Radiomics-Based Method for Predicting the Glioma Subtype as Defined by Tumor Grade, IDH Mutation, and 1p/19q Codeletion
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Datasets
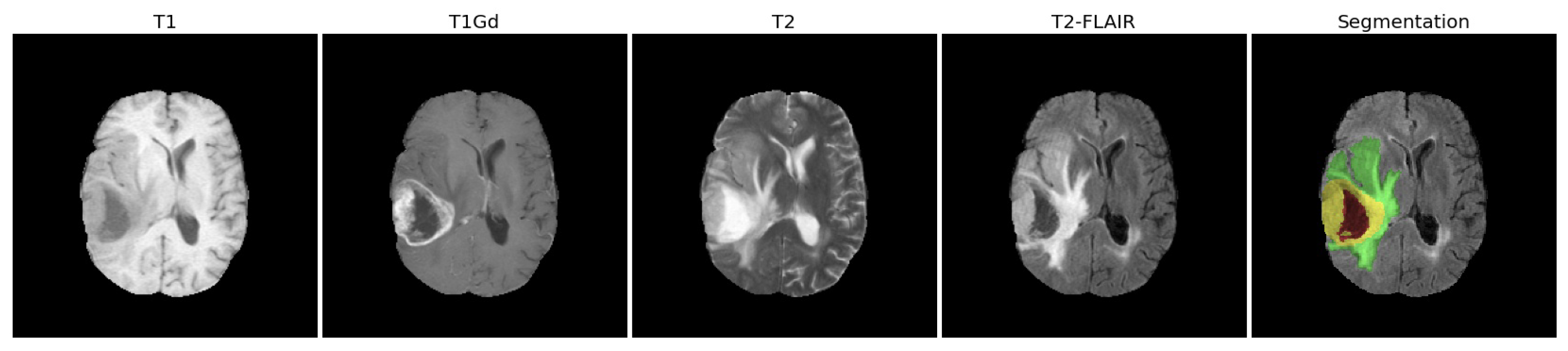
2.1.1. Clinical MRI Scans
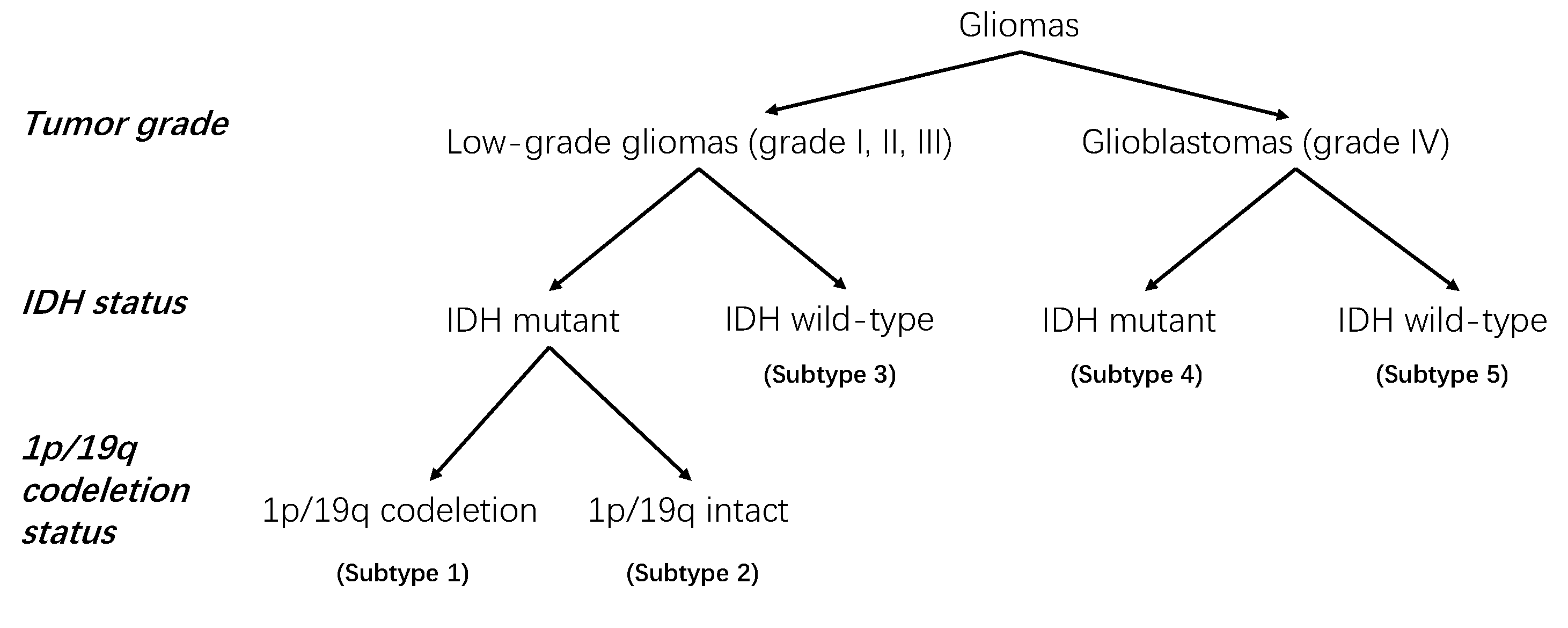
2.1.2. Clinical Information and Molecular Status
2.1.3. Data Cleaning
- Dropped two patients (“TCGA-06-0177” and “TCGA-CS-4941”) because of their very poor image qualities and abnormal image histograms.
- Dropped patient “TCGA-EZ-7265A” because the patient ID could not be found in the TCGA platform.
- Dropped 23 patients who lacked IDH mutation statuses. Among the 23 patients, 22 patients were of the GBM type and 1 was of the LGG type.
- Dropped five patients who lacked the 1p/19q codeletion status. Among the five patients, four patients were GBM with the IDH wild type, one patient was GBM with the IDH mutant.
2.2. Image Preprocessing
2.3. Extract Radiomic Features
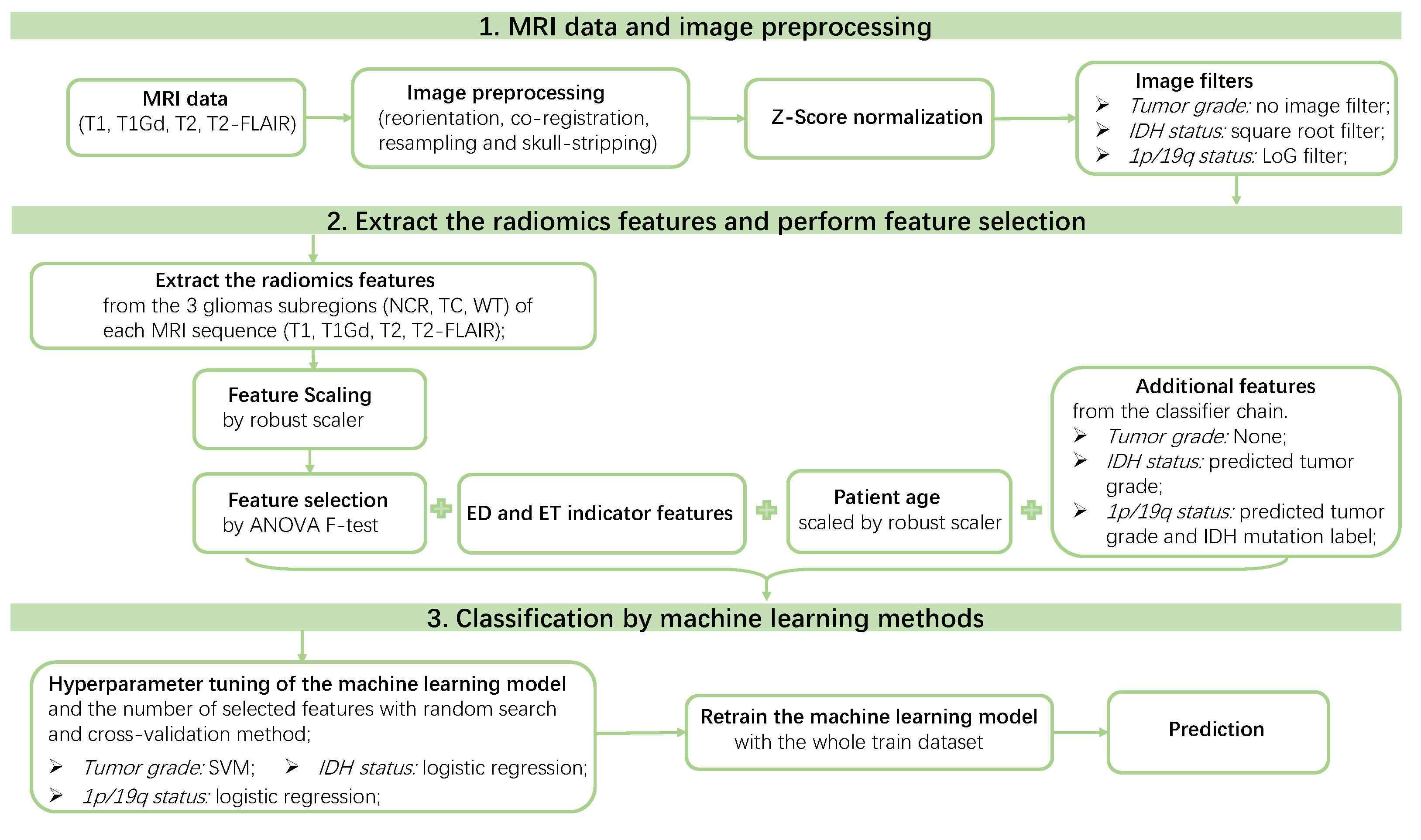
2.4. Basic Radiomics Pipeline
- (1)
- Medical images: we used the clinical MRI scans without any intensity normalization or image filters for the radiomics feature extraction.
- (2)
- Feature extraction: for each patient, we extracted the radiomic features from the whole tumor (WT) volume in the four MRI scans (T1, T1Gd, T2, T2-FLAIR).
- (3)
- Feature scaling: we applied a robust scaler [45] to scale features robustly to outliers.
- (4)
- Feature selection: we used the ANOVA F-test as the feature selection method because of its high efficiency and good performance. We set the number of selected features as a hyperparameter to be tuned from set by random search and five-fold cross validation. The highly correlated features were not "dropped out", because this step significantly increased the training time without obviously improving the performance in our experiments.
