A Mixture of Endocrine Disrupting Chemicals Associated with Lower Birth Weight in Children Induces Adipogenesis and DNA Methylation Changes in Human Mesenchymal Stem Cells
Abstract
1. Introduction
2. Results
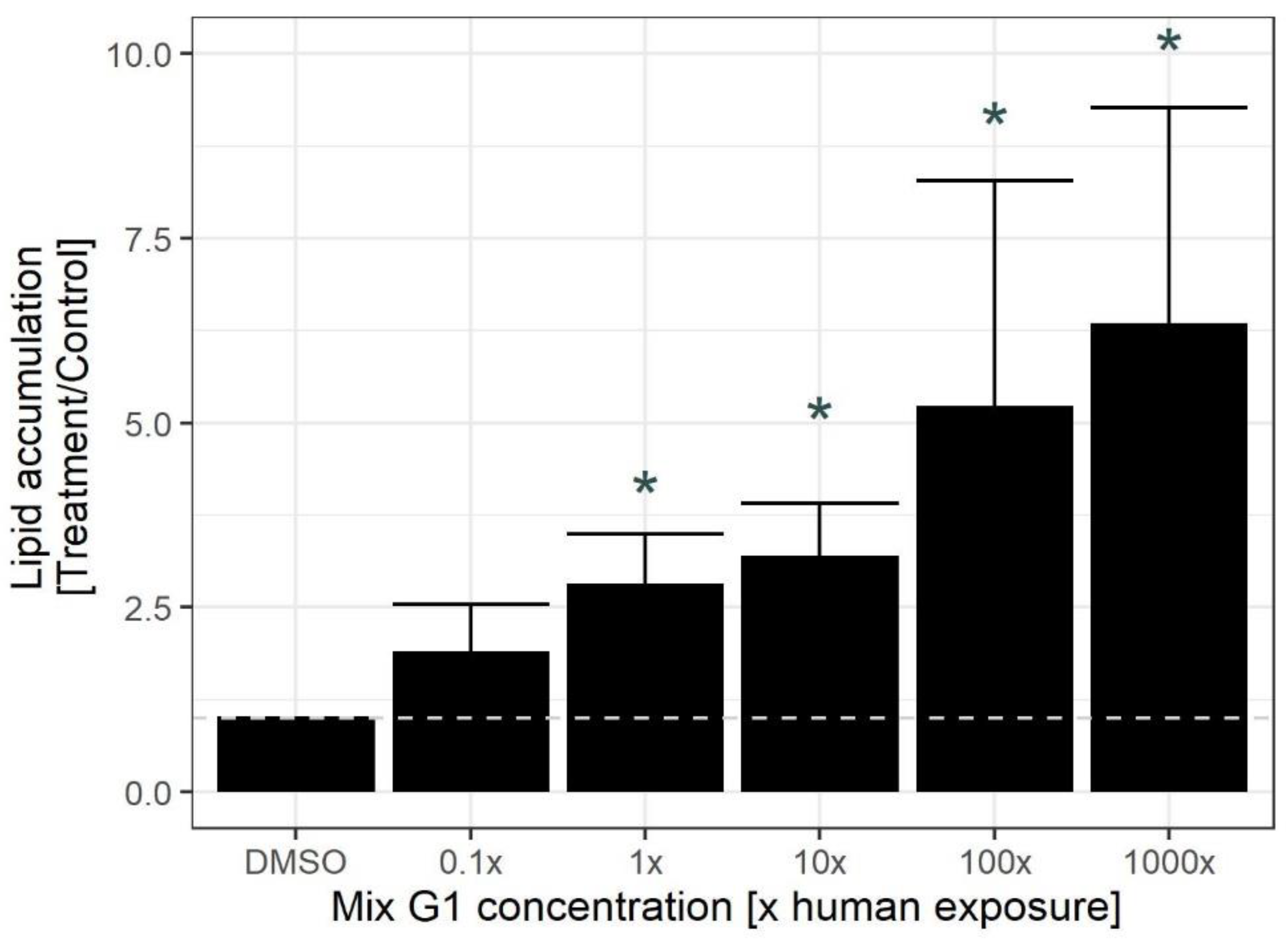
2.1. Exposure to Human Relevant Concentrations of Mix G1 Induces Adipogenesis in hMSCs
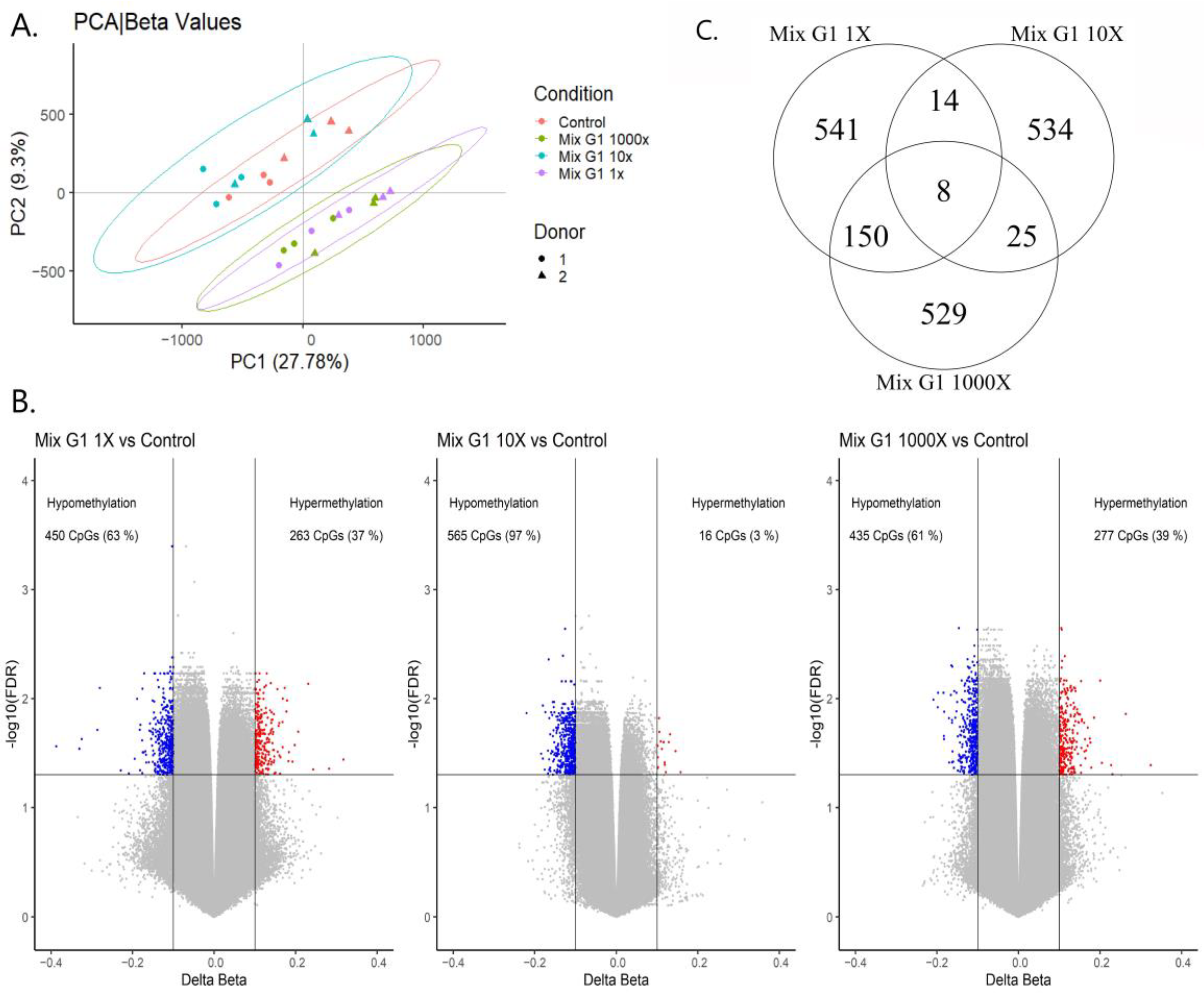
2.2. Exposure to Human Relevant Concentrations of Mix G1 Induces Altered DNA Methylation Profiles
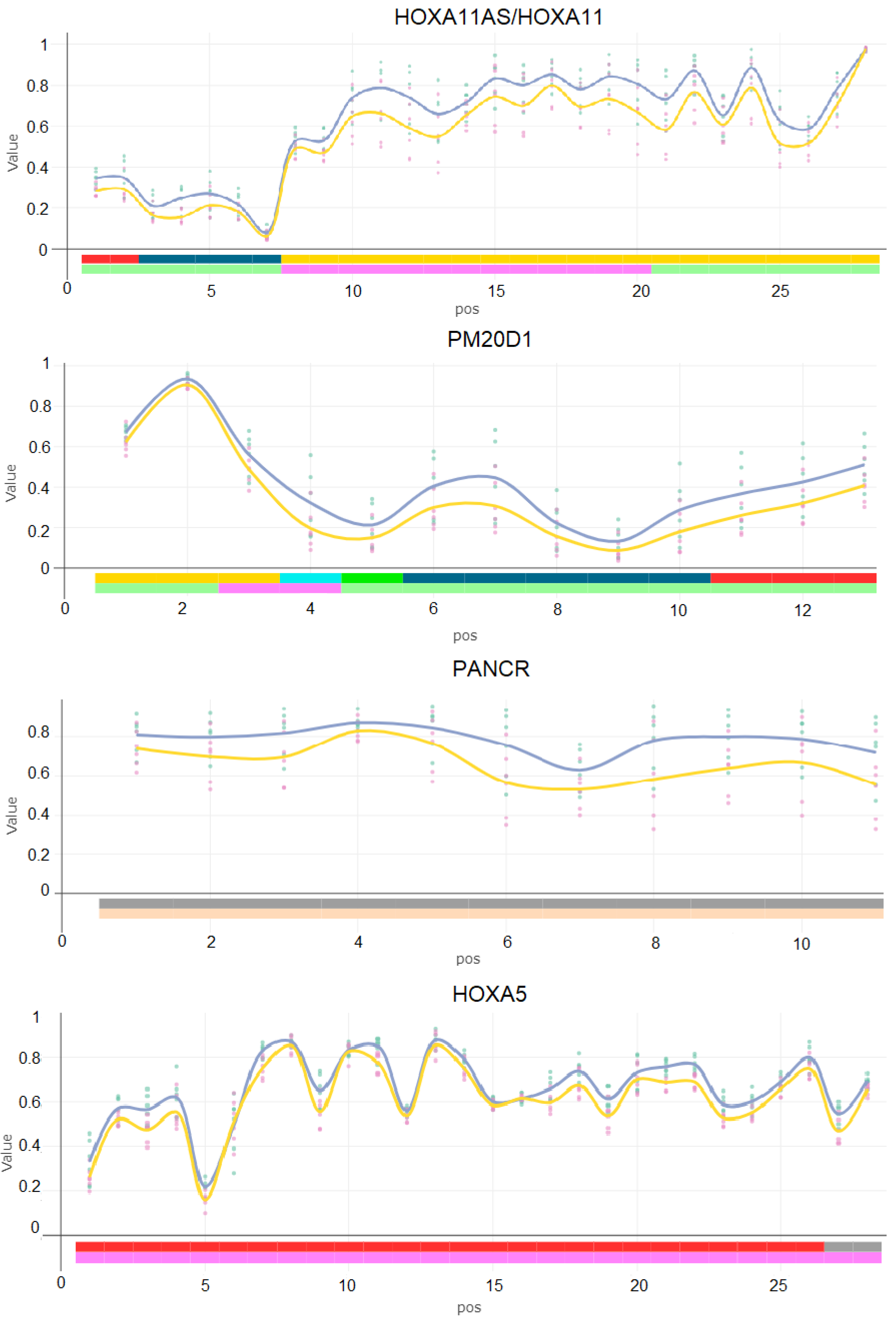
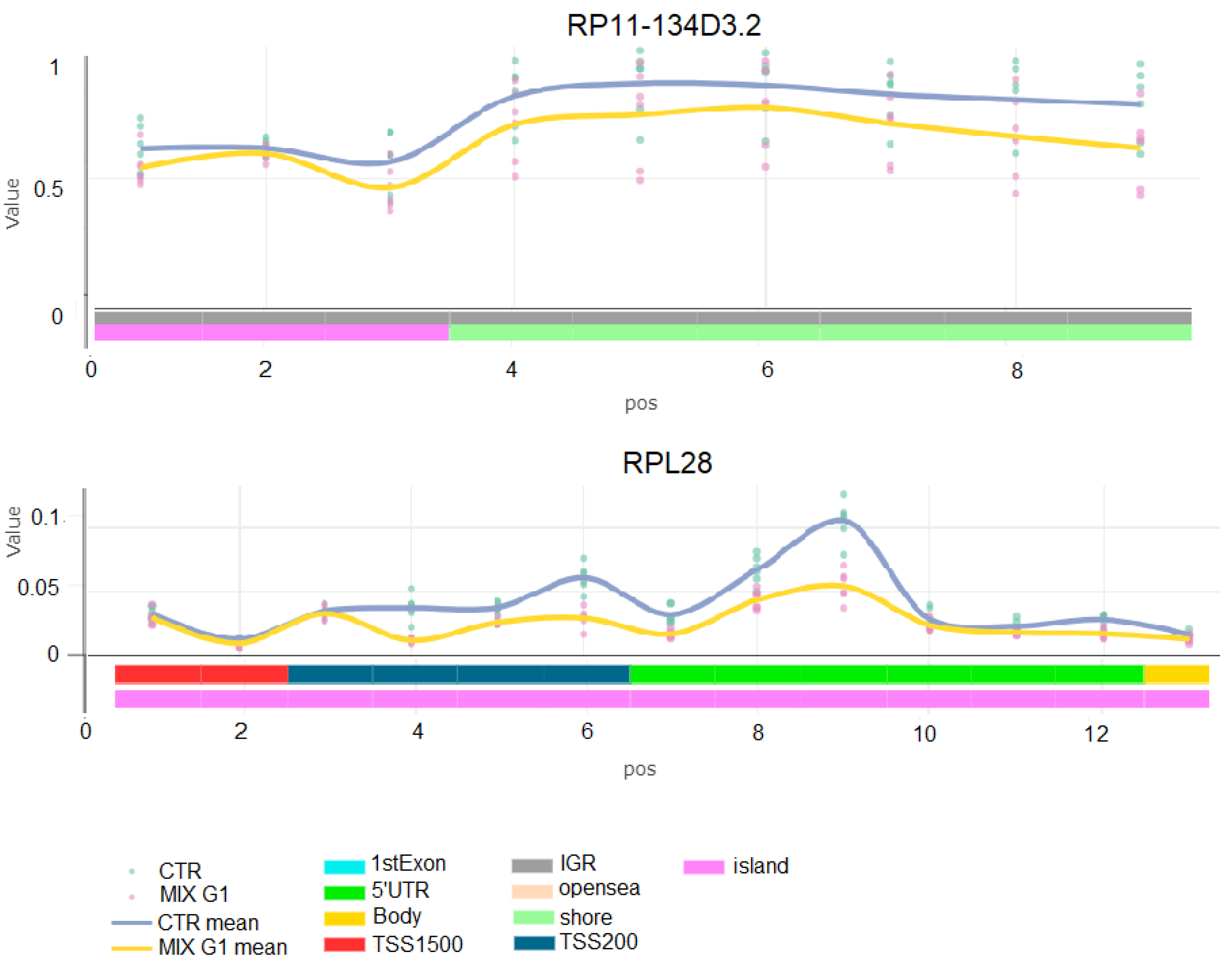
2.3. Differentially Methylated Regions Induced by Mix G1 Treatment Are Linked to Adipogenesis and Metabolic Functions
2.4. Mix G1-Induced DMRs Are Enriched at Genes Linked to Metabolism Related Pathways
3. Discussion
4. Materials and Methods
4.1. Identification and Preparation of Mix G1
4.2. Cell Culture
4.3. Lipid Droplet Accumulation
4.4. DNA Extraction and Genome-Wide DNA Methylation Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Diamanti-Kandarakis, E.; Bourguignon, J.-P.; Giudice, L.C.; Hauser, R.; Prins, G.S.; Soto, A.M.; Zoeller, R.T.; Gore, A.C. Endocrine-Disrupting Chemicals: An Endocrine Society Scientific Statement. Endocr. Rev. 2009, 30, 293–342. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, B.; Terekeci, H.; Sandal, S.; Kelestimur, F. Endocrine Disrupting Chemicals: Exposure, effects on human health, mechanism of action, models for testing and strategies for prevention. Rev. Endocr. Metab. Disord. 2020, 21, 127–147. [Google Scholar] [CrossRef] [PubMed]
- Woodruff, T.J.; Zota, A.R.; Schwartz, J.M. Environmental Chemicals in Pregnant Women in the United States: NHANES 2003–2004. Environ. Health Perspect. 2011, 119, 878–885. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Sarma, D.K.; Shubham, S.; Kumawat, M.; Verma, V.; Prakash, A.; Tiwari, R. Environmental Endocrine-Disrupting Chemical Exposure: Role in Non-Communicable Diseases. Front. Public Health 2020, 8, 549. [Google Scholar] [CrossRef] [PubMed]
- Engdahl, E.; Rüegg, J. Prenatal Exposure to Endocrine Di.isrupting Chemicals and Their Effect on Health Later in Life. In Beyond Our Genes: Pathophysiology of Gene and Environment Interaction and Epigenetic Inheritance; Teperino, R., Ed.; Springer International Publishing: Cham, Switzerland, 2020; pp. 53–77. ISBN 978-3-030-35213-4. [Google Scholar]
- Gancz, D.; Gilboa, L. Hormonal Control of Stem Cell Systems. Annu. Rev. Cell Dev. Biol. 2013, 29, 137–162. [Google Scholar] [CrossRef] [PubMed]
- Kopchick, J.J.; Berryman, D.E.; Puri, V.; Lee, K.Y.; Jorgensen, J.O.L. The effects of growth hormone on adipose tissue: Old observations, new mechanisms. Nat. Rev. Endocrinol. 2020, 16, 135–146. [Google Scholar] [CrossRef] [PubMed]
- de Sá, P.M.; Richard, A.J.; Hang, H.; Stephens, J.M. Transcriptional Regulation of Adipogenesis. In Comprehensive Physiology; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2017; pp. 635–674. ISBN 978-0-470-65071-4. [Google Scholar]
- Braun, J.M. Early-life exposure to EDCs: Role in childhood obesity and neurodevelopment. Nat. Rev. Endocrinol. 2017, 13, 161–173. [Google Scholar] [CrossRef] [PubMed]
- Cohen, I.C.; Cohenour, E.R.; Harnett, K.G.; Schuh, S.M. BPA, BPAF and TMBPF Alter Adipogenesis and Fat Accumulation in Human Mesenchymal Stem Cells, with Implications for Obesity. Int. J. Mol. Sci 2021, 22, 5363. [Google Scholar] [CrossRef]
- Hack, M.; Klein, N.K.; Taylor, H.G. Long-term developmental outcomes of low birth weight infants. Future Child. 1995, 5, 176–196. [Google Scholar] [CrossRef]
- McGuire, S.F. Understanding the Implications of Birth Weight. Nurs. Women’s Health 2017, 21, 45–49. [Google Scholar] [CrossRef][Green Version]
- Gluckman, P.D.; Hanson, M.A.; Cooper, C.; Thornburg, K.L. Effect of in utero and early-life conditions on adult health and disease. N. Engl. J. Med. 2008, 359, 61–73. [Google Scholar] [CrossRef]
- Barker, D.J.P.; Osmond, C. Infant mortality, childhood nutrition, and ischaemic heart disease in England and Wales. Lancet 1986, 327, 1077–1081. [Google Scholar] [CrossRef]
- Roberts, G.; Cheong, J.L.Y. Long-term growth and general health for the tiniest or most immature infants. Semin. Fetal Neonatal Med. 2014, 19, 118–124. [Google Scholar] [CrossRef] [PubMed]
