Transcriptomic Analysis Reveals Functional Interaction of mRNA–lncRNA–miRNA in Steroidogenesis and Spermatogenesis of Gynogenetic Japanese Flounder (Paralichthys olivaceus)
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experiment Material and Sample Preparation
2.2. RNA Extraction, Library Construction and Sequencing
2.3. Transcriptome Analysis and lncRNA Characterization
2.4. Differential Expression of lncRNA and mRNA with Functional Annotation
2.5. miRNA Identification, Differential Expression and Functional Annotation
2.6. Construction of DEmRNA–DEmiRNA–DElncRNA Network
2.7. miRNA Cluster Identification
2.8. qRT-PCR Analysis
2.9. Vector Construction and Dual-Luciferase Activity Assay
3. Results and Discussion
3.1. Transcriptome Profiles of Gynogenetic P. olivaceus Gonads
3.2. Differential Expression of mRNAs, lncRNAs and miRNAs in Gynogenetic P. olivaceus Gonads
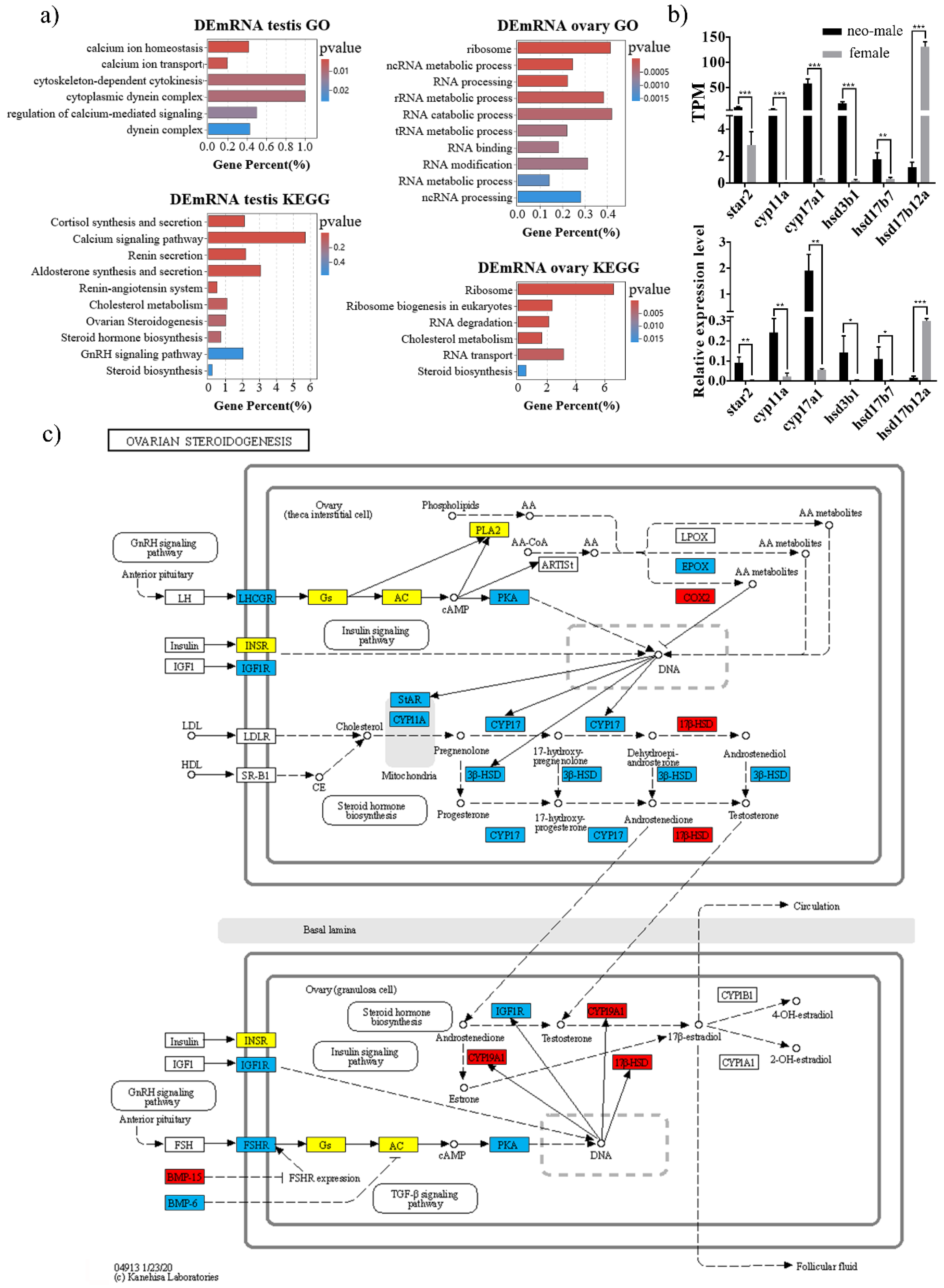
3.3. DEmRNAs Participate in the Steroid Hormone Biogenesis of Gynogenetic P. olivaceus
3.4. Sperm Motility-Related mRNA–miRNA–lncRNA Interaction in Gynogenetic P. olivaceus
3.5. Clustered miRNAs and miRNA Host lncRNAs in Gynogenetic P. olivaceus
3.6. Regulation of Steroidogenesis Pathway by let-7/miR-125b in P. olivaceus
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Smith, C.; Wootton, R.J. The remarkable reproductive diversity of teleost fishes. Fish Fish. 2016, 17, 1208–1215. [Google Scholar] [CrossRef]
- Rastetter, R.H.; Smith, C.A.; Wilhelm, D. The role of non-coding RNAs in male sex determination and differentiation. Reproduction 2015, 150, R93–R107. [Google Scholar] [CrossRef] [PubMed]
- Robles, V.; Valcarce, D.; Riesco, M. Non-coding RNA regulation in reproduction: Their potential use as biomarkers. Noncoding RNA Res. 2019, 4, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Reza, A.M.M.T.; Choi, Y.; Han, S.; Song, H.; Park, C.; Hong, K.; Kim, J. Roles of microRNAs in mammalian reproduction: From the commitment of germ cells to peri-implantation embryos. Biol. Rev. 2019, 94, 415–438. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.; Abdelmohsen, K.; Gorospe, M. Functional interactions among microRNAs and long noncoding RNAs. Semin. Cell Dev. Biol. 2014, 34, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Thomson, D.W.; Dinger, M.E. Endogenous microRNA sponges: Evidence and controversy. Nat. Rev. Genet. 2016, 17, 272–283. [Google Scholar] [CrossRef]
- Du, P.; Wang, L.; Sliz, P.; Gregory, R.I. A biogenesis step upstream of microprocessor controls miR-17-92 expression. Cell 2015, 162, 885–899. [Google Scholar] [CrossRef]
- Luo, L.; Hou, C.; Yang, W. Small non-coding RNAs and their associated proteins in spermatogenesis. Gene 2016, 578, 141–157. [Google Scholar] [CrossRef]
- Garcia-Lopez, J.; Alonso, L.; Cardenas, D.B.; Artaza-Alvarez, H.; Hourcade, J.; Martinez, S.; Brieno-Enriquez, M.A.; Del Mazo, J. Diversity and functional convergence of small noncoding RNAs in male germ cell differentiation and fertilization. RNA 2015, 21, 946–962. [Google Scholar] [CrossRef]
- Salilew-Wondim, D.; Gebremedhn, S.; Hoelker, M.; Tholen, E.; Hailay, T.; Tesfaye, D. The role of microRNAs in mammalian fertility: From gametogenesis to embryo implantation. Int. J. Mol. Sci. 2020, 21, 585. [Google Scholar] [CrossRef]
- Hurtado, A.; Palomino, R.; Georg, I.; Lao, M.; Real, F.M.; Carmona, F.D.; Burgos, M.; Jimenez, R.; Barrionuevo, F.J. Deficiency of the onco-miRNA cluster, miR-106b-25, causes oligozoospermia and the cooperative action of miR-106b-25 and miR-17-92 is required to maintain male fertility. Mol. Hum. Reprod. 2020, 26, 389–401. [Google Scholar] [CrossRef] [PubMed]