- (5)
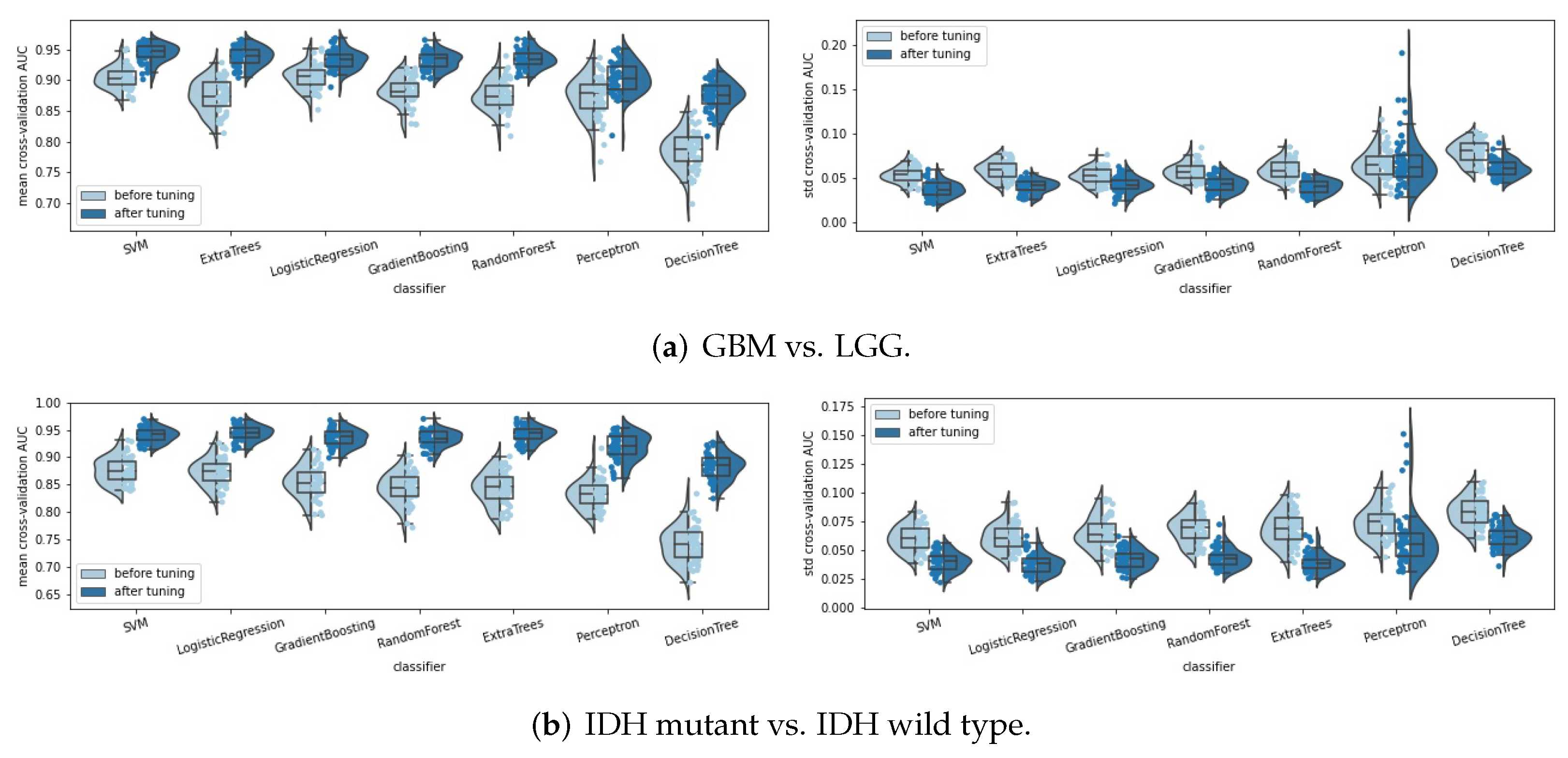
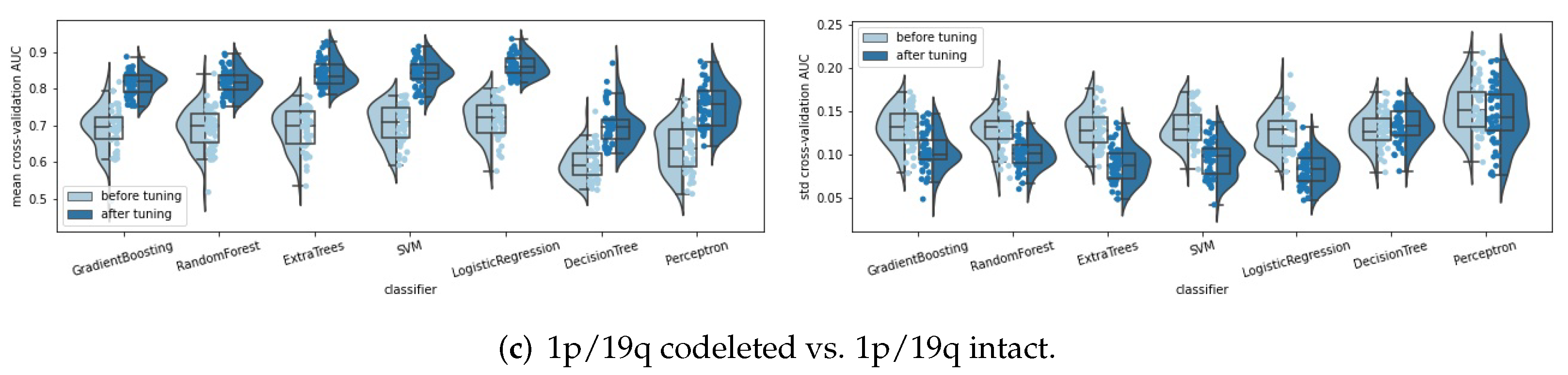
- Classifiers: seven classifiers were compared, namely, support vector machine (SVM), perceptron, logistic regression, decision tree, random forest, extra trees, and gradient boosting. The hyperparameters of each classifier were tuned automatically by a random search method (50 parameter settings were randomly sampled) and five-fold cross validation. Some specific hyperparameters were included in this hyperparameter tuning process to avoid overfitting, such as the regularization methods and the regularization parameters used in SVM, perceptron, and logistic regression, and the parameters, such as “max_depth”, “min_samples_split” and “min_samples_leaf” for the tree-based methods. The area under the receiver operating characteristic curve (ROC AUC, or AUC for simplicity) was used as the score metric in the random search. The classifier that achieved the highest mean cross-validation AUC among the seven classifiers was chosen as the final classifier.
2.5. Tuning the Radiomics Pipeline
3. Results
3.1. Pipeline Tuning
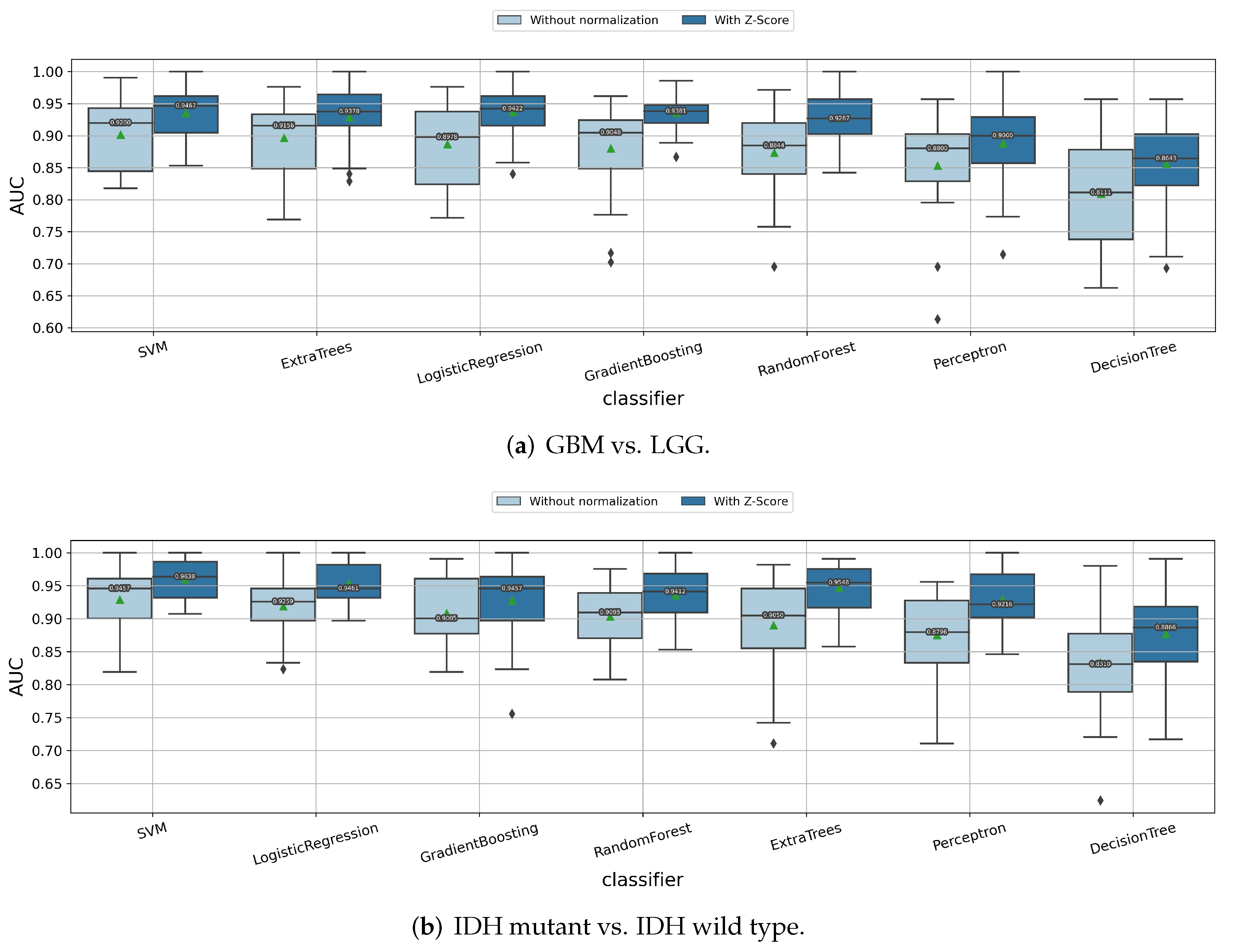
3.1.1. Impact of Z-Score Intensity Normalization
3.1.2. Impact of the Feature Extraction Strategy
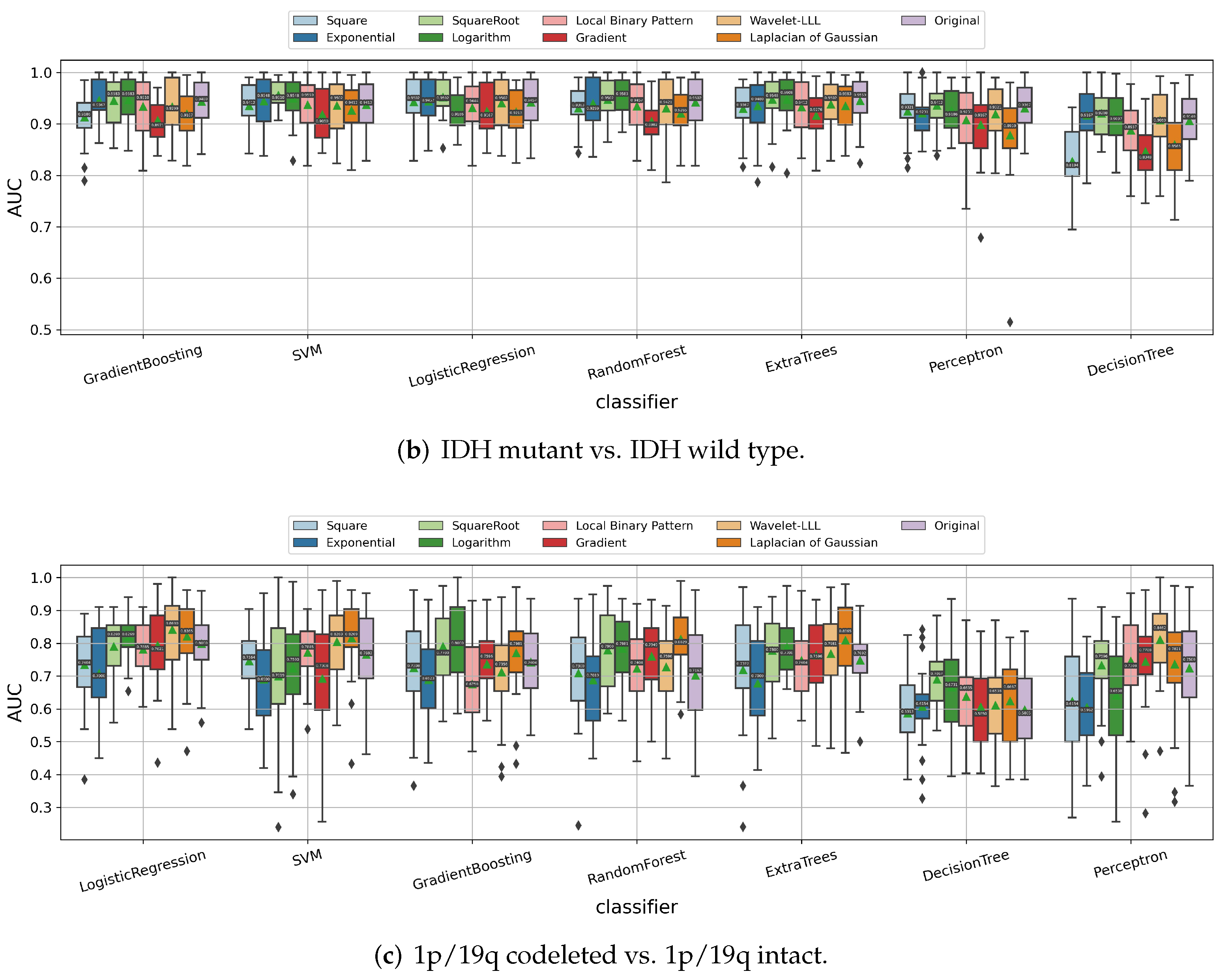
3.1.3. Impact of Image Filters
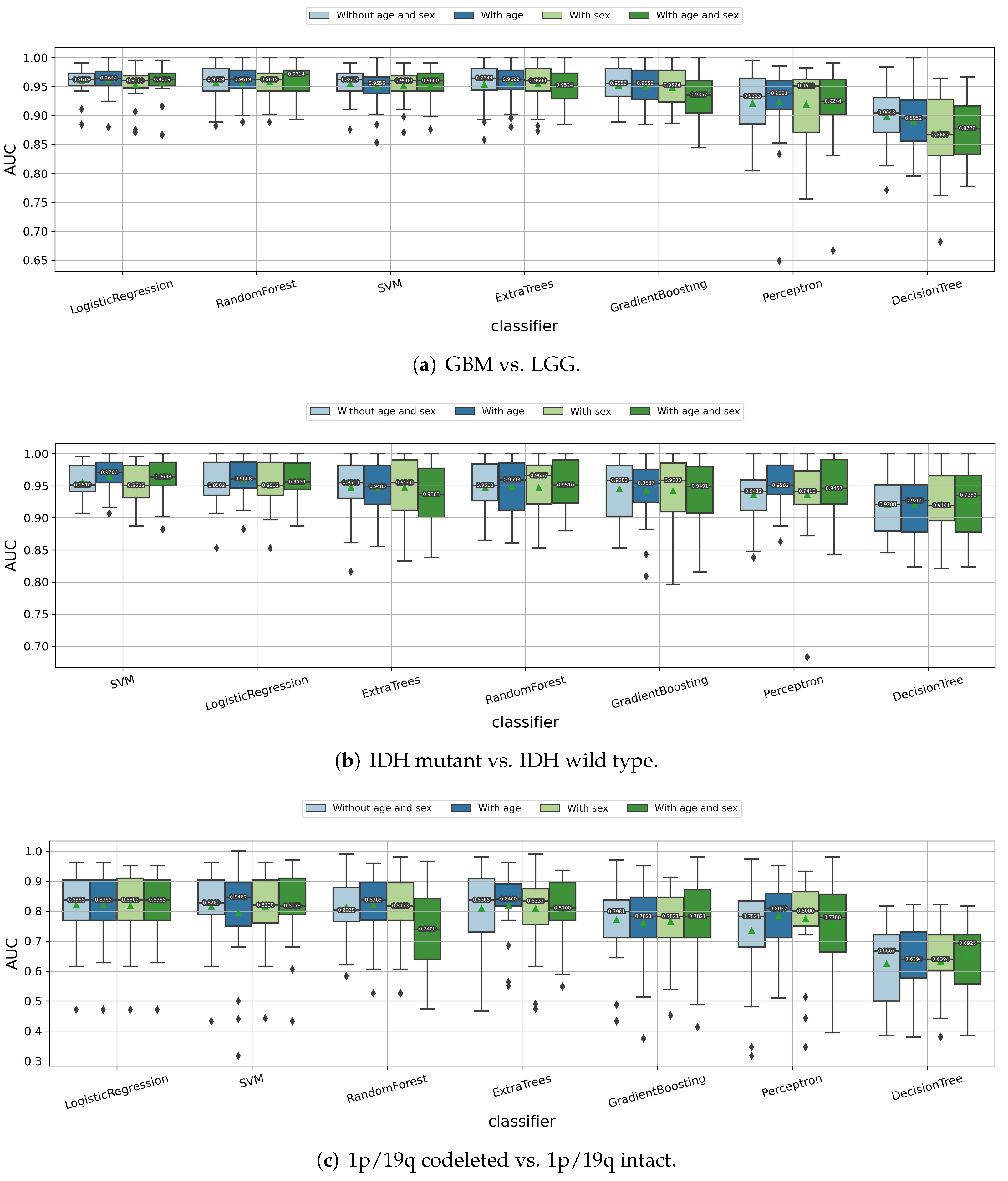
3.1.4. Impact of the Age and Sex Information
3.1.5. Impact of ComBat Harmonization
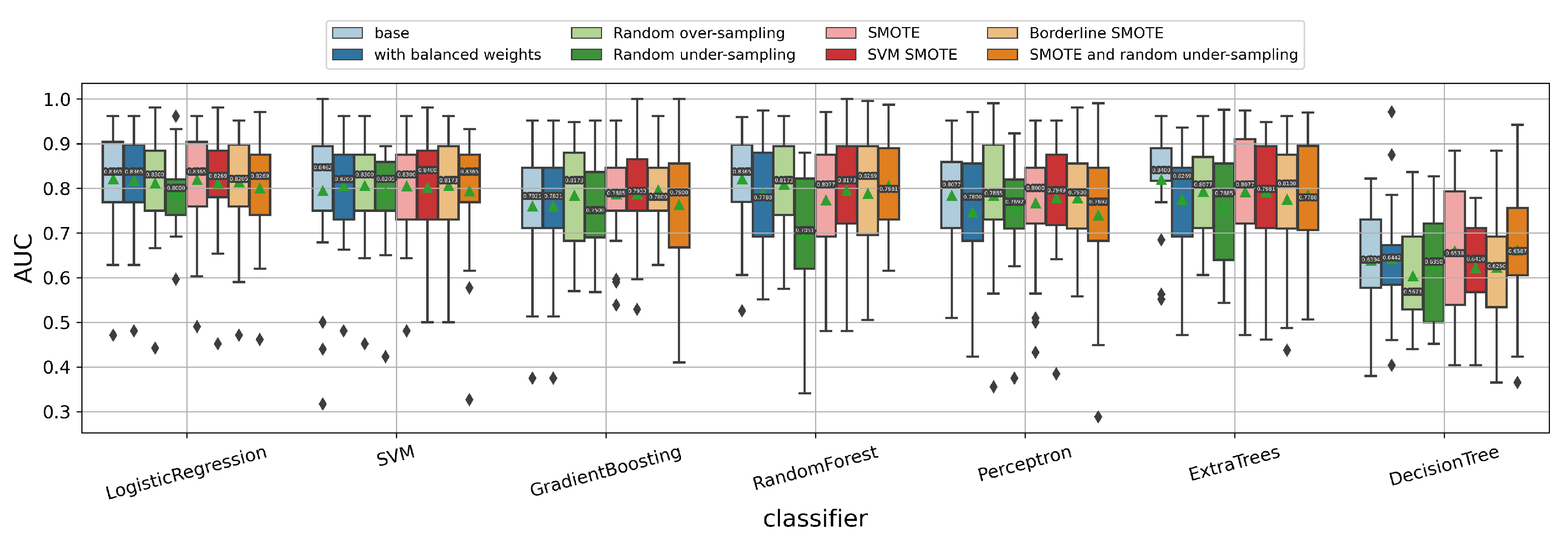
3.1.6. Impact of Data Imbalance Strategy
3.1.7. Impact of Using Classifier Chain Idea
3.2. Summarized Pipeline
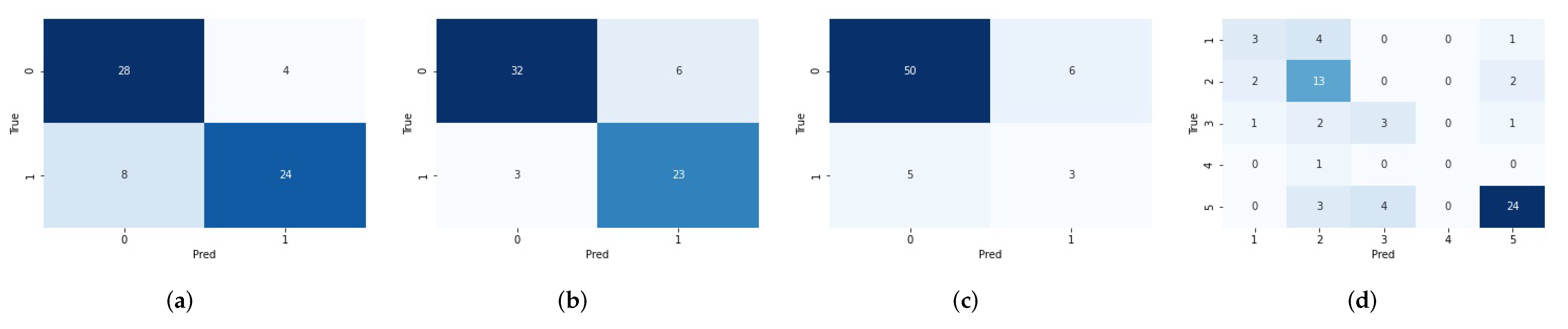
3.3. Predict Glioma Subtype by the Final Pipeline
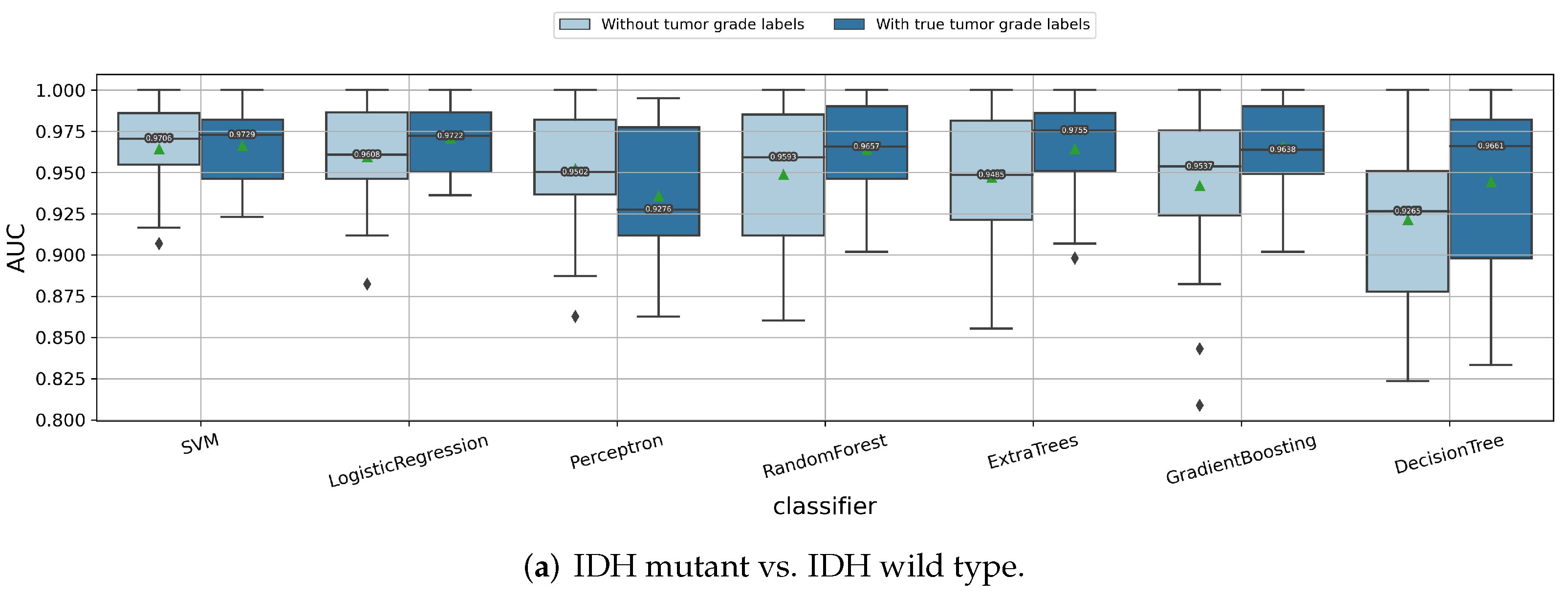
3.4. Interpretability of the Radiomics Models
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CNS | central nervous system |
| WHO | World Health Organization |
| 2016 CNS WHO | 2016 World Health Organization Classification of Tumors of the |
| Central Nervous System | |
| TCGA | The Cancer Genome Atlas |
| TCIA | The Cancer Imaging Archive |
| LGG | low-grade glioma |
| GBM | glioblastoma multiforme |
| TCGA-LGG | The Cancer Genome Atlas Low Grade Glioma |
| TCGA-GBM | The Cancer Genome Atlas Glioblastoma Multiforme |
| IDH | isocitrate dehydrogenase |
| MGMT | O6-methylguanine-DNA methyltransferase |
| MRI | magnetic resonance imaging |
| FLAIR | fluid attenuated inversion recovery |
| VOI | volume of interest |
| ROC AUC | area under the receiver operating characteristic curve |
| ROC | receiver operating characteristic |
| AUC | area under the ROC curve |
| EB | empirical Bayes |
| BraTS | brain tumor segmentation |
| CI | confidence interval |
| SVM | support vector machine |
| GLRLM | gray level run length matrix |
| GLSZM | gray level size zone matrix |
| GLDM | gray level dependence matrix |
| NGTDM | neighbouring gray tone difference matrix |
| GLCM | gray level co-occurrence matrix |
Appendix A
Appendix A.1. Summary of the Train and Test Data
| Tumor Subtypes | Train Data | Test Data | Total |
|---|---|---|---|
| 1—LGG, IDH mutant, 1p/19q codeleted | 19 | 8 | 27 |
