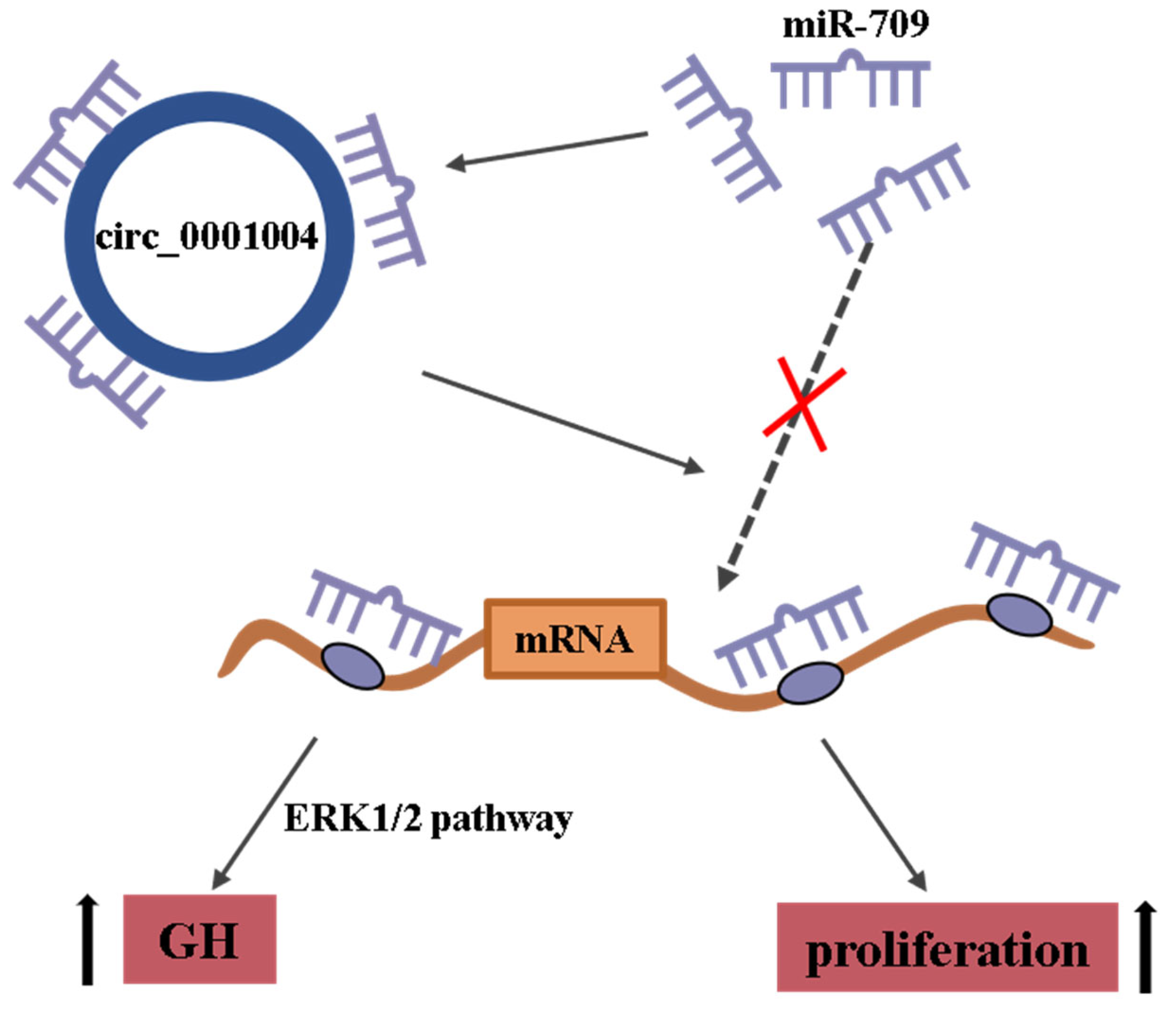
Rno_circ_0001004 Acts as a miR-709 Molecular Sponge to Regulate the Growth Hormone Synthesis and Cell Proliferation