- Barker, D.J.P. The origins of the developmental origins theory. J. Intern. Med. 2007, 261, 412–417. [Google Scholar] [CrossRef] [PubMed]
- Kopec, G.; Shekhawat, P.S.; Mhanna, M.J. Prevalence of diabetes and obesity in association with prematurity and growth restriction. DMSO 2017, 10, 285–295. [Google Scholar] [CrossRef]
- Wikström, S.; Lin, P.-I.; Lindh, C.H.; Shu, H.; Bornehag, C.-G. Maternal serum levels of perfluoroalkyl substances in early pregnancy and offspring birth weight. Pediatr. Res. 2020, 87, 1093–1099. [Google Scholar] [CrossRef]
- Marks, K.J.; Cutler, A.J.; Jeddy, Z.; Northstone, K.; Kato, K.; Hartman, T.J. Maternal serum concentrations of perfluoroalkyl substances and birth size in British boys. Int. J. Hyg. Environ. Health 2019, 222, 889–895. [Google Scholar] [CrossRef]
- Gyllenhammar, I.; Diderholm, B.; Gustafsson, J.; Berger, U.; Ridefelt, P.; Benskin, J.P.; Lignell, S.; Lampa, E.; Glynn, A. Perfluoroalkyl acid levels in first-time mothers in relation to offspring weight gain and growth. Environ. Int. 2018, 111, 191–199. [Google Scholar] [CrossRef]
- Lenters, V.; Portengen, L.; Rignell-Hydbom, A.; Jönsson, B.A.G.; Lindh, C.H.; Piersma, A.H.; Toft, G.; Bonde, J.P.; Heederik, D.; Rylander, L.; et al. Prenatal Phthalate, Perfluoroalkyl Acid, and Organochlorine Exposures and Term Birth Weight in Three Birth Cohorts: Multi-Pollutant Models Based on Elastic Net Regression. Environ. Health Perspect. 2016, 124, 365–372. [Google Scholar] [CrossRef]
- Birks, L.; Casas, M.; Garcia, A.M.; Alexander, J.; Barros, H.; Bergström, A.; Bonde, J.P.; Burdorf, A.; Costet, N.; Danileviciute, A.; et al. Occupational Exposure to Endocrine-Disrupting Chemicals and Birth Weight and Length of Gestation: A European Meta-Analysis. Environ. Health Perspect. 2016, 124, 1785–1793. [Google Scholar] [CrossRef]
- Ghassabian, A.; Vandenberg, L.; Kannan, K.; Trasande, L. Endocrine-Disrupting Chemicals and Child Health. Annu. Rev. Pharmacol. Toxicol. 2021, 22, 573–594. [Google Scholar] [CrossRef] [PubMed]
- Starling, A.P.; Adgate, J.L.; Hamman, R.F.; Kechris, K.; Calafat, A.M.; Dabelea, D. Prenatal exposure to per- and polyfluoroalkyl substances and infant growth and adiposity: The Healthy Start Study. Environ. Int. 2019, 131, 104983. [Google Scholar] [CrossRef] [PubMed]
- Halldorsson, T.I.; Rytter, D.; Haug, L.S.; Bech, B.H.; Danielsen, I.; Becher, G.; Henriksen, T.B.; Olsen, S.F. Prenatal Exposure to Perfluorooctanoate and Risk of Overweight at 20 Years of Age: A Prospective Cohort Study. Environ. Health Perspect. 2012, 120, 668–673. [Google Scholar] [CrossRef] [PubMed]
- Braun, J.M.; Chen, A.; Romano, M.E.; Calafat, A.M.; Webster, G.M.; Yolton, K.; Lanphear, B.P. Prenatal Perfluoroalkyl Substance Exposure and Child Adiposity at 8 Years of Age: The HOME Study. Obesity 2016, 24, 231–237. [Google Scholar] [CrossRef]
- Mora, A.M.; Oken, E.; Rifas-Shiman, S.L.; Webster, T.F.; Gillman, M.W.; Calafat, A.M.; Ye, X.; Sagiv, S.K. Prenatal Exposure to Perfluoroalkyl Substances and Adiposity in Early and Mid-Childhood. Environ. Health Perspect. 2017, 125, 467–473. [Google Scholar] [CrossRef]
- Howard, S.G. Developmental Exposure to Endocrine Disrupting Chemicals and Type 1 Diabetes Mellitus. Front. Endocrinol. 2018, 9, 513. [Google Scholar] [CrossRef]
- Blake, B.E.; Fenton, S.E. Early life exposure to per- and polyfluoroalkyl substances (PFAS) and latent health outcomes: A review including the placenta as a target tissue and possible driver of peri- and postnatal effects. Toxicology 2020, 443, 152565. [Google Scholar] [CrossRef]
- Heindel, J.J.; Blumberg, B.; Cave, M.; Machtinger, R.; Mantovani, A.; Mendez, M.A.; Nadal, A.; Palanza, P.; Panzica, G.; Sargis, R.; et al. Metabolism Disrupting Chemicals and Metabolic Disorders. Reprod. Toxicol. 2017, 68, 3–33. [Google Scholar] [CrossRef]
- Papalou, O.; Kandaraki, E.A.; Papadakis, G.; Diamanti-Kandarakis, E. Endocrine Disrupting Chemicals: An Occult Mediator of Metabolic Disease. Front. Endocrinol. 2019, 10, 112. [Google Scholar] [CrossRef]
- Wu, H.; Sun, Y.E. Epigenetic Regulation of Stem Cell Differentiation. Pediatr. Res. 2006, 59, 21–25. [Google Scholar] [CrossRef]
- Atlasi, Y.; Stunnenberg, H.G. The interplay of epigenetic.c marks during stem cell differentiation and development. Nat. Rev. Genet. 2017, 18, 643–658. [Google Scholar] [CrossRef] [PubMed]