| 2—LGG, IDH mutant, 1p/19q intact | 40 | 17 | 57 |
| 3—LGG, IDH wild type | 14 | 7 | 21 |
| 4—GBM, IDH mutant | 3 | 1 | 4 |
| 5—GBM, IDH wild type | 72 | 31 | 103 |
| Data | Tumor Grade | IDH Mutation | 1p/19q Codeletion |
|---|---|---|---|
| Train data (148 patients) | 75 GBM vs. 73 LGG | 62 IDH mutant vs. 86 IDH wild type | 129 1p/19q codeleted vs. 19 1p/19q intact |
| Test data (64 patients) | 32 GBM vs. 32 LGG | 26 IDH mutant vs. 38 IDH wild type | 56 1p/19q codeleted vs. 8 1p/19q intact |
Appendix A.2. Image Histograms before and after Normalization
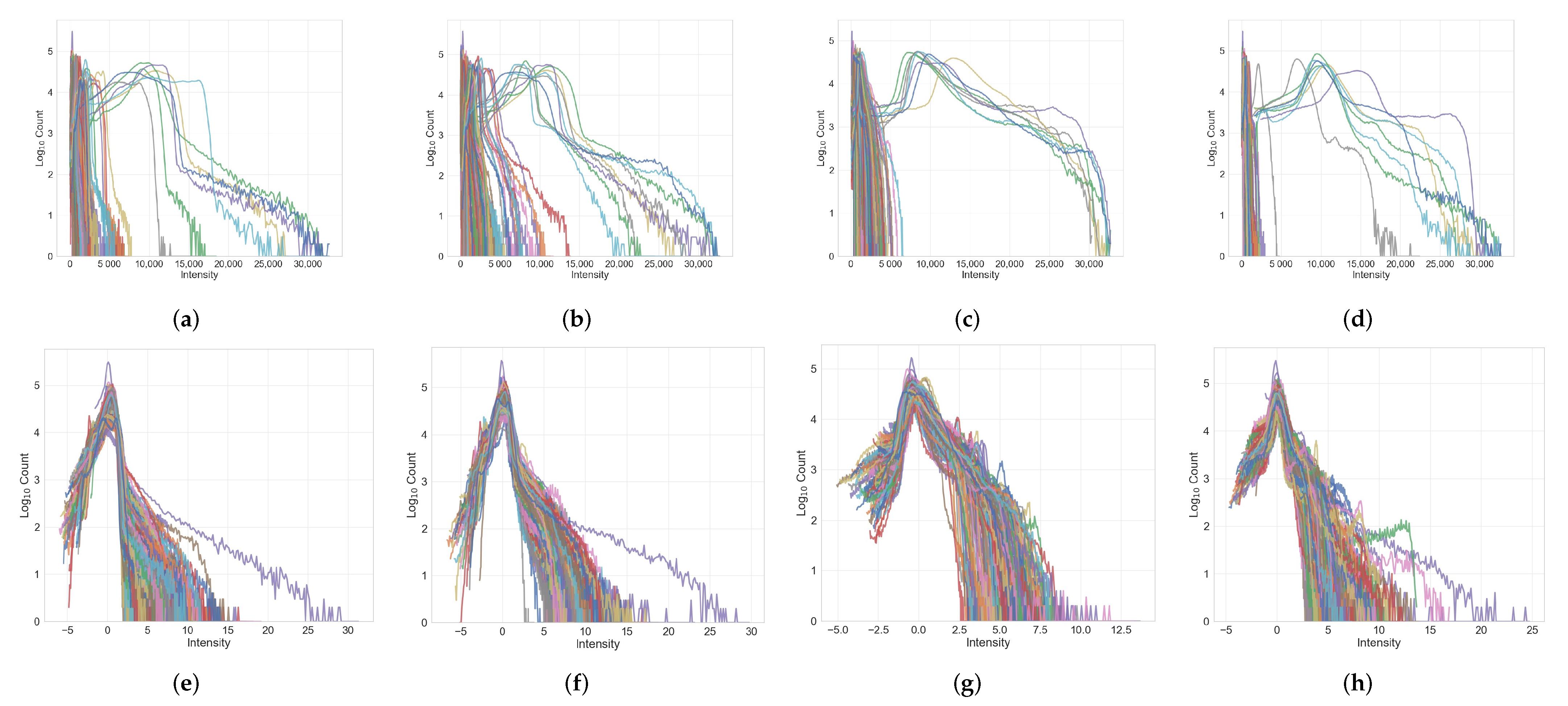
References
- Walker, C.; Baborie, A.; Crooks, D.; Wilkins, S.; Jenkinson, M. Biology, genetics and imaging of glial cell tumours. Br. J. Radiol. 2011, 84, S90–S106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Louis, D.N.; Perry, A.; Reifenberger, G.; Von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization classification of tumors of the central nervous system: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delev, D.; Heiland, D.H.; Franco, P.; Reinacher, P.; Mader, I.; Staszewski, O.; Lassmann, S.; Grau, S.; Schnell, O. Surgical management of lower-grade glioma in the spotlight of the 2016 WHO classification system. J. Neuro-Oncol. 2019, 141, 223–233. [Google Scholar] [CrossRef]
- Chai, R.; Li, G.; Liu, Y.; Zhang, K.; Zhao, Z.; Wu, F.; Chang, Y.; Pang, B.; Li, J.; Li, Y.; et al. Predictive value of MGMT promoter methylation on the survival of TMZ treated IDH-mutant glioblastoma. Cancer Biol. Med. 2021, 18, 272–282. [Google Scholar] [CrossRef] [PubMed]
- Qi, S.; Yu, L.; Gui, S.; Ding, Y.; Han, H.; Zhang, X.; Wu, L.; Yao, F. IDH mutations predict longer survival and response to temozolomide in secondary glioblastoma. Cancer Sci. 2012, 103, 269–273. [Google Scholar]
- Tandel, G.S.; Biswas, M.; Kakde, O.G.; Tiwari, A.; Suri, H.S.; Turk, M.; Laird, J.R.; Asare, C.K.; Ankrah, A.A.; Khanna, N.; et al. A review on a deep learning perspective in brain cancer classification. Cancers 2019, 11, 111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, V.; Gu, Y.; Basu, S.; Berglund, A.; Eschrich, S.A.; Schabath, M.B.; Forster, K.; Aerts, H.J.; Dekker, A.; Fenstermacher, D.; et al. Radiomics: The process and the challenges. Magn. Reson. Imaging 2012, 30, 1234–1248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images are more than pictures, they are data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lambin, P.; Leijenaar, R.T.; Deist, T.M.; Peerlings, J.; De Jong, E.E.; Van Timmeren, J.; Sanduleanu, S.; Larue, R.T.; Even, A.J.; Jochems, A.; et al. Radiomics: The bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 2017, 14, 749–762. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.B.; Liu, Z.; Zhang, H.; Shen, C.; Wang, X.C.; Tan, Y.; Wang, S.; Wu, X.F.; Tian, J. Grading of gliomas by using radiomic features on multiple magnetic resonance imaging (MRI) sequences. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2017, 23, 2168–2178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ditmer, A.; Zhang, B.; Shujaat, T.; Pavlina, A.; Luibrand, N.; Gaskill-Shipley, M.; Vagal, A. Diagnostic accuracy of MRI texture analysis for grading gliomas. J. Neuro-Oncol. 