- Tong, M.; Mitchell, D.; McGowan, S.; Evanoff, R.; Griswold, M. Two miRNA cluster, Mir-17-92 (Mirc1) and Mir-106b-25 (Mirc3), are involved in the regulation of spermatogonial differentiation in mice. Biol. Reprod. 2012, 86, 72. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.; Zhou, Y.; Zhao, H.; Wu, J.; Yu, H.; Yang, Y.; Yang, Y.; Zhao, H.; Li, H. Comprehensive transcriptome analysis of hypothalamus reveals genes associated with disorders of sex development in pigs. J. Steroid Biochem. Mol. Biol. 2021, 210, 105875. [Google Scholar] [CrossRef] [PubMed]
- Bhat, R.A.; Priyam, M.; Foysal, M.J.; Gupta, S.K.; Sundaray, J.K. Role of sex-biased miRNAs in teleosts—A review. Rev. Aquac. 2021, 13, 269–281. [Google Scholar] [CrossRef]
- Lan, T.; Chen, Y.L.; Gul, Y.; Zhao, B.W.; Gao, Z.X. Comparative expression analysis of let-7 microRNAs during ovary development in Megalobrama amblycephala. Fish Physiol. Biochem. 2019, 45, 1101–1115. [Google Scholar] [CrossRef]
- Liu, J.; Luo, M.; Sheng, Y.; Hong, Q.; Cheng, H.; Zhou, R. Dynamic evolution and biogenesis of small RNAs during sex reversal. Sci. Rep. 2015, 5, 9999. [Google Scholar] [CrossRef]
- Fan, Z.; You, F.; Wang, L.; Weng, S.; Wu, Z.; Hu, J.; Zou, Y.; Tan, X.; Zhang, P. Gonadal transcriptome analysis of male and female olive flounder (Paralichthys olivaceus). BioMed Res. Int. 2014, 2014, 291067. [Google Scholar] [CrossRef]
- Xu, K.; Duan, W.; Xiao, J.; Tao, M.; Zhang, C.; Liu, Y.; Liu, S. Development and application of biological technologies in fish genetic breeding. Sci. China-Life Sci. 2015, 58, 187–201. [Google Scholar] [CrossRef]
- Manan, H.; Hidayati, A.B.N.; Lyana, N.A.; Amin-Safwan, A.; Ma, H.; Kasan, N.A.; Ikhwanuddin, M. A review of gynogenesis manipulation in aquatic animals. Aquac. Fish. 2020, 7, 1–6. [Google Scholar] [CrossRef]
- Wu, Z.; Wang, L.; Wu, Q.; Lu, Y.; Song, Z.; Li, J.; You, F. Study on artificial induction, growth and gamete quality of mitogynogenetic turbot Scophthalmus maximus. Aquaculture 2020, 515, 734585. [Google Scholar] [CrossRef]
- Ma, D.; Weng, S.; Sun, P.; Li, J.; Zhang, P.; You, F. Histological observation on adult gonads from meiogynogenetic olive flounder Paralichthys olivaceus. Int. J. Agric. Biol. 2018, 20, 689–694. [Google Scholar] [CrossRef]