Abstract
:1. Introduction
2. Results
2.1. Characterization of Rno_circ_0001004 in GH3 Cells
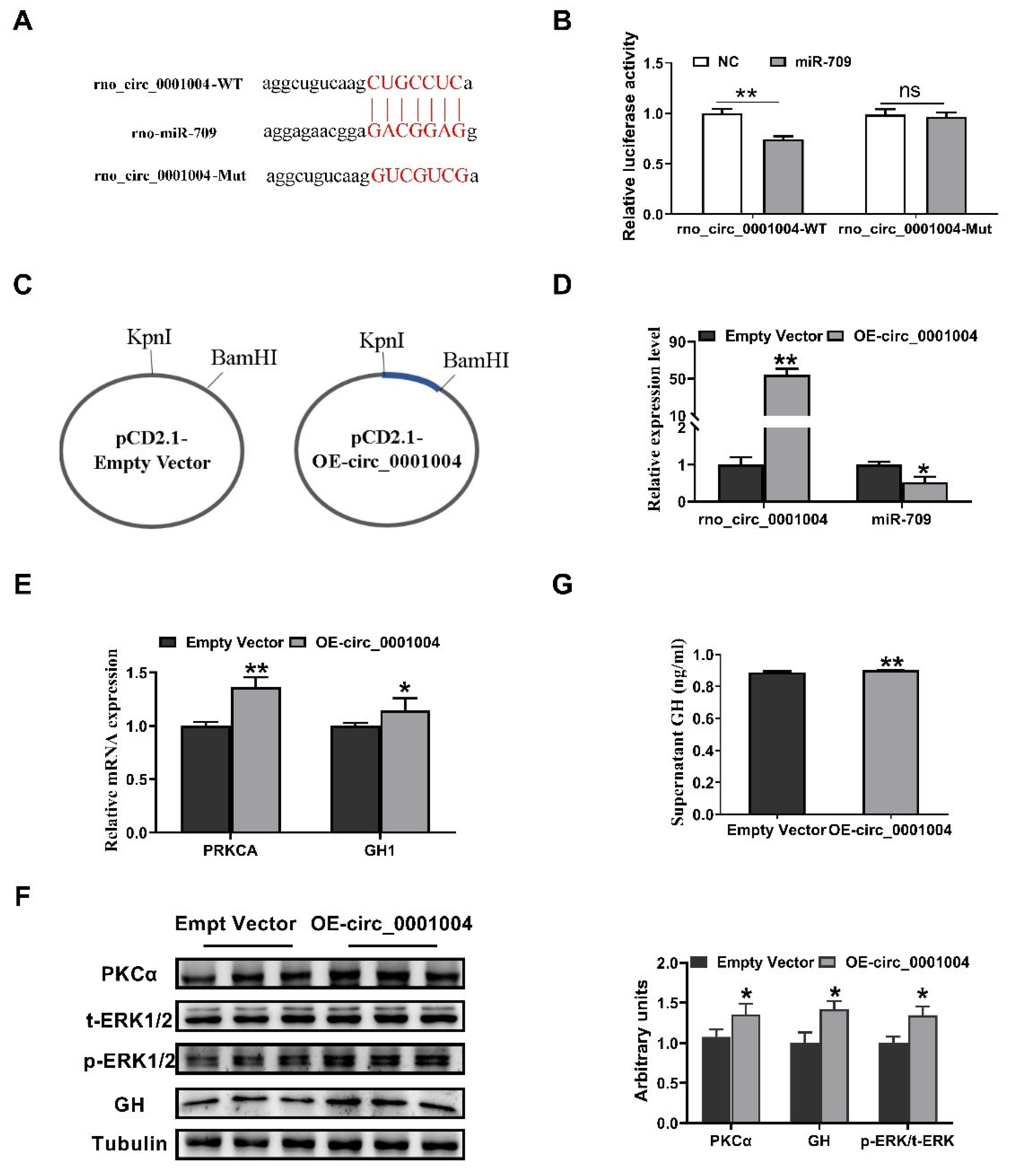