2018, 140, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.H.; Lee, S.H.; Kim, J.; Park, H. Classification of the glioma grading using radiomics analysis. PeerJ 2018, 6, e5982. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, Q.; Yan, L.F.; Zhang, X.; Zhang, X.; Hu, Y.C.; Han, Y.; Liu, Z.C.; Nan, H.Y.; Sun, Q.; Sun, Y.Z.; et al. Radiomics strategy for glioma grading using texture features from multiparametric MRI. J. Magn. Reson. Imaging 2018, 48, 1518–1528. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Liu, B.; Peng, S.; Sun, J.; Qiao, X. Computer-aided grading of gliomas combining automatic segmentation and radiomics. Int. J. Biomed. Imaging 2018, 2018, 2512037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, P.; Wang, D.; Mok, V.C.; Shi, L. Comparison of feature selection methods and machine learning classifiers for radiomics analysis in glioma grading. IEEE Access 2019, 7, 102010–102020. [Google Scholar] [CrossRef]
- Park, Y.W.; Choi, Y.S.; Ahn, S.S.; Chang, J.H.; Kim, S.H.; Lee, S.K. Radiomics MRI phenotyping with machine learning to predict the grade of lower-grade gliomas: A study focused on nonenhancing tumors. Korean J. Radiol. 2019, 20, 1381–1389. [Google Scholar] [CrossRef] [Green Version]
- Cinarer, G.; Emiroglu, B.G. Classification of brain tumours using radiomic features on MRI. New Trends Issues Proc. Adv. Pure Appl. Sci. 2020, 12, 80–90. [Google Scholar] [CrossRef]
- Xiao, T.; Hua, W.; Li, C.; Wang, S. Glioma grading prediction by exploring radiomics and deep learning features. In Proceedings of the Third International Symposium on Image Computing and Digital Medicine, Xi’an, China, 24–26 August 2019; pp. 208–213. [Google Scholar]
- Zhang, X.; Tian, Q.; Wang, L.; Liu, Y.; Li, B.; Liang, Z.; Gao, P.; Zheng, K.; Zhao, B.; Lu, H. Radiomics strategy for molecular subtype stratification of lower-grade glioma: Detecting IDH and TP53 mutations based on multimodal MRI. J. Magn. Reson. Imaging 2018, 48, 916–926. [Google Scholar] [CrossRef]
- Kim, M.; Jung, S.Y.; Park, J.E.; Jo, Y.; Park, S.Y.; Nam, S.J.; Kim, J.H.; Kim, H.S. Diffusion-and perfusion-weighted MRI radiomics model may predict isocitrate dehydrogenase (IDH) mutation and tumor aggressiveness in diffuse lower grade glioma. Eur. Radiol. 2020, 30, 2142–2151. [Google Scholar] [CrossRef]
- Arita, H.; Kinoshita, M.; Kawaguchi, A.; Takahashi, M.; Narita, Y.; Terakawa, Y.; Tsuyuguchi, N.; Okita, Y.; Nonaka, M.; Moriuchi, S.; et al. Lesion location implemented magnetic resonance imaging radiomics for predicting IDH and TERT promoter mutations in grade II/III gliomas. Sci. Rep. 2018, 8, 11773. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Li, Y.; Li, S.; Fan, X.; Sun, Z.; Yang, Z.; Wang, K.; Zhang, Z.; Jiang, T.; Liu, Y.; et al. IDH mutation-specific radiomic signature in lower-grade gliomas. Aging 2019, 11, 673. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.S.; Bae, S.; Chang, J.H.; Kang, S.G.; Kim, S.H.; Kim, J.; Rim, T.H.; Choi, S.H.; Jain, R.; Lee, S.K. Fully automated hybrid approach to predict the IDH mutation status of gliomas via deep learning and radiomics. Neuro-Oncology 2021, 23, 304–313. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Meng, J.; Yu, Q.; Li, P.; Fu, S. Radiomics-based machine learning methods for isocitrate dehydrogenase genotype prediction of diffuse gliomas. J. Cancer Res. Clin. Oncol. 2019, 145, 543–550. [Google Scholar] [CrossRef] [Green Version]
