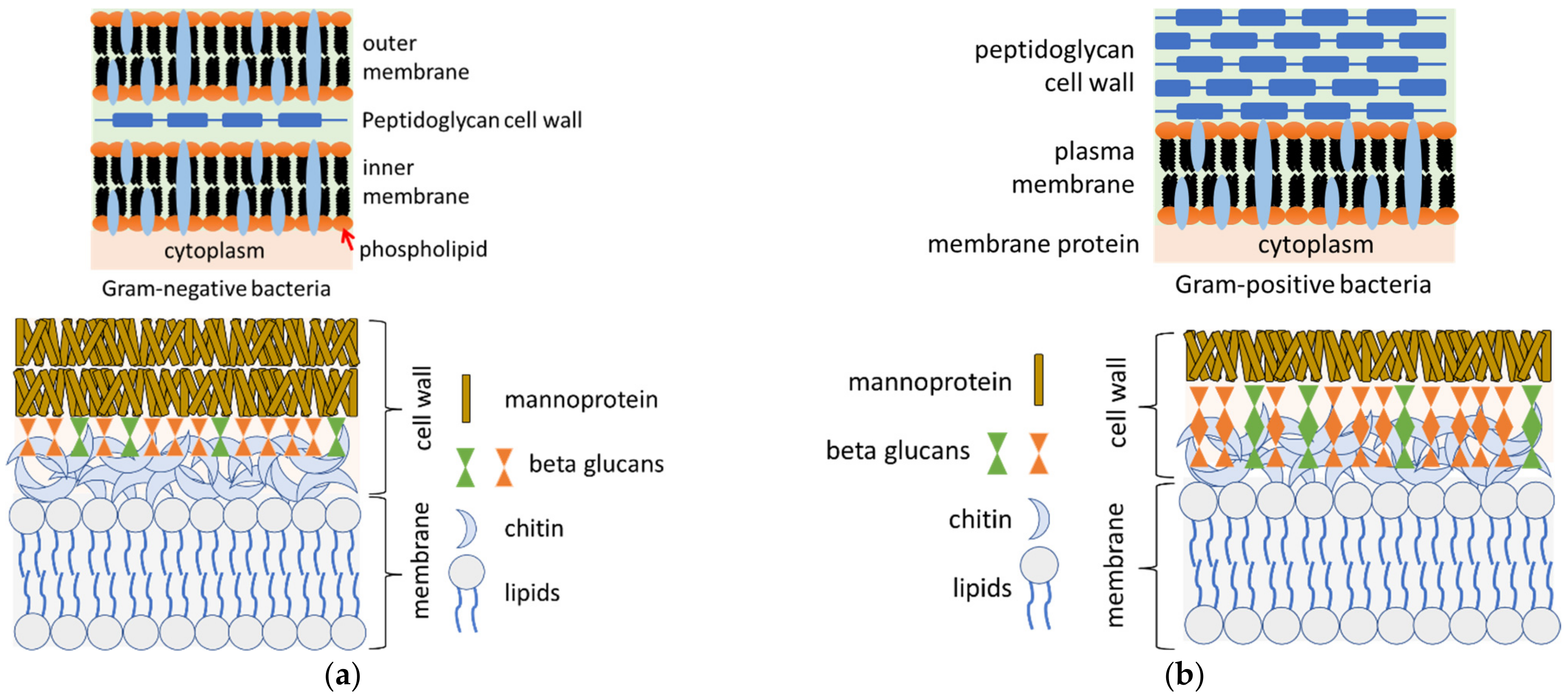




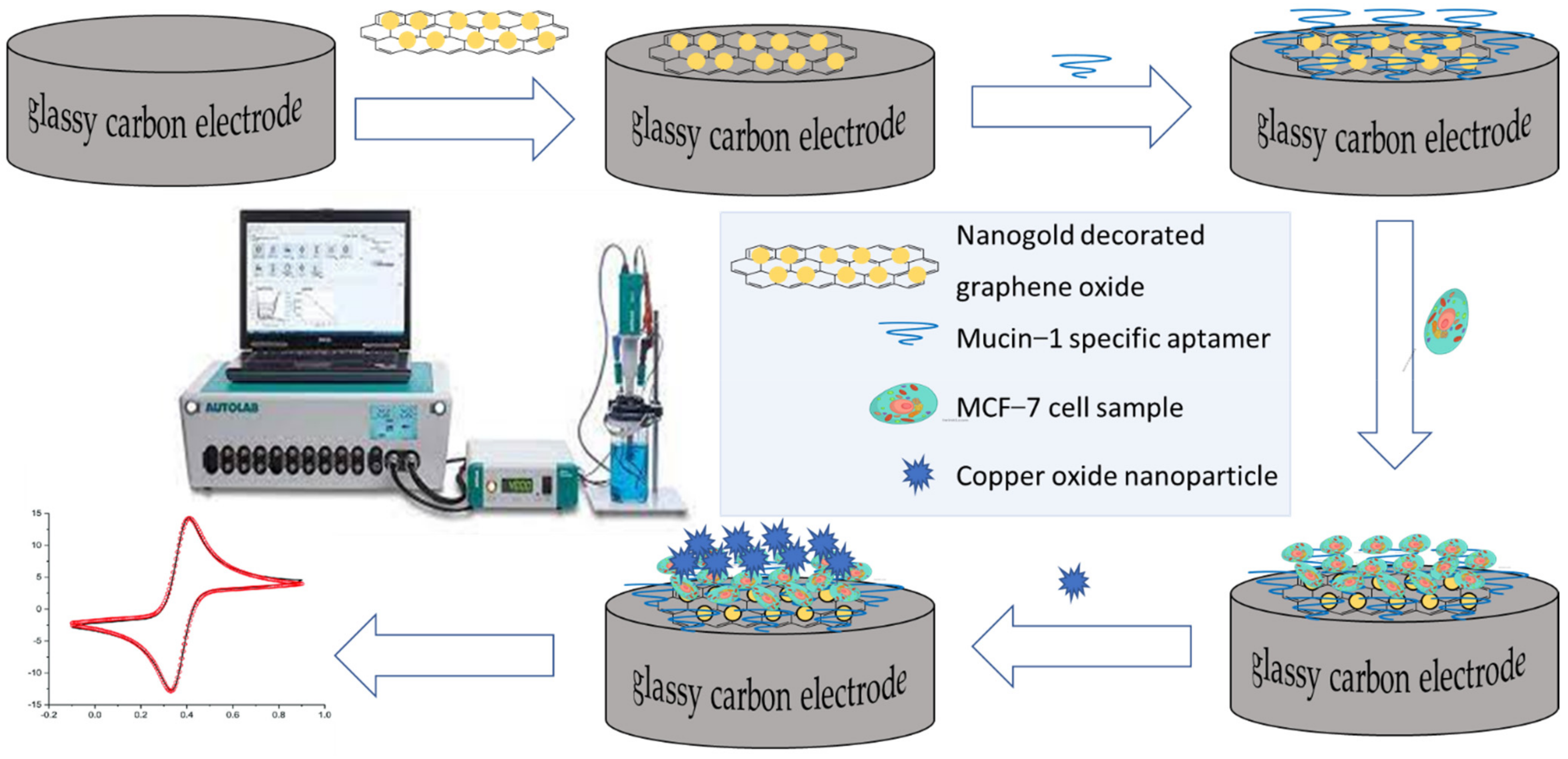

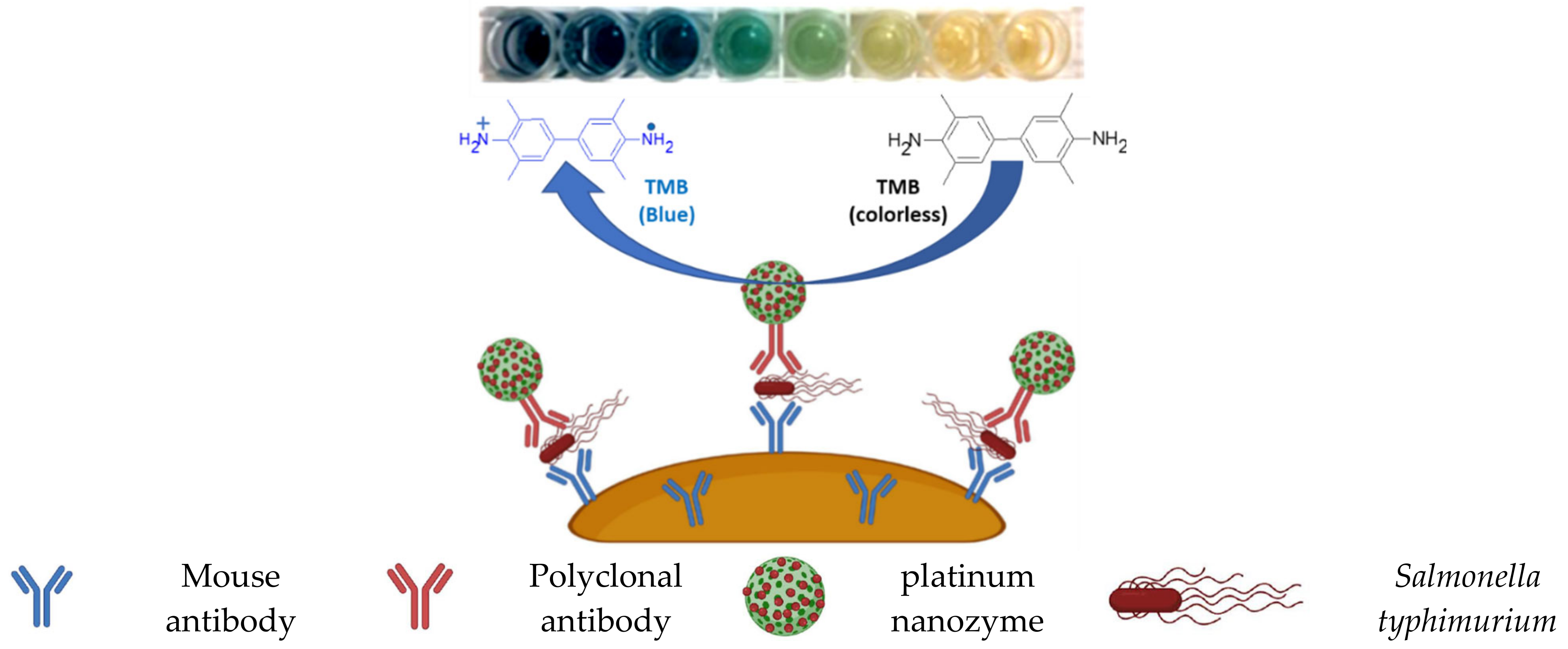




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Songca, S.P. Applications of Nanozymology in the Detection and Identification of Viral, Bacterial and Fungal Pathogens. Int. J. Mol. Sci. 2022, 23, 4638. https://doi.org/10.3390/ijms23094638
Songca SP. Applications of Nanozymology in the Detection and Identification of Viral, Bacterial and Fungal Pathogens. International Journal of Molecular Sciences. 2022; 23(9):4638. https://doi.org/10.3390/ijms23094638
Chicago/Turabian StyleSongca, Sandile Phinda. 2022. "Applications of Nanozymology in the Detection and Identification of Viral, Bacterial and Fungal Pathogens" International Journal of Molecular Sciences 23, no. 9: 4638. https://doi.org/10.3390/ijms23094638
APA StyleSongca, S. P. (2022). Applications of Nanozymology in the Detection and Identification of Viral, Bacterial and Fungal Pathogens. International Journal of Molecular Sciences, 23(9), 4638. https://doi.org/10.3390/ijms23094638