2.2. Rno_circ_0001004 Antagonizes miR-709-Mediated Repression of GH Synthesis and Secretion
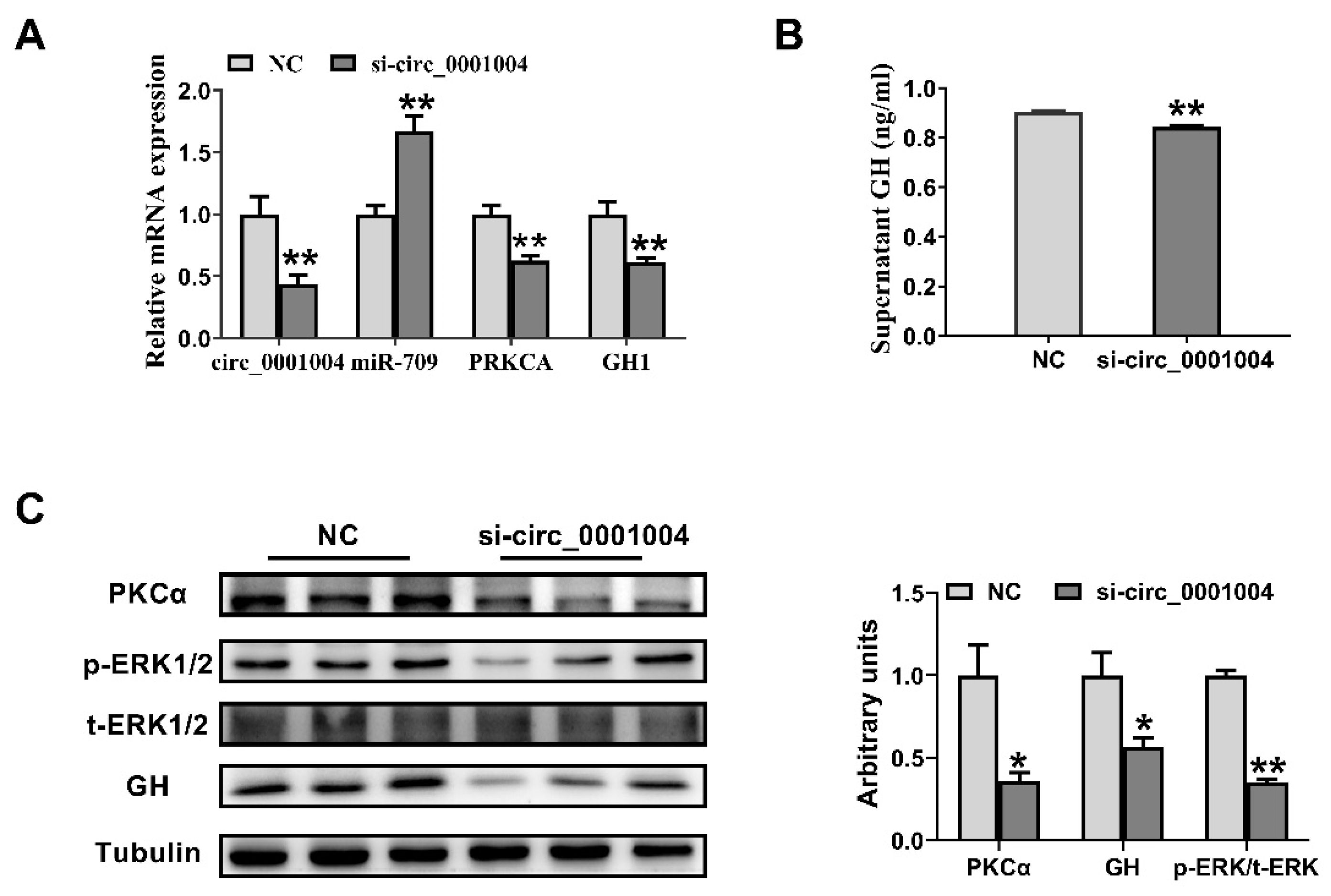
2.3. Knockdown of Rno_circ_0001004 