- Tan, Y.; Zhang, S.T.; Wei, J.W.; Dong, D.; Wang, X.C.; Yang, G.Q.; Tian, J.; Zhang, H. A radiomics nomogram may improve the prediction of IDH genotype for astrocytoma before surgery. Eur. Radiol. 2019, 29, 3325–3337. [Google Scholar] [CrossRef] [PubMed]
- Chougule, T.; Shinde, S.; Santosh, V.; Saini, J.; Ingalhalikar, M. On Validating Multimodal MRI Based Stratification of IDH Genotype in High Grade Gliomas Using CNNs and Its Comparison to Radiomics. In International Workshop on Radiomics and Radiogenomics in Neuro-Oncology; Springer: Cham, Switzerland, 2019; pp. 53–60. [Google Scholar]
- Yu, J.; Shi, Z.; Lian, Y.; Li, Z.; Liu, T.; Gao, Y.; Wang, Y.; Chen, L.; Mao, Y. Noninvasive IDH1 mutation estimation based on a quantitative radiomics approach for grade II glioma. Eur. Radiol. 2017, 27, 3509–3522. [Google Scholar] [CrossRef]
- Li, Z.C.; Bai, H.; Sun, Q.; Zhao, Y.; Lv, Y.; Zhou, J.; Liang, C.; Chen, Y.; Liang, D.; Zheng, H. Multiregional radiomics profiling from multiparametric MRI: Identifying an imaging predictor of IDH1 mutation status in glioblastoma. Cancer Med. 2018, 7, 5999–6009. [Google Scholar] [CrossRef] [Green Version]
- Gore, S.; Chougule, T.; Saini, J.; Ingalhalikar, M.; Jagtap, J. Local Binary and Ternary Patterns Based Quantitative Texture Analysis for Assessment of IDH Genotype in Gliomas on Multi-modal MRI. In Machine Learning in Clinical Neuroimaging and Radiogenomics in Neuro-Oncology; Springer: Cham, Switzerland, 2020; pp. 240–248. [Google Scholar]
- Shofty, B.; Artzi, M.; Bashat, D.B.; Liberman, G.; Haim, O.; Kashanian, A.; Bokstein, F.; Blumenthal, D.T.; Ram, Z.; Shahar, T. MRI radiomics analysis of molecular alterations in low-grade gliomas. Int. J. Comput. Assist. Radiol. Surg. 2018, 13, 563–571. [Google Scholar] [CrossRef]
- van der Voort, S.R.; Incekara, F.; Wijnenga, M.M.; Kapas, G.; Gardeniers, M.; Schouten, J.W.; Starmans, M.P.; Tewarie, R.N.; Lycklama, G.J.; French, P.J.; et al. Predicting the 1p/19q codeletion status of presumed low-grade glioma with an externally validated machine learning algorithm. Clin. Cancer Res. 2019, 25, 7455–7462. [Google Scholar] [CrossRef] [Green Version]
- Kocak, B.; Durmaz, E.S.; Ates, E.; Sel, I.; Gunes, S.T.; Kaya, O.K.; Zeynalova, A.; Kilickesmez, O. Radiogenomics of lower-grade gliomas: Machine learning-based MRI texture analysis for predicting 1p/19q codeletion status. Eur. Radiol. 2020, 30, 877–886. [Google Scholar] [CrossRef]
- Kong, Z.; Jiang, C.; Zhang, Y.; Liu, S.; Liu, D.; Liu, Z.; Chen, W.; Liu, P.; Yang, T.; Lyu, Y.; et al. Thin-Slice Magnetic Resonance Imaging-Based Radiomics Signature Predicts Chromosomal 1p/19q Co-deletion Status in Grade II and III Gliomas. Front. Neurol. 2020, 11, 1304. [Google Scholar] [CrossRef]