- Kiefer, J.C. Epigenetics in development. Dev. Dyn. 2007, 236, 1144–1156. [Google Scholar] [CrossRef] [PubMed]
- Nicoglou, A.; Merlin, F. Epigenetics: A way to bridge the gap between biological fields. Stud. Hist. Philos. Biol. Biomed. Sci. 2017, 66, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, M.N.; Marczylo, E.L.; Guerrero-Bosagna, C.; Rüegg, J. Marked for Life: Epigenetic Effects of Endocrine Disrupting Chemicals. Annu. Rev. Environ. Resour. 2017, 42, 105–160. [Google Scholar] [CrossRef]
- Zhu, Z.; Cao, F.; Li, X. Epigenetic Programming and Fetal Metabolic Programming. Front. Endocrinol. 2019, 10, 764. [Google Scholar] [CrossRef]
- Moore, L.D.; Le, T.; Fan, G. DNA Methylation and Its Basic Function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef]
- Vilahur, N.; Bustamante, M.; Morales, E.; Motta, V.; Fernandez, M.F.; Salas, L.A.; Escaramis, G.; Ballester, F.; Murcia, M.; Tardon, A.; et al. Prenatal exposure to mixtures of xenoestrogens and genome-wide DNA methylation in human placenta. Epigenomics 2016, 8, 43–54. [Google Scholar] [CrossRef]
- Wadhwa, P.D.; Buss, C.; Entringer, S.; Swanson, J.M. Developmental Origins of Health and Disease: Brief History of the Approach and Current Focus on Epigenetic Mechanisms. Semin. Reprod. Med. 2009, 27, 358–368. [Google Scholar] [CrossRef]
- Kortenkamp, A. Low dose mixture effects of endocrine disrupters and their implications for regulatory thresholds in chemical risk assessment. Curr. Opin. Pharmacol. 2014, 19, 105–111. [Google Scholar] [CrossRef]
- Hernández, A.F.; Tsatsakis, A.M. Human exposure to chemical mixtures: Challenges for the integration of toxicology with epidemiology data in risk assessment. Food Chem. Toxicol. 2017, 103, 188–193. [Google Scholar] [CrossRef]
- Ribeiro, E.; Ladeira, C.; Viegas, S. EDCs Mixtures: A Stealthy Hazard for Human Health? Toxics 2017, 5, 5. [Google Scholar] [CrossRef] [PubMed]
- Bornehag, C.-G.; Kitraki, E.; Stamatakis, A.; Panagiotidou, E.; Rudén, C.; Shu, H.; Lindh, C.; Ruegg, J.; Gennings, C. A Novel Approach to Chemical Mixture Risk Assessment—Linking Data from Population-Based Epidemiology and Experimental Animal Tests. Risk Anal. 2019, 39, 2259–2271. [Google Scholar] [CrossRef] [PubMed]
- Mentor, A.; Brunström, B.; Mattsson, A.; Jönsson, M. Developmental exposure to a human relevant mixture of endocrine disruptors alters metabolism and adipogenesis in zebrafish (Danio rerio). Chemosphere 2020, 238, 124584. [Google Scholar] [CrossRef] [PubMed]
- Repouskou, A.; Panagiotidou, E.; Panagopoulou, L.; Bisting, P.L.; Tuck, A.R.; Sjödin, M.O.D.; Lindberg, J.; Bozas, E.; Rüegg, J.; Gennings, C.; et al. Gestational exposure to an epidemiologically defined mixture of phthalates leads to gonadal dysfunction in mouse offspring of both sexes. Sci. Rep. 2019, 9, 6424. [Google Scholar] [CrossRef]
- Caporale, N.; Leemans, M.; Birgersson, L.; Germain, P.-L.; Cheroni, C.; Borbély, G.; Engdahl, E.; Lindh, C.; Bressan, R.B.; Cavallo, F.; et al. From cohorts to molecules: Adverse impacts of endocrine disrupting mixtures. Science 2022, 375, 6852. [Google Scholar] [CrossRef]
- Poissonnet, C.M.; Burdi, A.R.; Bookstein, F.L. Growth and development of human adipose tissue during early gestation. Early Hum. Dev. 1983, 8, 1–11. [Google Scholar] [CrossRef]
- Orsso, C.E.; Colin-Ramirez, E.; Field, C.J.; Madsen, K.L.; Prado, C.M.; Haqq, A.M. Adipose Tissue Development and Expansion from the Womb to Adolescence: An Overview. Nutrients 2020, 12, 2735. [Google Scholar] [CrossRef]
- Chen, Y.-S.; Wu, R.; Yang, X.; Kou, S.; MacDougald, O.A.; Yu, L.; Shi, H.; Xue, B. Inhibiting DNA methylation switches adipogenesis to osteoblastogenesis by activating Wnt10a. Sci. Rep. 2016, 6, 25283. [Google Scholar] [CrossRef]
- Shamsi, F.; Xue, R.; Huang, T.L.; Lundh, M.; Liu, Y.; Leiria, L.O.; Lynes, M.D.; Kempf, E.; Wang, C.-H.; Sugimoto, S.; et al. FGF6 and FGF9 regulate UCP1 expression independent of brown adipogenesis. Nat. Commun. 2020, 11, 1421. [Google Scholar] [CrossRef]
- Nozato, Y.; Takami, Y.; Yamamoto, K.; Nagasawa, M.; Nozato, S.; Imaizumi, Y.; Takeshita, H.; Wang, C.; Ito, Y.; Takeda, S.; et al. Novel properties of myoferlin in glucose metabolism via pathways involving modulation of adipose functions. FASEB J. 2020, 34, 2792–2811. [Google Scholar] [CrossRef]