- Zhang, X.; Hou, J.; Wang, G.; Jiang, H.; Wang, Y.; Sun, Z.; Jiang, X.; Yu, Q.; Liu, H. Gonadal transcriptome analysis in sterile double haploid Japanese flounder. PLoS ONE 2015, 10, e0143204. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhao, H.; Du, X.; Zhang, K.; Liu, Y.; Wang, Y.; Liu, J.; He, Y.; Wang, X.; Zhang, Q. Weighted correlation network analysis (WGCNA) of Japanese flounder (Paralichthys olivaceus) embryo transcriptome provides crucial gene sets for understanding haploid syndrome and rescue by diploidization. J. Ocean Univ. China 2018, 17, 1441–1450. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Zhang, Y.; Ye, Z.; Liu, X.; Zhao, S.; Wei, L.; Gao, G. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007, 36, W345–W349. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Luo, H.; Bu, D.; Zhao, G.; Yu, K.; Zhang, C.; Liu, Y.; Chen, R.; Zhao, Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013, 41, e166. [Google Scholar] [CrossRef]
- Wang, L.; Jung, P.H.; Surendra, D.; Wang, S.; Kocher, J.; Li, W. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013, 41, e74. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef]
- John, B.; Enright, A.J.; Aravin, A.; Tuschl, T.; Sander, C.; Marks, D.S. Human microRNA targets. PLoS Biol. 2004, 2, 1863–1879. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Sun, L. Evaluation of housekeeping genes as references for quantitative real time RT-PCR analysis of gene expression in Japanese flounder (Paralichthys olivaceus). Fish Shellfish Immunol. 2011, 30, 638–645. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Song, H.; Liu, Z.; Lu, W.; Zhang, Q.; Cheng, J. Selection of References for microRNA Quantification in Japanese Flounder (Paralichthys olivaceus) Normal Tissues and Edwardsiella tarda-Infected Livers. Genes 2022, 13, 175. [Google Scholar] [CrossRef]