- Zhou, H.; Vallières, M.; Bai, H.X.; Su, C.; Tang, H.; Oldridge, D.; Zhang, Z.; Xiao, B.; Liao, W.; Tao, Y.; et al. MRI features predict survival and molecular markers in diffuse lower-grade gliomas. Neuro-Oncology 2017, 19, 862–870. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.F.; Hsu, F.T.; Hsieh, K.L.C.; Kao, Y.C.J.; Cheng, S.J.; Hsu, J.B.K.; Tsai, P.H.; Chen, R.J.; Huang, C.C.; Yen, Y.; et al. Machine learning-based radiomics for molecular subtyping of gliomas. Clin. Cancer Res. 2018, 24, 4429–4436. [Google Scholar] [CrossRef] [Green Version]
- Bakas, S.; Akbari, H.; Sotiras, A.; Bilello, M.; Rozycki, M.; Kirby, J.; Freymann, J.; Farahani, K.; Davatzikos, C. Segmentation labels for the pre-operative scans of the TCGA-GBM collection [Data set]. Cancer Imaging Arch. 2017. [Google Scholar] [CrossRef]
- Bakas, S.; Akbari, H.; Sotiras, A.; Bilello, M.; Rozycki, M.; Kirby, J.S.; Freymann, J.B.; Farahani, K.; Davatzikos, C. Advancing the cancer genome atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci. Data 2017, 4, 170117. [Google Scholar] [CrossRef] [Green Version]
- Bakas, S.; Akbari, H.; Sotiras, A.; Bilello, M.; Rozycki, M.; Kirby, J.; Freymann, J.; Farahani, K.; Davatzikos, C. Segmentation labels and radiomic features for the pre-operative scans of the TCGA-LGG collection [Data Set]. Cancer Imaging Arch. 2017. [Google Scholar] [CrossRef]
- Clark, K.; Vendt, B.; Smith, K.; Freymann, J.; Kirby, J.; Koppel, P.; Moore, S.; Phillips, S.; Maffitt, D.; Pringle, M.; et al. The Cancer Imaging Archive (TCIA): Maintaining and operating a public information repository. J. Digit. Imaging 2013, 26, 1045–1057. [Google Scholar] [CrossRef] [Green Version]
- Rohlfing, T.; Zahr, N.M.; Sullivan, E.V.; Pfefferbaum, A. The SRI24 multichannel atlas of normal adult human brain structure. Hum. Brain Mapp. 2010, 31, 798–819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bakas, S.; Zeng, K.; Sotiras, A.; Rathore, S.; Akbari, H.; Gaonkar, B.; Rozycki, M.; Pati, S.; Davatzizkos, C. GLISTRboost: Combining multimodal MRI segmentation, registration, and biophysical tumor growth modeling with gradient boosting machines for glioma segmentation. In BrainLes 2015; Springer: Cham, Switzerland, 2015; pp. 144–155. [Google Scholar]
- Baid, U.; Ghodasara, S.; Mohan, S.; Bilello, M.; Calabrese, E.; Colak, E.; Farahani, K.; Kalpathy-Cramer, J.; Kitamura, F.C.; Pati, S.; et al. The rsna-asnr-miccai brats 2021 benchmark on brain tumor segmentation and radiogenomic classification. arXiv 2021, arXiv:2107.02314. [Google Scholar]
- Li, Y.; Ammari, S.; Balleyguier, C.; Lassau, N.; Chouzenoux, E. Impact of Preprocessing and Harmonization Methods on the Removal of Scanner Effects in Brain MRI Radiomic Features. Cancers 2021, 13, 3000. [Google Scholar] [CrossRef]
- Van Griethuysen, J.J.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.; Fillion-Robin, J.C.; Pieper, S.; Aerts, H.J. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Johnson, W.E.; Li, C.; Rabinovic, A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007, 8, 118–127. [Google Scholar] [CrossRef] [PubMed]