- Chen, N.; Wang, J. Wnt/β-Catenin Signaling and Obesity. Front. Physiol. 2018, 9, 792. [Google Scholar] [CrossRef] [PubMed]
- Ellis, J.M.; Bowman, C.E.; Wolfgang, M.J. Metabolic and Tissue-Specific Regulation of Acyl-CoA Metabolism. PLoS ONE 2015, 10, e0116587. [Google Scholar] [CrossRef] [PubMed]
- Płatek, T.; Polus, A.; Góralska, J.; Raźny, U.; Gruca, A.; Kieć-Wilk, B.; Zabielski, P.; Kapusta, M.; Słowińska-Solnica, K.; Solnica, B.; et al. DNA methylation microarrays identify epigenetically regulated lipid related genes in obese patients with hypercholesterolemia. Mol. Med. 2020, 26, 93. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Li, W.-D.; Zhang, C.K.; Wang, Z.; Glessner, J.T.; Grant, S.F.A.; Zhao, H.; Hakonarson, H.; Price, R.A. A Genome-Wide Association Study on Obesity and Obesity-Related Traits. PLoS ONE 2011, 6, e18939. [Google Scholar] [CrossRef]
- Alvine, T.; Dhasarathy, A.; Bundy, A.; Bhattacharya, A.; Darland, D.; Hur, J.; Perley, D.; Johnson, L.; Rusten, M.; Roemmich, J.; et al. RBMS1 Methylation and mRNA Expression Are Differentially Regulated in Placenta Tissue from Obese Women (P11-131-19). Curr. Dev. Nutr. 2019, 3, nzz048.P11-131-19. [Google Scholar] [CrossRef]
- Bradley, D.; Yin, Z.; Liu, J.Z.; Blaszczak, A.M.; Wong, S.T.; Hsueh, W. Adipocyte EGFL6 Expression from Subcutaneous Adipose Tissue Alters Glucose Homeostasis and Affects Human Obesity. Diabetes 2018, 67, 1751. [Google Scholar] [CrossRef]
- Klenke, S.; Tan, S.; Hahn, S.; Mann, K.; Hauner, H.; Manthey, I.; Peters, J.; Siffert, W.; Frey, U.H. A functional GNAQ promoter haplotype is associated with altered Gq expression and with insulin resistance and obesity in women with polycystic ovary syndrome. Pharm. Genom. 2010, 20, 476–484. [Google Scholar] [CrossRef]
- Krüger, J.; Brachs, S.; Trappiel, M.; Kintscher, U.; Meyborg, H.; Wellnhofer, E.; Thöne-Reineke, C.; Stawowy, P.; Östman, A.; Birkenfeld, A.L.; et al. Enhanced insulin signaling in den.nsity-enhanced phosphatase-1 (DEP-1) knockout mice. Mol. Metab. 2015, 4, 325–336. [Google Scholar] [CrossRef]
- Liu, L.; Chen, L.; Li, Z.; Li, L.; Wang, M.; Qu, J.; Xue, J. Association of genetic variants in TOMM7 gene and gene environment interaction with type 2 diabetes in Chinese Dong population. Zhong Nan Da Xue Xue Bao Yi Xue Ban 2015, 40, 31–38. [Google Scholar] [CrossRef]
- Iván, J.; Major, E.; Sipos, A.; Kovács, K.; Horváth, D.; Tamás, I.; Bay, P.; Dombrádi, V.; Lontay, B. The Short-Chain Fatty Acid Propionate Inhibits Adipogenic Differentiation of Human Chorion-Derived Mesenchymal Stem Cells Through the Free Fatty Acid Receptor 2. Stem. Cells Dev. 2017, 26, 1724–1733. [Google Scholar] [CrossRef]
- Miki, H.; Yamauchi, T.; Suzuki, R.; Komeda, K.; Tsuchida, A.; Kubota, N.; Terauchi, Y.; Kamon, J.; Kaburagi, Y.; Matsui, J.; et al. Essential Role of Insulin Receptor Substrate 1 (IRS-1) and IRS-2 in Adipocyte Differentiation. Mol. Cell Biol. 2001, 21, 2521–2532. [Google Scholar] [CrossRef] [PubMed]
- Serria, M.S.; Ikeda, H.; Omoteyama, K.; Hirokawa, J.; Nishi, S.; Sakai, M. Regulation and differential expression of the c-maf gene in differentiating cultured cells. Biochem. Biophys. Res. Commun. 2003, 310, 318–326. [Google Scholar] [CrossRef] [PubMed]
- Voermans, N.C.; Preisler, N.; Madsen, K.L.; Janssen, M.C.H.; Kusters, B.; Abu Bakar, N.; Conte, F.; Lamberti, V.M.L.; Nusman, F.; van Engelen, B.G.; et al. PGM1 deficiency: Substrate use during exercise and effect of treatment with galactose. Neuromuscul. Disord. 2017, 27, 370–376. [Google Scholar] [CrossRef] [PubMed]
- Haller, R.; Heinicke, K.; Newby, M.; Wyrick, P.; Dimitrov, I.; Mancias, P.; Cohen, J. Phosphoglucomutase (PGM 1) Deficiency: A Novel Defect of Muscle Glycogen Degradation and Synthesis (P07.200). Neurology 2012, 78, P07.200. [Google Scholar] [CrossRef]
- Nuermaimaiti, N.; Liu, J.; Liang, X.; Jiao, Y.; Zhang, D.; Liu, L.; Meng, X.; Guan, Y. Effect of lncRNA HOXA11-AS1 on adipocyte differentiation in human adipose-derived stem cells. Biochem. Biophys Res. Commun. 2018, 495, 1878–1884. [Google Scholar] [CrossRef]
- Cao, W.; Xu, Y.; Luo, D.; Saeed, M.; Sun, C. Hoxa5 Promotes Adipose Differentiation via Increasing DNA Methylation Level and Inhibiting PKA/HSL Signal Pathway in Mice. CPB 2018, 45, 1023–1033. [Google Scholar] [CrossRef]