- Finkelstein, M.; Etkovitz, N.; Breitbart, H. Ca2+ signaling in mammalian spermatozoa. Mol. Cell Endocrinol. 2020, 516, 110953. [Google Scholar] [CrossRef] [PubMed]
- Rajakumar, A.; Senthilkumaran, B. Steroidogenesis and its regulation in teleost-a review. Fish Physiol. Biochem. 2020, 46, 803–818. [Google Scholar] [CrossRef]
- Tokarz, J.; Moller, G.; de Angelis, M.; Adamski, J. Steroids in teleost fishes: A functional point of view. Steroids 2015, 103, 123–144. [Google Scholar] [CrossRef]
- Li, Y.R.; Yang, W.X. Myosin superfamily: The multi-functional and irreplaceable factors in spermatogenesis and testicular tumors. Gene 2016, 576, 195–207. [Google Scholar] [CrossRef]
- Lie, P.P.; Mruk, D.D.; Lee, W.M.; Cheng, C.Y. Cytoskeletal dynamics and spermatogenesis. Philos. Trans. R. Soc. B Biol. Sci. 2010, 365, 1581–1592. [Google Scholar] [CrossRef]
- Kardon, J.R.; Vale, R.D. Regulators of the cytoplasmic dynein motor. Nat. Rev. Mol. Cell Biol. 2009, 10, 854–865. [Google Scholar] [CrossRef] [PubMed]
- Hu, F.; Xu, K.; Zhou, Y.; Wu, C.; Wang, S.; Xiao, J.; Wen, M.; Zhao, R.; Luo, K.; Tao, M.; et al. Different expression patterns of sperm motility-related genes in testis of diploid and tetraploid cyprinid fish. Biol. Reprod. 2017, 96, 907–920. [Google Scholar] [CrossRef] [PubMed]
- Rashid, S.; Breckle, R.; Hupe, M.; Geisler, S.; Doerwald, N.; Neesen, J. The murine Dnali1 gene encodes a flagellar protein that interacts with the cytoplasmic dynein heavy chain 1. Mol. Reprod. Dev. 2006, 73, 784–794. [Google Scholar] [CrossRef] [PubMed]
- Zuccarello, D.; Ferlin, A.; Cazzadore, C.; Pepe, A.; Garolla, A.; Moretti, A.; Cordeschi, G.; Francavilla, S.; Foresta, C. Mutations in dynein genes in patients affected by isolated non-syndromic asthenozoospermia. Hum. Reprod. 2008, 23, 1957–1962. [Google Scholar] [CrossRef] [PubMed]