- Read, J.; Pfahringer, B.; Holmes, G.; Frank, E. Classifier chains for multi-label classification. Mach. Learn. 2011, 85, 333–359. [Google Scholar] [CrossRef] [Green Version]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic minority over-sampling technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Tang, Y.; Zhang, Y.Q.; Chawla, N.V.; Krasser, S. SVMs modeling for highly imbalanced classification. IEEE Trans. Syst. Man Cybern. Part B (Cybern.) 2008, 39, 281–288. [Google Scholar] [CrossRef] [Green Version]
- Han, H.; Wang, W.Y.; Mao, B.H. Borderline-SMOTE: A new over-sampling method in imbalanced data sets learning. In International Conference on Intelligent Computing; Springer: Berlin/Heidelberg, Germany, 2005; pp. 878–887. [Google Scholar]
- Orlhac, F.; Eertink, J.J.; Cottereau, A.S.; Zijlstra, J.M.; Thieblemont, C.; Meignan, M.; Boellaard, R.; Buvat, I. A guide to ComBat harmonization of imaging biomarkers in multicenter studies. J. Nucl. Med. 2022, 63, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Isensee, F.; Jaeger, P.F.; Kohl, S.A.; Petersen, J.; Maier-Hein, K.H. nnU-Net: A self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 2021, 18, 203–211. [Google Scholar] [CrossRef]
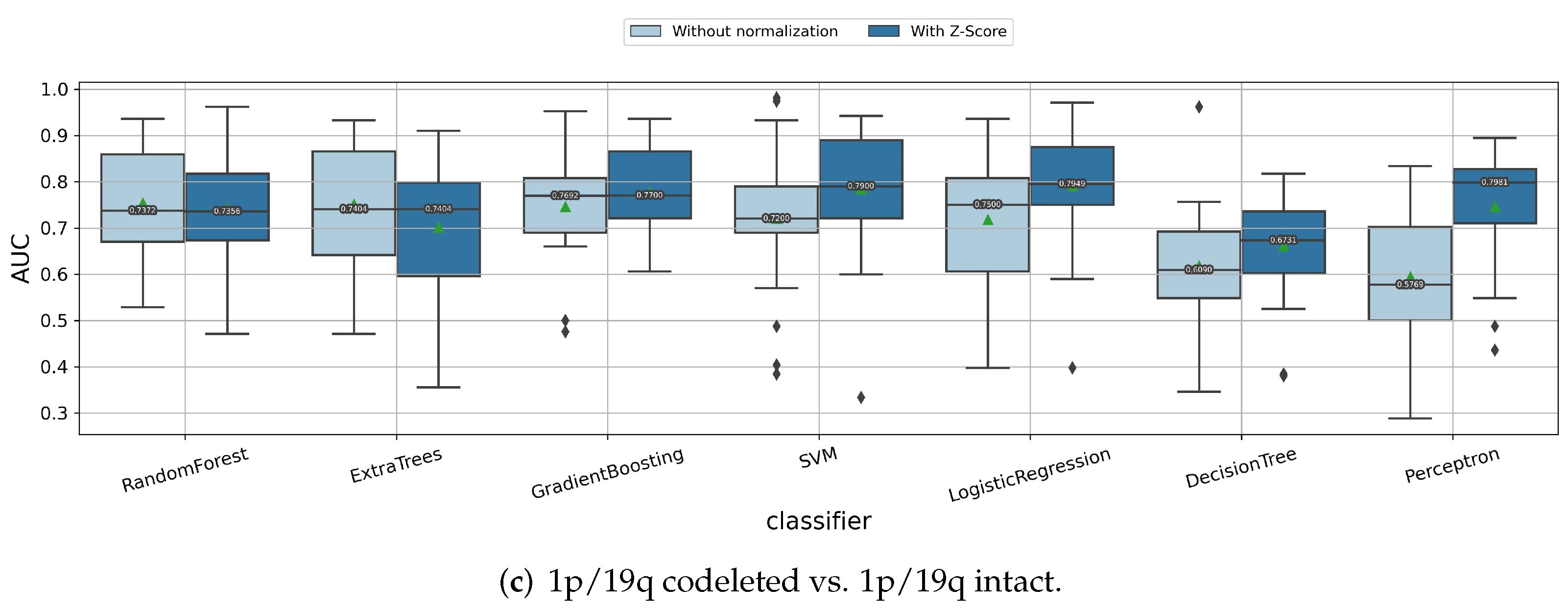
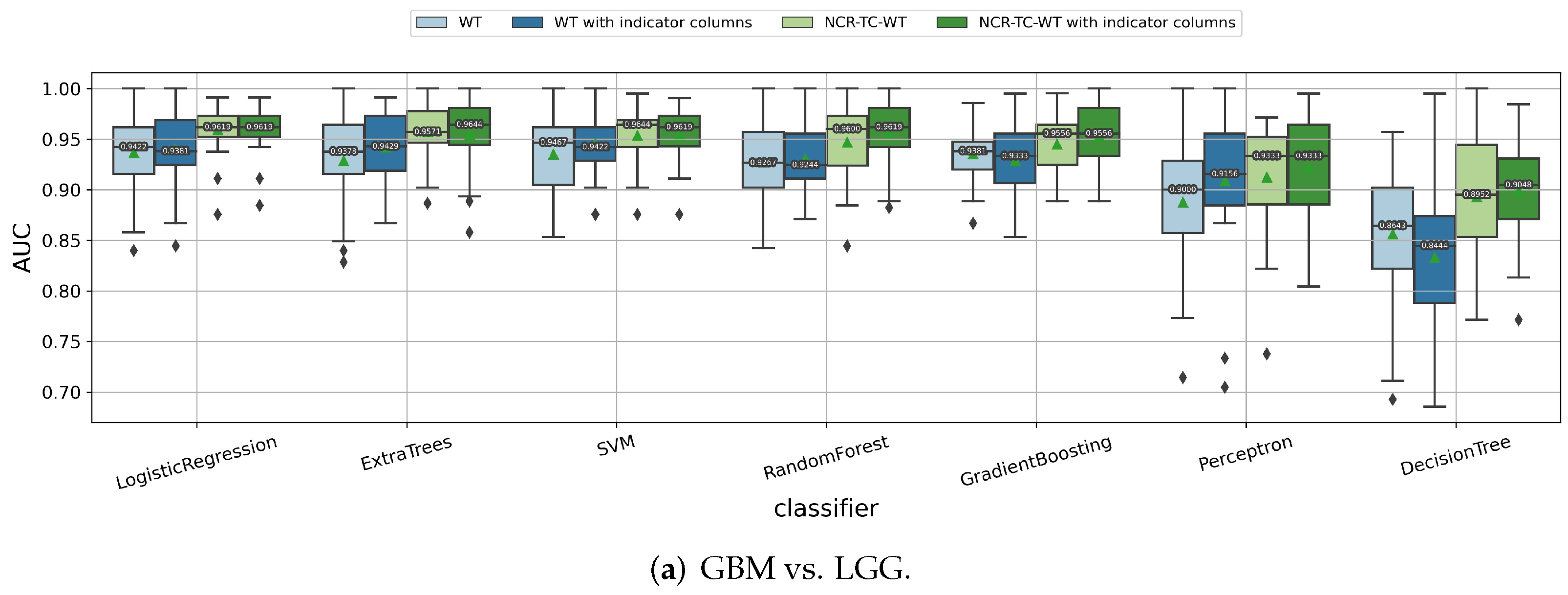
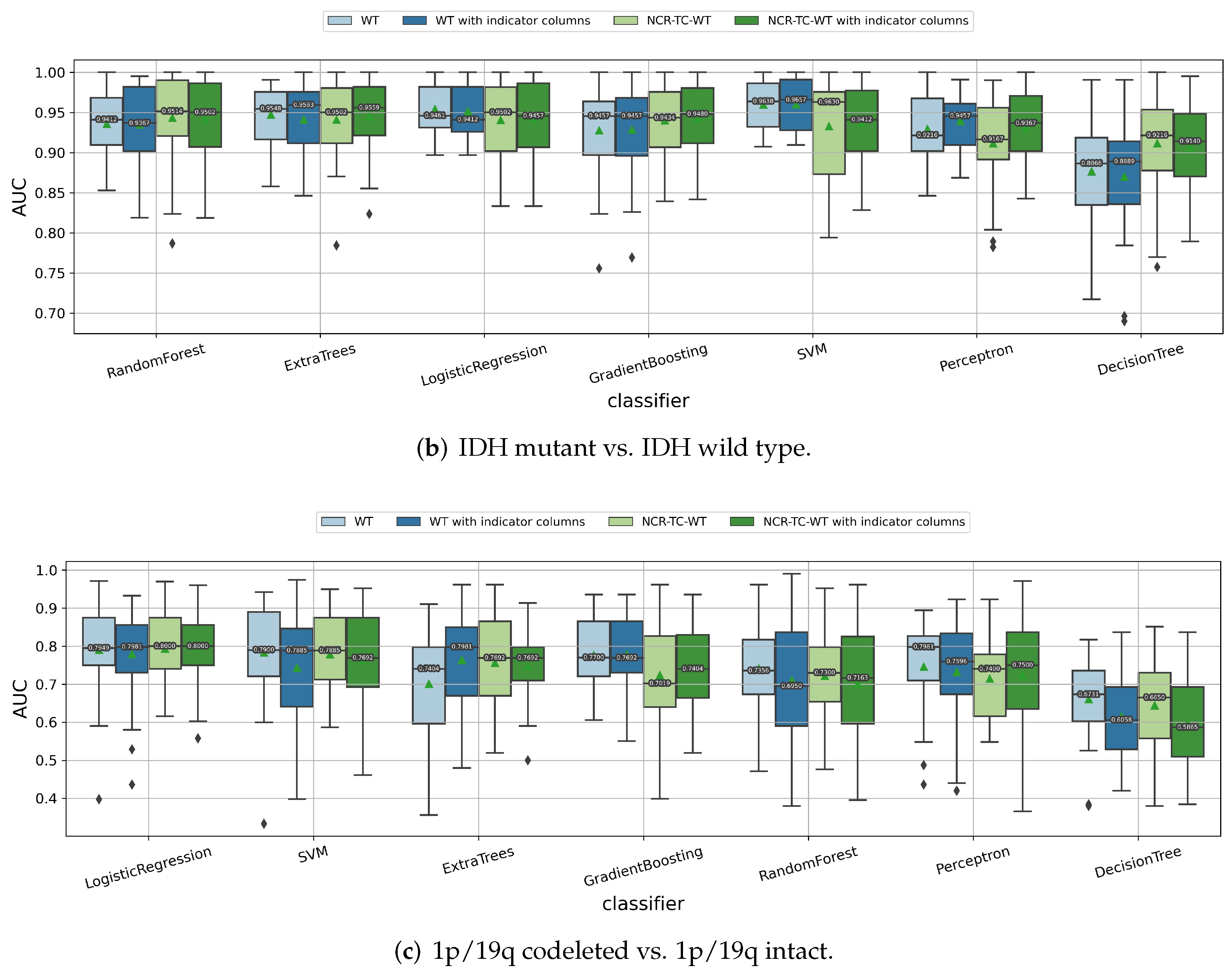
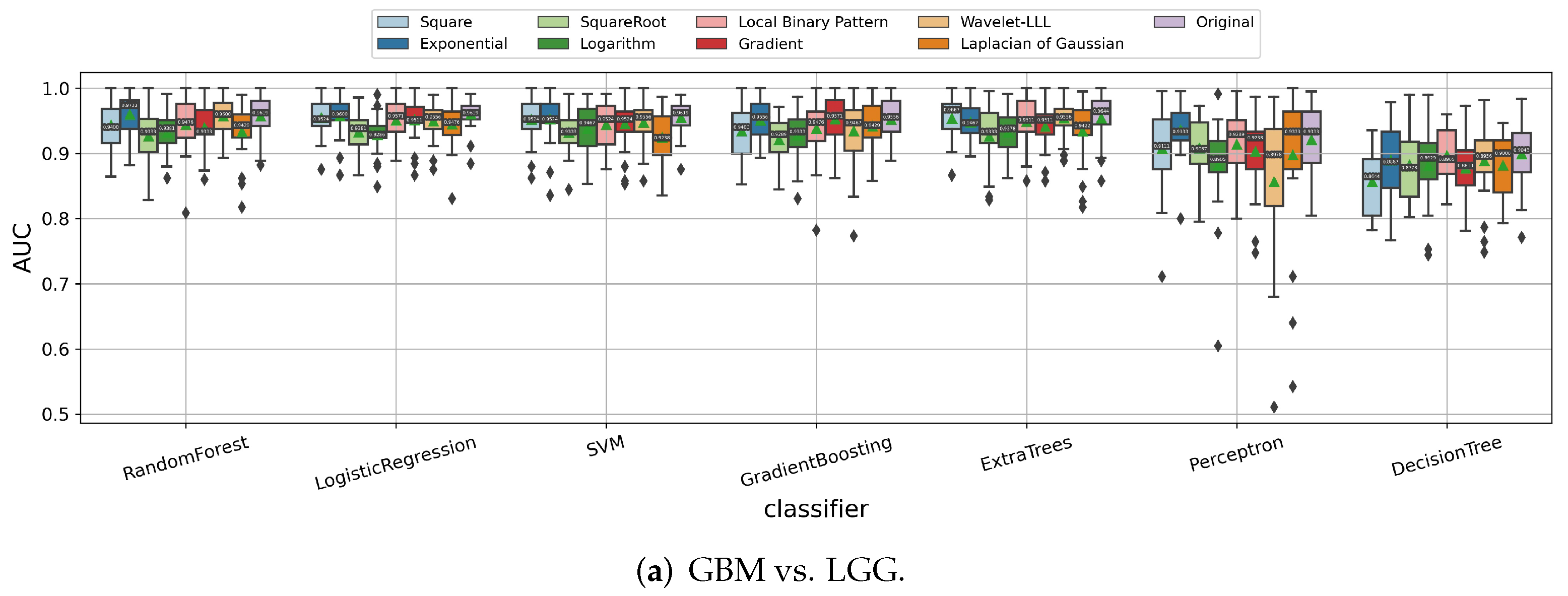


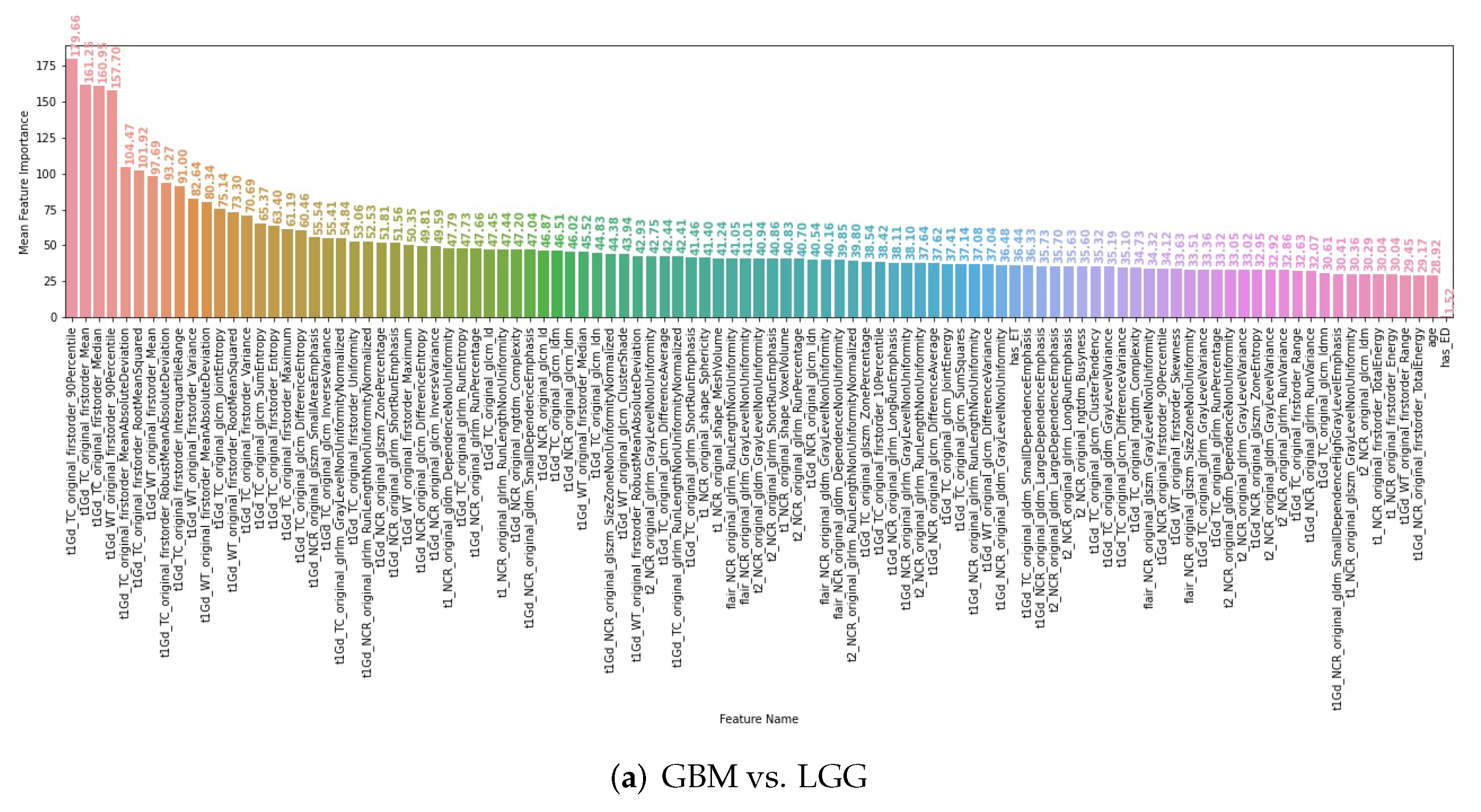
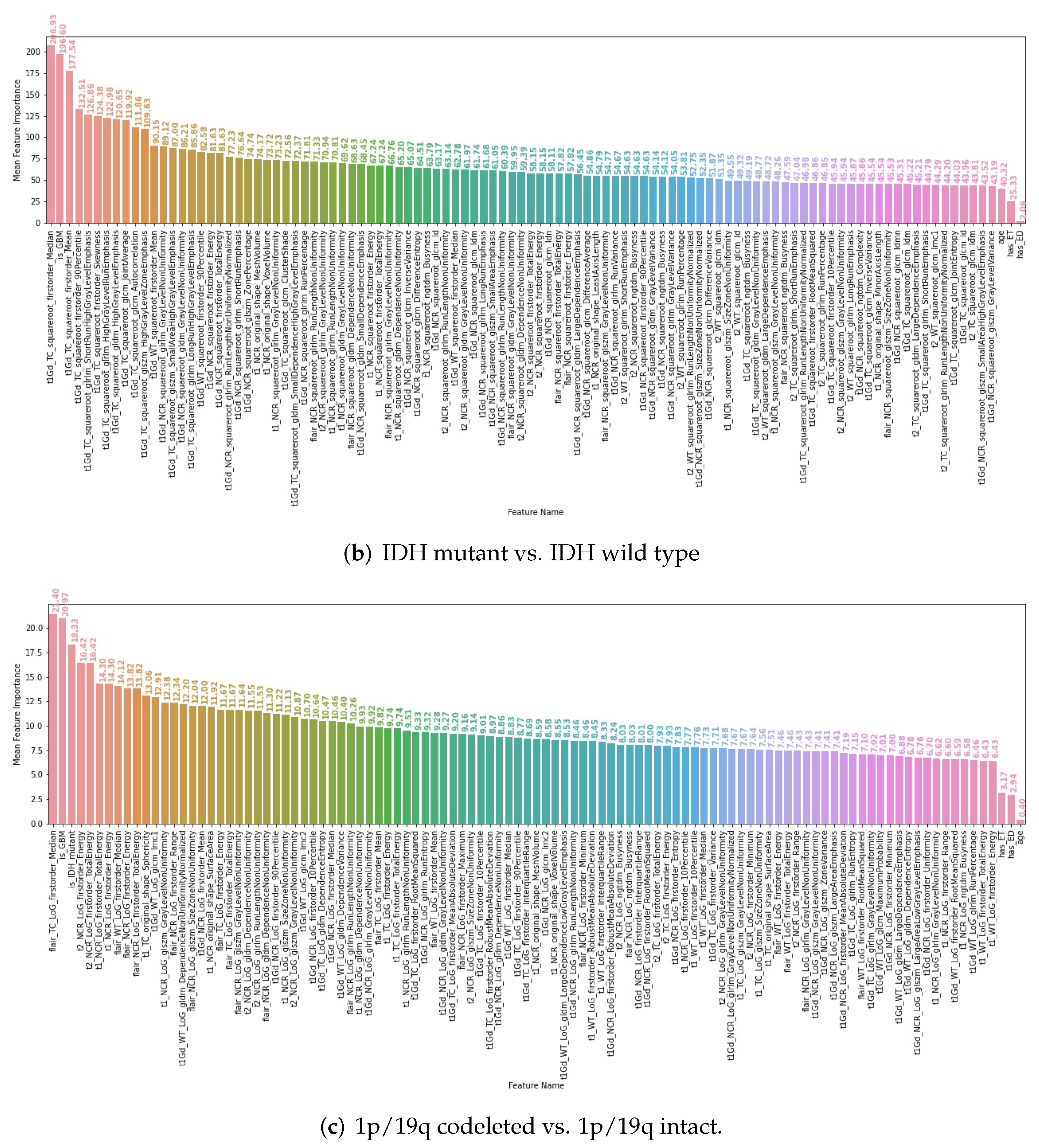
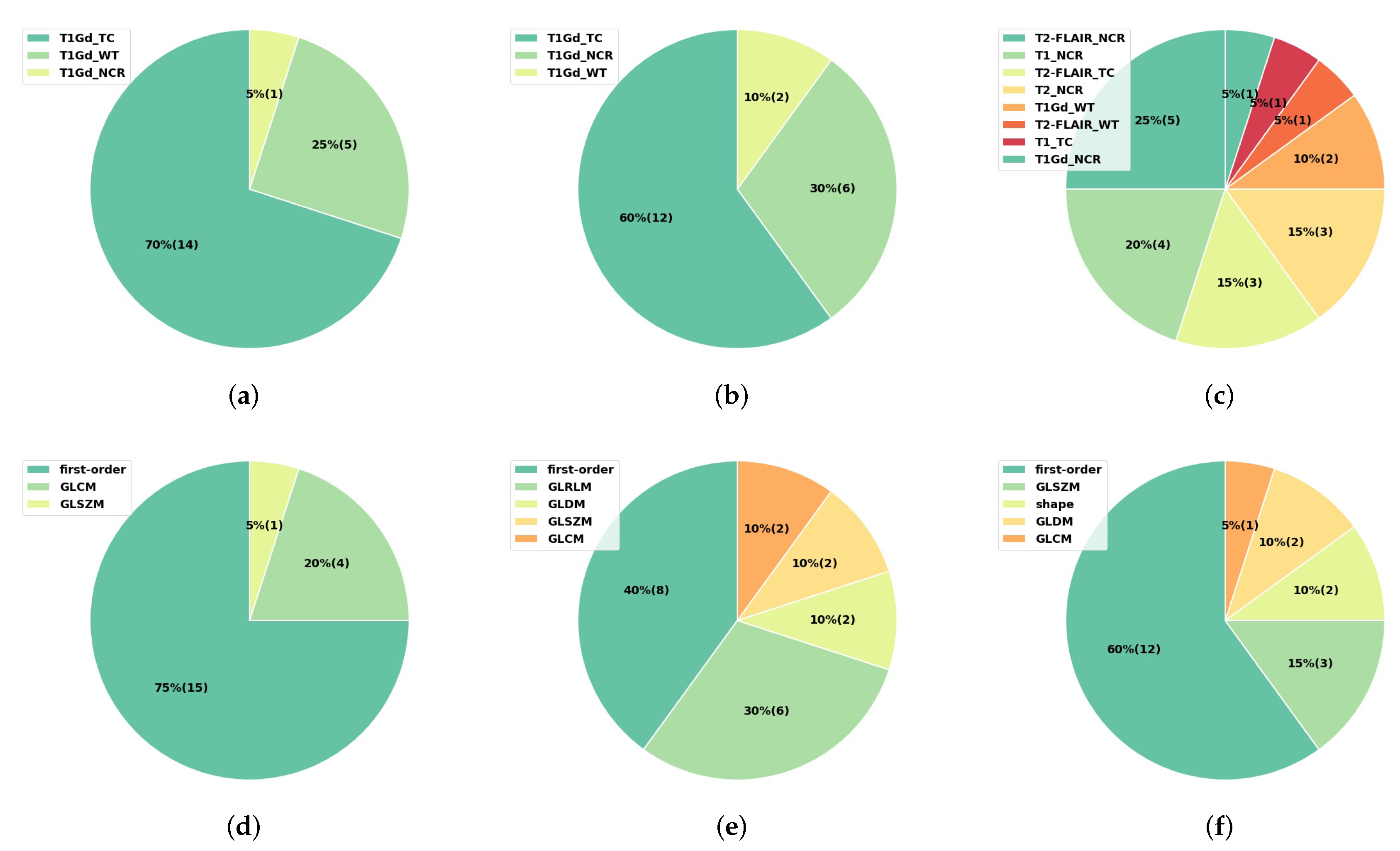
| IDH Mutant | IDH Wild Type | All | |||
|---|---|---|---|---|---|
| 1p/19q Codeletion | 1p/19q Intact | 1p/19q Codeletion | 1p/19q Intact | ||
| GBM | 0 | 4 | 0 | 103 | 107 |
| LGG | 27 | 57 | 0 | 21 | 105 |
| All | 27 | 61 | 0 | 124 | 212 |
| Classifier | Description | Tumor Grade | IDH Mutation Status | 1p/19q Codeletion Status | |||
|---|---|---|---|---|---|---|---|
| Mean CV AUC | Std CV AUC | Mean CV AUC | Std CV AUC | Mean CV AUC | Std CV AUC | ||
| SVM | Before tuning | 0.9039 [0.8991, 0.9087] | 0.0534 [0.0511, 0.0556] | 0.8769 [0.8703, 0.8834] | 0.0601 [0.0572, 0.0631] | 0.7048 [0.6906, 0.7190] | 0.1317 [0.1260, 0.1373] |
| After tuning | 0.9443 [0.9403, 0.9483] | 0.0372 [0.0348, 0.0395] | 0.9407 [0.9372, 0.9443] | 0.0398 [0.0376, 0.0421] | 0.8443 [0.8343, 0.8543] | 0.0945 [0.0887, 0.1003] | |
| Perceptron | Before tuning | 0.8723 [0.8632, 0.8813] | 0.0659 [0.0614, 0.0704] | 0.8328 [0.8256, 0.8400] | 0.0743 [0.0705, 0.0782] | 0.6370 [0.6198, 0.6541] | 0.1531 [0.1456, 0.1607] |
| After tuning | 0.9032 [0.8961, 0.9103] | 0.0686 [0.0605, 0.0768] | 0.9189 [0.9123, 0.9256] | 0.0587 [0.0517, 0.0658] | 0.7536 [0.7384, 0.7688] | 0.1450 [0.1358, 0.1543] | |
| Logistic Regression | Before tuning | 0.9048 [0.8994, 0.9102] | 0.0522 [0.0498, 0.0546] | 0.8732 [0.8666, 0.8799] | 0.0614 [0.0582, 0.0645] | 0.7176 [0.7036, 0.7317] | 0.1259 [0.1199, 0.1320] |
| After tuning | 0.9330 [0.9287, 0.9374] | 0.0421 [0.0397, 0.0445] | 0.9445 [0.9408, 0.9481] | 0.0379 [0.0356, 0.0403] | 0.8631 [0.8553, 0.8710] | 0.0831 [0.0783, 0.0879] | |