- Benson, K.K.; Hu, W.; Weller, A.H.; Bennett, A.H.; Chen, E.R.; Khetarpal, S.A.; Yoshino, S.; Bone, W.P.; Wang, L.; Rabinowitz, J.D.; et al. Natural human genetic variation determines basal and inducible expression of PM20D1, an obesity-associated gene. Proc. Natl. Acad. Sci. USA 2019, 116, 23232–23242. [Google Scholar] [CrossRef]
- Pant, R.; Firmal, P.; Shah, V.K.; Alam, A.; Chattopadhyay, S. Epigenetic Regulation of Adipogenesis in Development of Metabolic Syndrome. Front. Cell Dev. Biol. 2021, 8, 1766. [Google Scholar] [CrossRef]
- Henegar, C.; Tordjman, J.; Achard, V.; Lacasa, D.; Cremer, I.; Guerre-Millo, M.; Poitou, C.; Basdevant, A.; Stich, V.; Viguerie, N.; et al. Adipose tissue transcriptomic signature highlights the pathological relevance of extracellular matrix in human obesity. Genome. Biol. 2008, 9, R14. [Google Scholar] [CrossRef]
- Kim, J.; Sun, Q.; Yue, Y.; Yoon, K.S.; Whang, K.-Y.; Marshall Clark, J.; Park, Y. 4,4′-Dichlorodiphenyltrichloroethane (DDT) and 4,4′-dichlorodiphenyldichloroethylene (DDE) promote adipogenesis in 3T3-L1 adipocyte cell culture. Pestic. Biochem. Physiol. 2016, 131, 40–45. [Google Scholar] [CrossRef]
- Pesta, M.; Cedikova, M.; Dvorak, P.; Dvorakova, J.; Kulda, V.; Srbecka, K.; Muller, L.; Bouchalova, V.; Kralickova, M.; Babuska, V.; et al. Trends in gene expression changes during adipogenesis in human adipose derived mesenchymal stem cells under dichlorodiphenyldichloroethylene exposure. Mol. Cell. Toxicol. 2018, 14, 369–379. [Google Scholar] [CrossRef]
- Watt, J.; Schlezinger, J.J. Structurally-diverse, PPARγ-activating environmental toxicants induce adipogenesis and suppress osteogenesis in bone marrow mesenchymal stromal cells. Toxicology 2015, 331, 66–77. [Google Scholar] [CrossRef] [PubMed]
- Feige, J.N.; Gelman, L.; Rossi, D.; Zoete, V.; Métivier, R.; Tudor, C.; Anghel, S.I.; Grosdidier, A.; Lathion, C.; Engelborghs, Y.; et al. The Endocrine Disruptor Monoethyl-hexyl-phthalate Is a Selective Peroxisome Proliferator-activated Receptor γ Modulator That Promotes Adipogenesis. J. Biol. Chem. 2007, 282, 19152–19166. [Google Scholar] [CrossRef] [PubMed]
- Chiu, C.-Y.; Sun, S.-C.; Chiang, C.-K.; Wang, C.-C.; Chan, D.-C.; Chen, H.-J.; Liu, S.-H.; Yang, R.-S. Plasticizer di(2-ethylhexyl)phthalate interferes with osteoblastogenesis and adipogenesis in a mouse model. J. Orthop. Res. 2018, 36, 1124–1134. [Google Scholar] [CrossRef] [PubMed]
- Hurst, C.H.; Waxman, D.J. Activation of PPARα and PPARγ by Environmental Phthalate Monoesters. Toxicol. Sci. 2003, 74, 297–308. [Google Scholar] [CrossRef]
- Campioli, E.; Duong, T.B.; Deschamps, F.; Papadopoulos, V. Cyclohexane-1,2-dicarboxylic acid diisononyl ester and metabolite effects on rat epididymal stromal vascular fraction differentiation of adipose tissue. Environ. Res. 2015, 140, 145–156. [Google Scholar] [CrossRef]
- Watkins, A.M.; Wood, C.R.; Lin, M.T.; Abbott, B.D. The effects of perfluorinated chemicals on adipocyte differentiation in vitro. Mol. Cell. Endocrinol. 2015, 400, 90–101. [Google Scholar] [CrossRef]
- Bastos Sales, L.; Kamstra, J.H.; Cenijn, P.H.; van Rijt, L.S.; Hamers, T.; Legler, J. Effects of Endocrine Disrupting Chemicals on in vitro global DNA methylation and adipocyte differentiation. Toxicol. Vitr. 2013, 27, 1634–1643. [Google Scholar] [CrossRef]
- Guo, L.-W.; Wu, Q.; Green, B.; Nolen, G.; Shi, L.; LoSurdo, J.; Deng, H.; Bauer, S.; Fang, J.-L.; Ning, B. Cytotoxicity and inhibitory effects of low-concentration triclosan on adipogenic differentiation of human mesenchymal stem cells. Toxicol. Appl. Pharmacol. 2012, 262, 117–123. [Google Scholar] [CrossRef]
- Schmid, B.; Rippmann, J.F.; Tadayyon, M.; Hamilton, B.S. Inhibition of fatty acid synthase prevents preadipocyte differentiation. Biochem. Biophys. Res. Commun. 2005, 328, 1073–1082. [Google Scholar] [CrossRef]
- Yueh, M.-F.; He, F.; Chen, C.; Vu, C.; Tripathi, A.; Knight, R.; Karin, M.; Chen, S.; Tukey, R.H. Triclosan leads to dysregulation of the metabolic regulator FGF21 exacerbating high fat diet-induced nonalcoholic fatty liver disease. Proc. Natl. Acad. Sci. USA 2020, 117, 31259–31266. [Google Scholar] [CrossRef] [PubMed]
- Noer, A.; Sørensen, A.L.; Boquest, A.C.; Collas, P. Stable CpG Hypomethylation of Adipogenic Promoters in Freshly Isolated, Cultured, and Differentiated Mesenchymal Stem Cells from Adipose Tissue. MBoC 2006, 17, 3543–3556. [Google Scholar] [CrossRef] [PubMed]