- Neesen, J.; Kirschner, R.; Ochs, M.; Schmiedl, A.; Habermann, B.; Mueller, C.; Holstein, A.F.; Nuesslein, T.; Adham, I.; Engel, W. Disruption of an inner arm dynein heavy chain gene results in asthenozoospermia and reduced ciliary beat frequency. Hum. Mol. Genet. 2001, 10, 1117–1128. [Google Scholar] [CrossRef]
- Luo, S.; Gao, X.; Ding, J.; Liu, C.; Du, C.; Hou, C.; Zhu, J.; Lou, B. Transcriptome sequencing reveals the traits of spermatogenesis and testicular development in large yellow croaker (Larimichthys crocea). Genes 2019, 10, 958. [Google Scholar] [CrossRef]
- Wang, Y.; Luo, J.; Zhang, H.; Lu, J. microRNAs in the same clusters evolve to coordinately regulate functionally related genes. Mol. Biol. Evol. 2016, 33, 2232–2247. [Google Scholar] [CrossRef]
- Roush, S.; Slack, F.J. The let-7 family of microRNAs. Trends Cell Biol. 2008, 18, 505–516. [Google Scholar] [CrossRef]
- Hertel, J.; Bartschat, S.; Wintsche, A.; Otto, C.; The Students of the Bioinformatics Computer Lab 2011; Stadler, P.F. Evolution of the let-7 microRNA family. RNA Biol. 2012, 3, 231–241. [Google Scholar] [CrossRef]
- Lee, H.; Han, S.; Kwon, C.S.; Lee, D. Biogenesis and regulation of the let-7 miRNAs and their functional implications. Protein Cell 2016, 7, 100–113. [Google Scholar] [CrossRef]
- Tenugu, S.; Pranoty, A.; Mamta, S.K.; Senthilkumaran, B. Development and organisation of gonadal steroidogenesis in bony fishes—A review. Aquac. Fish. 2021, 6, 223–246. [Google Scholar] [CrossRef]
- Cochrane, D.R.; Cittelly, D.M.; Richer, J.K. Steroid receptors and microRNAs: Relationships revealed. Steroids 2011, 76, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Azhar, S.; Dong, D.; Shen, W.; Hu, Z.; Kraemer, F.B. The role of miRNAs in regulating adrenal and gonadal steroidogenesis. J. Mol. Endocrinol. 2020, 64, R21–R43. [Google Scholar] [CrossRef] [PubMed]
- Men, Y.; Fan, Y.; Shen, Y.; Lu, L.; Kallen, A.N. The steroidogenic acute regulatory protein (StAR) is regulated by the H19/let-7 axis. Endocrinology 2017, 158, 402–409. [Google Scholar] [CrossRef] [PubMed]