| Random Forest | Before tuning | 0.8758 [0.8690, 0.8825] | 0.0591 [0.0563, 0.0618] | 0.8437 [0.8361, 0.8513] | 0.0684 [0.0650, 0.0717] | 0.6924 [0.6770, 0.7077] | 0.1288 [0.1228, 0.1348] |
| After tuning | 0.9348 [0.9308, 0.9388] | 0.0401 [0.0380, 0.0421] | 0.9349 [0.9309, 0.9388] | 0.0431 [0.0408, 0.0454] | 0.8196 [0.8102, 0.8289] | 0.1000 [0.0956, 0.1044] | |
| Decision Tree | Before tuning | 0.7850 [0.7770, 0.7931] | 0.0794 [0.0761, 0.0826] | 0.7415 [0.7325, 0.7504] | 0.0835 [0.0801, 0.0869] | 0.5962 [0.5840, 0.6083] | 0.1285 [0.1232, 0.1339] |
| After tuning | 0.8745 [0.8682, 0.8807] | 0.0609 [0.0583, 0.0636] | 0.8828 [0.8764, 0.8892] | 0.0611 [0.0585, 0.0637] | 0.6991 [0.6842, 0.7139] | 0.1331 [0.1278, 0.1384] | |
| Extra Trees | Before tuning | 0.8739 [0.8668, 0.8811] | 0.0591 [0.0564, 0.0618] | 0.8438 [0.8357, 0.8520] | 0.0686 [0.0647, 0.0725] | 0.6901 [0.6747, 0.7055] | 0.1300 [0.1241, 0.1359] |
| After tuning | 0.9386 [0.9344, 0.9428] | 0.0403 [0.0383, 0.0423] | 0.9420 [0.9382, 0.9458] | 0.0393 [0.0371, 0.0415] | 0.8424 [0.8323, 0.8526] | 0.0879 [0.0824, 0.0934] | |
| Gradient Boosting | Before tuning | 0.8825 [0.8764, 0.8886] | 0.0572 [0.0547, 0.0598] | 0.8550 [0.8473, 0.8628] | 0.0655 [0.0618, 0.0691] | 0.6910 [0.6746, 0.7073] | 0.1310 [0.1250, 0.1369] |
| After tuning | 0.9316 [0.9277, 0.9355] | 0.0427 [0.0403, 0.0451] | 0.9354 [0.9312, 0.9396] | 0.0423 [0.0401, 0.0446] | 0.8151 [0.8062, 0.8240] | 0.1034 [0.0976, 0.1092] | |
| Tumor Grade | IDH Mutation Status | 1p/19q Codeletion Status | 5 Glioma Subtypes | ||||
|---|---|---|---|---|---|---|---|
| AUC | Accuracy | AUC | Accuracy | AUC | Accuracy | Accuracy | |
| Basic radiomics pipeline (Section 2.4) | 0.8935 (±0.0351) | 0.8171 (±0.0442) | 0.8676 (±0.0421) | 0.7840 (±0.0636) | 0.6473 (±0.1074) | 0.7209 (±0.2250) | 0.5772 (±0.0816) |
| Final radiomics pipeline (Section 3.2) | 0.9319 (±0.0386) | 0.8453 (±0.0691) | 0.9283 (±0.0333) | 0.8560 (±0.0333) | 0.8196 (±0.0702) | 0.8499 (±0.0351) | 0.6716 (±0.0655) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Ammari, S.; Lawrance, L.; Quillent, A.; Assi, T.; Lassau, N.; Chouzenoux, E. Radiomics-Based Method for Predicting the Glioma Subtype as Defined by Tumor Grade, IDH Mutation, and 1p/19q Codeletion. Cancers 2022, 14, 1778. https://doi.org/10.3390/cancers14071778
Li Y, Ammari S, Lawrance L, Quillent A, Assi T, Lassau N, Chouzenoux E. Radiomics-Based Method for Predicting the Glioma Subtype as Defined by Tumor Grade, IDH Mutation, and 1p/19q Codeletion. Cancers. 2022; 14(7):1778. https://doi.org/10.3390/cancers14071778
Chicago/Turabian StyleLi, Yingping, Samy Ammari, Littisha Lawrance, Arnaud Quillent, Tarek Assi, Nathalie Lassau, and Emilie Chouzenoux. 2022. "Radiomics-Based Method for Predicting the Glioma Subtype as Defined by Tumor Grade, IDH Mutation, and 1p/19q Codeletion" Cancers 14, no. 7: 1778. https://doi.org/10.3390/cancers14071778
APA StyleLi, Y., Ammari, S., Lawrance, L., Quillent, A., Assi, T., Lassau, N., & Chouzenoux, E. (2022). Radiomics-Based Method for Predicting the Glioma Subtype as Defined by Tumor Grade, IDH Mutation, and 1p/19q Codeletion. Cancers, 14(7), 1778. https://doi.org/10.3390/cancers14071778