- Fujiki, K.; Kano, F.; Shiota, K.; Murata, M. Expression of the peroxisome proliferator activated receptor γ gene is repressed by DNA methylation in visceral adipose tissue of mouse models of diabetes. BMC Biol. 2009, 7, 38. [Google Scholar] [CrossRef]
- Almamun, M.; Kholod, O.; Stuckel, A.J.; Levinson, B.T.; Johnson, N.T.; Arthur, G.L.; Davis, J.W.; Taylor, K.H. Inferring a role for methylation of intergenic DNA in the regulation of genes aberrantly expressed in precursor B-cell acute lymphoblastic leukemia. Leuk. Lymphoma 2017, 58, 2156–2164. [Google Scholar] [CrossRef] [PubMed]
- Weinberg, D.N.; Papillon-Cavanagh, S.; Chen, H.; Yue, Y.; Chen, X.; Rajagopalan, K.N.; Horth, C.; McGuire, J.T.; Xu, X.; Nikbakht, H.; et al. H3K36me2 recruits DNMT3A and shapes the intergenic DNA methylation landscape. Nature 2019, 573, 281–286. [Google Scholar] [CrossRef] [PubMed]
- van den Dungen, M.W.; Murk, A.J.; Kok, D.E.; Steegenga, W.T. Persistent organic pollutants alter DNA methylation during human adipocyte differentiation. Toxicol. Vitr. 2017, 40, 79–87. [Google Scholar] [CrossRef]
- Liu, Y.; Duong, W.; Krawczyk, C.; Bretschneider, N.; Borbély, G.; Varshney, M.; Zinser, C.; Schär, P.; Rüegg, J. Oestrogen receptor β regulates epigenetic patterns at specific genomic loci through interaction with thymine DNA glycosylase. Epigenet. Chromatin 2016, 9, 7. [Google Scholar] [CrossRef]
- He, Y.-F.; Li, B.-Z.; Li, Z.; Liu, P.; Wang, Y.; Tang, Q.; Ding, J.; Jia, Y.; Chen, Z.; Li, L.; et al. Tet-Mediated Formation of 5-Carboxylcytosine and Its Excision by TDG in Mammalian DNA. Science 2011, 333, 1303–1307. [Google Scholar] [CrossRef]
- Cortázar, D.; Kunz, C.; Saito, Y.; Steinacher, R.; Schär, P. The enigmatic thymine DNA glycosylase. DNA Repair 2007, 6, 489–504. [Google Scholar] [CrossRef]
- Vandenberg, L.N.; Colborn, T.; Hayes, T.B.; Heindel, J.J.; Jacobs, D.R.; Lee, D.-H.; Shioda, T.; Soto, A.M.; vom Saal, F.S.; Welshons, W.V.; et al. Hormones and Endocrine-Disrupting Chemicals: Low-Dose Effects and Nonmonotonic Dose Responses. Endocr. Rev. 2012, 33, 378–455. [Google Scholar] [CrossRef]
- Rhomberg, L.R.; Goodman, J.E. Low-dose effects and nonmonotonic dose–responses of Endocrine Disrupting Chemicals: Has the case been made? Regul. Toxicol. Pharmacol. 2012, 64, 130–133. [Google Scholar] [CrossRef] [PubMed]
- Gore-Panter, S.R.; Hsu, J.; Barnard, J.; Moravec, C.S.; Van Wagoner, D.R.; Chung, M.K.; Smith, J.D. PANCR, the PITX2 Adjacent Noncoding RNA, Is Expressed in Human Left Atria and Regulates PITX2c Expression. Circ. Arrhythm. Electrophysiol. 2016, 9, e003197. [Google Scholar] [CrossRef] [PubMed]
- Okubo, T.; Hayashi, R.; Shibata, S.; Kudo, Y.; Ishikawa, Y.; Inoue, S.; Kobayashi, Y.; Honda, A.; Honma, Y.; Kawasaki, S.; et al. Generation and validation of a PITX2–EGFP reporter line of human induced pluripotent stem cells enables isolation of periocular mesenchymal cells. J. Biol. Chem. 2020, 295, 3456–3465. [Google Scholar] [CrossRef] [PubMed]
- Long, J.Z.; Svensson, K.J.; Bateman, L.A.; Lin, H.; Kamenecka, T.; Lokurkar, I.A.; Lou, J.; Rao, R.R.; Chang, M.R.; Jedrychowski, M.P.; et al. The secreted enzyme PM20D1 regulates lipidated amino acid uncouplers of mitochondria. Cell 2016, 166, 424–435. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Mut, J.V.; Heyn, H.; Silva, B.A.; Dixsaut, L.; Garcia-Esparcia, P.; Vidal, E.; Sayols, S.; Glauser, L.; Monteagudo-Sánchez, A.; Perez-Tur, J.; et al. PM20D1 is a quantitative trait locus associated with Alzheimer’s disease. Nat. Med. 2018, 24, 598–603. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Chen, Y.; Readhead, B.; Chen, K.; Su, Y.; Reiman, E.M.; Dudley, J.T. Longitudinal data in peripheral blood confirm that PM20D1 is a quantitative trait locus (QTL) for Alzheimer’s disease and implicate its dynamic role in disease progression. Clin. Epigenet. 2020, 12, 189. [Google Scholar] [CrossRef]
- Haworth, K.E.; Farrell, W.E.; Emes, R.D.; Ismail, K.M.; Carroll, W.D.; Hubball, E.; Rooney, A.; Yates, A.M.; Mein, C.; Fryer, A.A. Methylation of the FGFR2 gene is associated with high birth weight centile in humans. Epigenomics 2014, 6, 477–491. [Google Scholar] [CrossRef]