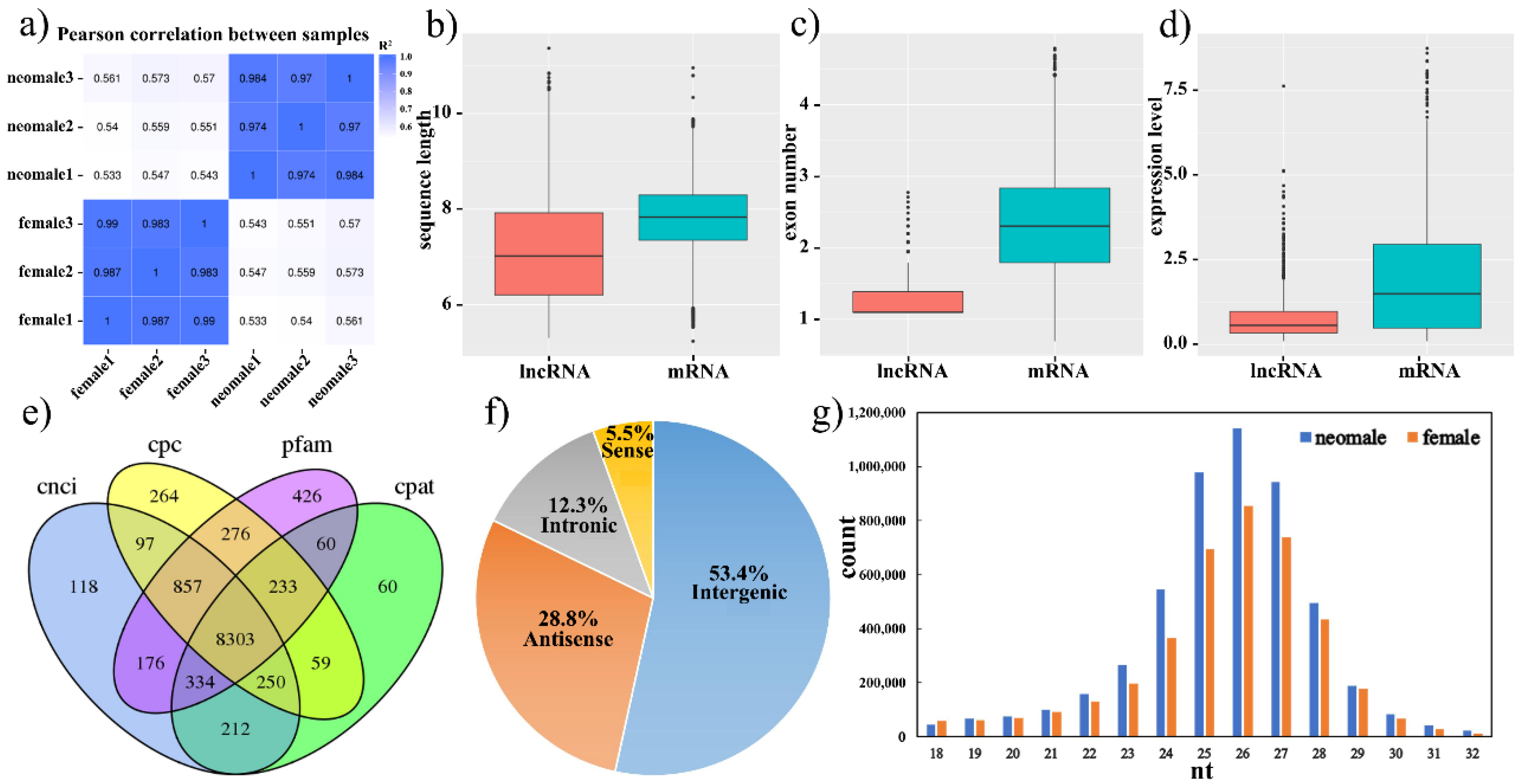
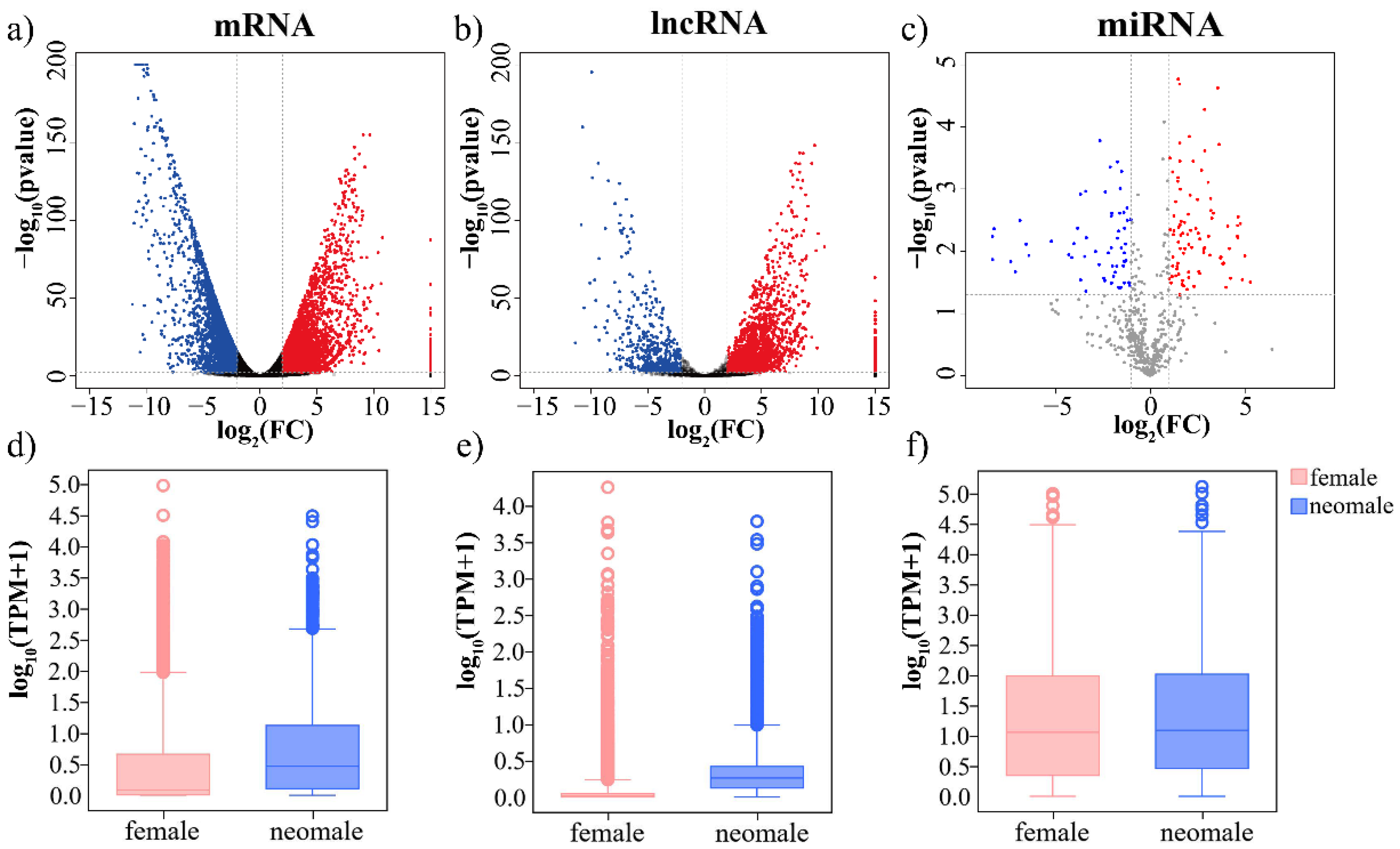



| DE RNAs | mRNAs | lncRNAs | miRNAs | |||
|---|---|---|---|---|---|---|
| Up | Down | Up | Down | Up | Down | |
| neo-male vs. female | 3541 (15.5%) | 3231 (14.1%) | 1870 (22.5%) | 414 (5.0%) | 146 (17.7%) | 98 (11.9%) |
| Species | miRNAs | miRNA Clusters | miRNAs in Clusters | miRNAs in Clusters/miRNAs |
|---|---|---|---|---|
| P. olivaceus | 824 | 106 | 427 | 52% |
| N. furzeri | 754 | 83 | 213 | 28% |
| D. rerio | 765 | 96 | 305 | 40% |
| O. latipes | 366 | 58 | 151 | 41% |
| G. aculeatus | 504 | 68 | 299 | 59% |
| T. rubripes | 337 | 59 | 143 | 42% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, J.; Yang, F.; Liu, S.; Zhao, H.; Lu, W.; Zhang, Q. Transcriptomic Analysis Reveals Functional Interaction of mRNA–lncRNA–miRNA in Steroidogenesis and Spermatogenesis of Gynogenetic Japanese Flounder (Paralichthys olivaceus). Biology 2022, 11, 213. https://doi.org/10.3390/biology11020213
Cheng J, Yang F, Liu S, Zhao H, Lu W, Zhang Q. Transcriptomic Analysis Reveals Functional Interaction of mRNA–lncRNA–miRNA in Steroidogenesis and Spermatogenesis of Gynogenetic Japanese Flounder (Paralichthys olivaceus). Biology. 2022; 11(2):213. https://doi.org/10.3390/biology11020213
Chicago/Turabian StyleCheng, Jie, Fan Yang, Saisai Liu, Haitao Zhao, Wei Lu, and Quanqi Zhang. 2022. "Transcriptomic Analysis Reveals Functional Interaction of mRNA–lncRNA–miRNA in Steroidogenesis and Spermatogenesis of Gynogenetic Japanese Flounder (Paralichthys olivaceus)" Biology 11, no. 2: 213. https://doi.org/10.3390/biology11020213
APA StyleCheng, J., Yang, F., Liu, S., Zhao, H., Lu, W., & Zhang, Q. (2022). Transcriptomic Analysis Reveals Functional Interaction of mRNA–lncRNA–miRNA in Steroidogenesis and Spermatogenesis of Gynogenetic Japanese Flounder (Paralichthys olivaceus). Biology, 11(2), 213. https://doi.org/10.3390/biology11020213