- Bai, Y.; Fang, N.; Gu, T.; Kang, Y.; Wu, J.; Yang, D.; Zhang, H.; Suo, Z.; Ji, S. HOXA11 gene is hypermethylation and aberrant expression in gastric cancer. Cancer Cell Int. 2014, 14, 79. [Google Scholar] [CrossRef]
- Parrillo, L.; Costa, V.; Raciti, G.A.; Longo, M.; Spinelli, R.; Esposito, R.; Nigro, C.; Vastolo, V.; Desiderio, A.; Zatterale, F.; et al. Hoxa5 undergoes dynamic DNA methylation and transcriptional repression in the adipose tissue of mice exposed to high-fat diet. Int. J. Obes. 2016, 40, 929–937. [Google Scholar] [CrossRef]
- Li, Q.; Chen, C.; Ren, X.; Sun, W. DNA methylation profiling identifies the HOXA11 gene as an early diagnostic and prognostic molecular marker in human lung adenocarcinoma. Oncotarget 2017, 8, 33100–33109. [Google Scholar] [CrossRef][Green Version]
- Xu, Y.; Wu, D.; Liu, J.; Ma, Z.; Hui, B.; Wang, J.; Chen, Y.; Wang, S.; Lian, Y.; Sun, L. Down-regulated Long Noncoding RNA HOXA11-AS affects trophoblast cell proliferation and migration by regulating RND3 and HOXA7 expression in preeclampsia. BioRxiv 2018, 12, 195–206. [Google Scholar] [CrossRef]
- Day, S.E.; Coletta, R.L.; Kim, J.Y.; Garcia, L.A.; Campbell, L.E.; Benjamin, T.R.; Roust, L.R.; De Filippis, E.A.; Mandarino, L.J.; Coletta, D.K. Potential epigenetic biomarkers of obesity-related insulin resistance in human whole-blood. Epigenetics 2017, 12, 254–263. [Google Scholar] [CrossRef] [PubMed]
- Gudbjartsson, D.F.; Arnar, D.O.; Helgadottir, A.; Gretarsdottir, S.; Holm, H.; Sigurdsson, A.; Jonasdottir, A.; Baker, A.; Thorleifsson, G.; Kristjansson, K.; et al. Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature 2007, 448, 353–357. [Google Scholar] [CrossRef] [PubMed]
- Franco, D.; Campione, M. The role of Pitx2 during cardiac development. Linking left-right signaling and congenital heart diseases. Trends Cardiovasc. Med. 2003, 13, 157–163. [Google Scholar] [CrossRef]
- Larsson, S.C.; Drca, N.; Jensen-Urstad, M.; Wolk, A. Incidence of atrial fibrillation in relation to birth weight and preterm birth. Int. J. Cardiol. 2015, 178, 149–152. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.; Qi, W.; Clark, J.M.; Park, Y. Permethrin potentiates adipogenesis via intracellular calcium and endoplasmic reticulum stress-mediated mechanisms in 3T3-L1 adipocytes. Food Chem. Toxicol. 2017, 109, 123–129. [Google Scholar] [CrossRef]
- Rüegg, J.; Cai, W.; Karimi, M.; Kiss, N.B.; Swedenborg, E.; Larsson, C.; Ekström, T.J.; Pongratz, I. Epigenetic Regulation of Glucose Transporter 4 by Estrogen Receptor β. Mol. Endocrinol. 2011, 25, 2017–2028. [Google Scholar] [CrossRef]
- Bornehag, C.-G.; Moniruzzaman, S.; Larsson, M.; Lindström, C.B.; Hasselgren, M.; Bodin, A.; von Kobyletzkic, L.B.; Carlstedt, F.; Lundin, F.; Nånberg, E.; et al. The SELMA Study: A Birth Cohort Study in Sweden Following More Than 2000 Mother–Child Pairs. Paediatr. Perinat. Epidemiol. 2012, 26, 456–467. [Google Scholar] [CrossRef]
- Carrico, C.; Gennings, C.; Wheeler, D.C.; Factor-Litvak, P. Characterization of Weighted Quantile Sum Regression for Highly Correlated Data in a Risk Analysis Setting. JABES 2015, 20, 100–120. [Google Scholar] [CrossRef]
- Koch, H.M.; Becker, K.; Wittassek, M.; Seiwert, M.; Angerer, J.; Kolossa-Gehring, M. Di-n-butylphthalate and butylbenzylphthalate—urinary metabolite levels and estimated daily intakes: Pilot study for the German Environmental Survey on children. J. Expo. Sci. Environ. Epidemiol. 2007, 17, 378–387. [Google Scholar] [CrossRef]
- Fromme, H.; Schlummer, M.; Möller, A.; Gruber, L.; Wolz, G.; Ungewiss, J.; Böhmer, S.; Dekant, W.; Mayer, R.; Liebl, B.; et al. Exposure of an Adult Population to Perfluorinated Substances Using Duplicate Diet Portions and Biomonitoring Data. Environ. Sci. Technol. 2007, 41, 7928–7933. [Google Scholar] [CrossRef] [PubMed]
- Vemuri, M.C.; Chase, L.G.; Rao, M.S. Mesenchymal Stem Cell Assays and Applications. In Mesenchymal Stem Cell Assays and Applications. Methods in Molecular Biology (Methods and Protocols); Vemuri, M., Chase, L., Rao, M., Eds.; Humana Press: Totowa, NJ, USA, 2011; p. 698. [Google Scholar] [CrossRef]
- Bibikova, M.; Lin, Z.; Zhou, L.; Chudin, E.; Garcia, E.W.; Wu, B.; Doucet, D.; Thomas, N.J.; Wang, Y.; Vollmer, E.; et al. High-throughput DNA methylation profiling using universal bead arrays. Genome. Res. 2006, 16, 383–393. [Google Scholar] [CrossRef] [PubMed]
- Du, P.; Zhang, X.; Huang, C.-C.; Jafari, N.; Kibbe, W.A.; Hou, L.; Lin, S.M. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinform. 2010, 11, 587. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. Toolbox. In ggplot2: Elegant Graphics for Data Analysis; Wickham, H., Ed.; Springer: New York, NY, USA, 2009; pp. 65–90. ISBN 978-0-387-98141-3. [Google Scholar]
- Jaffe, A.E.; Murakami, P.; Lee, H.; Leek, J.T.; Fallin, M.D.; Feinberg, A.P.; Irizarry, R.A. Bump hunting to identify differentially methylated regions in epigenetic epidemiology studies. Int. J. Epidemiol. 2012, 41, 200–209. [Google Scholar] [CrossRef] [PubMed]
- Campagna, M.P.; Xavier, A.; Lechner-Scott, J.; Maltby, V.; Scott, R.J.; Butzkueven, H.; Jokubaitis, V.G.; Lea, R.A. Epigenome-wide association studies: Current knowledge, strategies and recommendations. Clin. Epigenet. 2021, 13, 214. [Google Scholar] [CrossRef] [PubMed]
- Lent, S.; Xu, H.; Wang, L.; Wang, Z.; Sarnowski, C.; Hivert, M.-F.; Dupuis, J. Comparison of novel and existing methods for detecting differentially methylated regions. BMC Genet. 2018, 19, 84. [Google Scholar] [CrossRef]


| Chemical Class | Parent Compound 1 | Mix G1 Compound 2 | Full Compound Name | Concentration (nM) 3 | CAS Number |
|---|---|---|---|---|---|
| Phthalates | DEP | MEP | Monoethyl phthalate | 29.7 | 2306-33-4 |
| DBP | MBP | Monobutyl phthalate | 26.4 | 131-70-4 | |
| BBzP | MBzP | Monobenzyl phthalate | 5.3 | 2528-16-7 | |
| DEHP | MEHP | Mono-(2-ethylhexyl) phthalate | 19.0 | 4376-20-9 | |
| Plasticizer | DiNCH | MINCH | 2–4-Methyl-7-oxyooctyl-oxycarbonyl-cyclohexane carboxylic acid | 0.5 | 1588520-62-0 |
| TTP | DPP | Diphenylphosphate | 0.5 | 838-85-7 | |
| Antibacterial | Triclosan | 0.3 | 3380-34-5 | ||
| PAH | 2OHPH | 2-Hydroxyphenanthrene | 1.3 | 605-55-0 | |
| Pesticide | Pyrethroids | 3-PBA | 3-Phenoxybenzoic acid | 0.1 | 3739-38-6 |
| PFAS | PFOA | Perfluorooctanoic acid | 3.6 | 335-67-1 | |
| PFOS | Perfluorooctane sulfonate | 9.7 | 1763-23-1 | ||
| PFHxS | Perfluorohexane sulfonate | 3.0 | 355-46-4 | ||
| Organo-chlorine pesticide | HCB | Hexachlorobenzene | 0.1 | 118-74-1 | |
| DDT | p,p′ DDE | Dichlorodiphenyltrichloroethane | 0.5 | 50-29-3 |
| Gene | CHR 1 | Start 2 | End 3 | Length | Number of CpGs | FWER 4 |
|---|---|---|---|---|---|---|
| HOXA11AS/HOXA11 | 7 | 27,224,700 | 27,226,329 | 1629 | 28 | 0.096 |
| PM20D1 | 1 | 205,818,484 | 205,819,609 | 1125 | 13 | 0.096 |
| PANCR | 16 | 87,101,534 | 87,102,691 | 1157 | 11 | 0.12 |
| HOXA5 | 7 | 27,183,643 | 27,184,853 | 1210 | 28 | 0.144 |
| RP11-134D3.2 | 4 | 111,532,996 | 111,533,951 | 955 | 9 | 0.152 |
| RPL28 | 19 | 55,896,842 | 55,897,819 | 977 | 13 | 0.152 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lizunkova, P.; Engdahl, E.; Borbély, G.; Gennings, C.; Lindh, C.; Bornehag, C.-G.; Rüegg, J. A Mixture of Endocrine Disrupting Chemicals Associated with Lower Birth Weight in Children Induces Adipogenesis and DNA Methylation Changes in Human Mesenchymal Stem Cells. Int. J. Mol. Sci. 2022, 23, 2320. https://doi.org/10.3390/ijms23042320
Lizunkova P, Engdahl E, Borbély G, Gennings C, Lindh C, Bornehag C-G, Rüegg J. A Mixture of Endocrine Disrupting Chemicals Associated with Lower Birth Weight in Children Induces Adipogenesis and DNA Methylation Changes in Human Mesenchymal Stem Cells. International Journal of Molecular Sciences. 2022; 23(4):2320. https://doi.org/10.3390/ijms23042320
Chicago/Turabian StyleLizunkova, Polina, Elin Engdahl, Gábor Borbély, Chris Gennings, Christian Lindh, Carl-Gustaf Bornehag, and Joëlle Rüegg. 2022. "A Mixture of Endocrine Disrupting Chemicals Associated with Lower Birth Weight in Children Induces Adipogenesis and DNA Methylation Changes in Human Mesenchymal Stem Cells" International Journal of Molecular Sciences 23, no. 4: 2320. https://doi.org/10.3390/ijms23042320
APA StyleLizunkova, P., Engdahl, E., Borbély, G., Gennings, C., Lindh, C., Bornehag, C.-G., & Rüegg, J. (2022). A Mixture of Endocrine Disrupting Chemicals Associated with Lower Birth Weight in Children Induces Adipogenesis and DNA Methylation Changes in Human Mesenchymal Stem Cells. International Journal of Molecular Sciences, 23(4), 2320. https://doi.org/10.3390/ijms23042320