Suppresses the GH Synthesis and Secretion in GH3 Cell
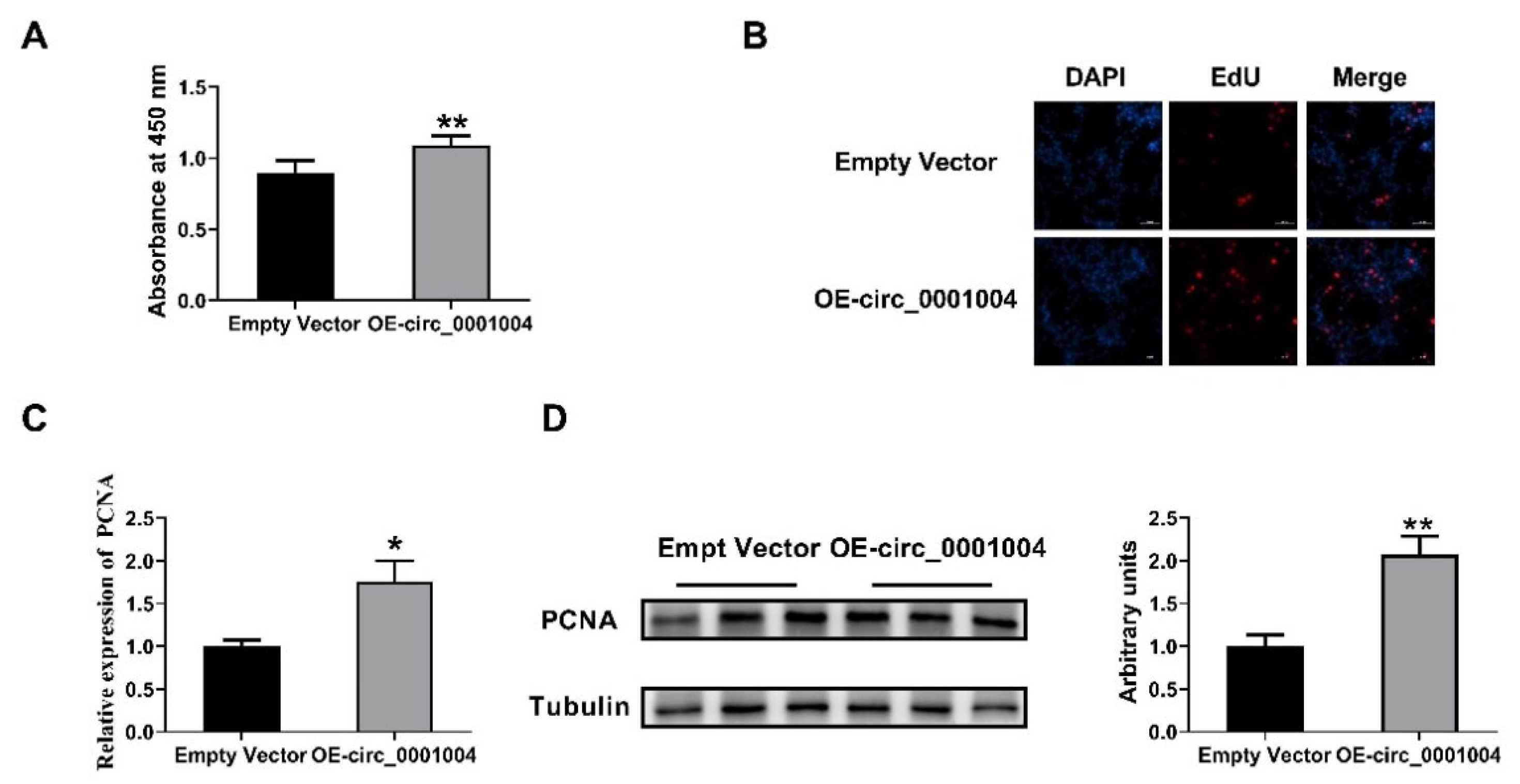
2.4. Rno_circ_0001004 Promoted the Viability of GH3 Cells
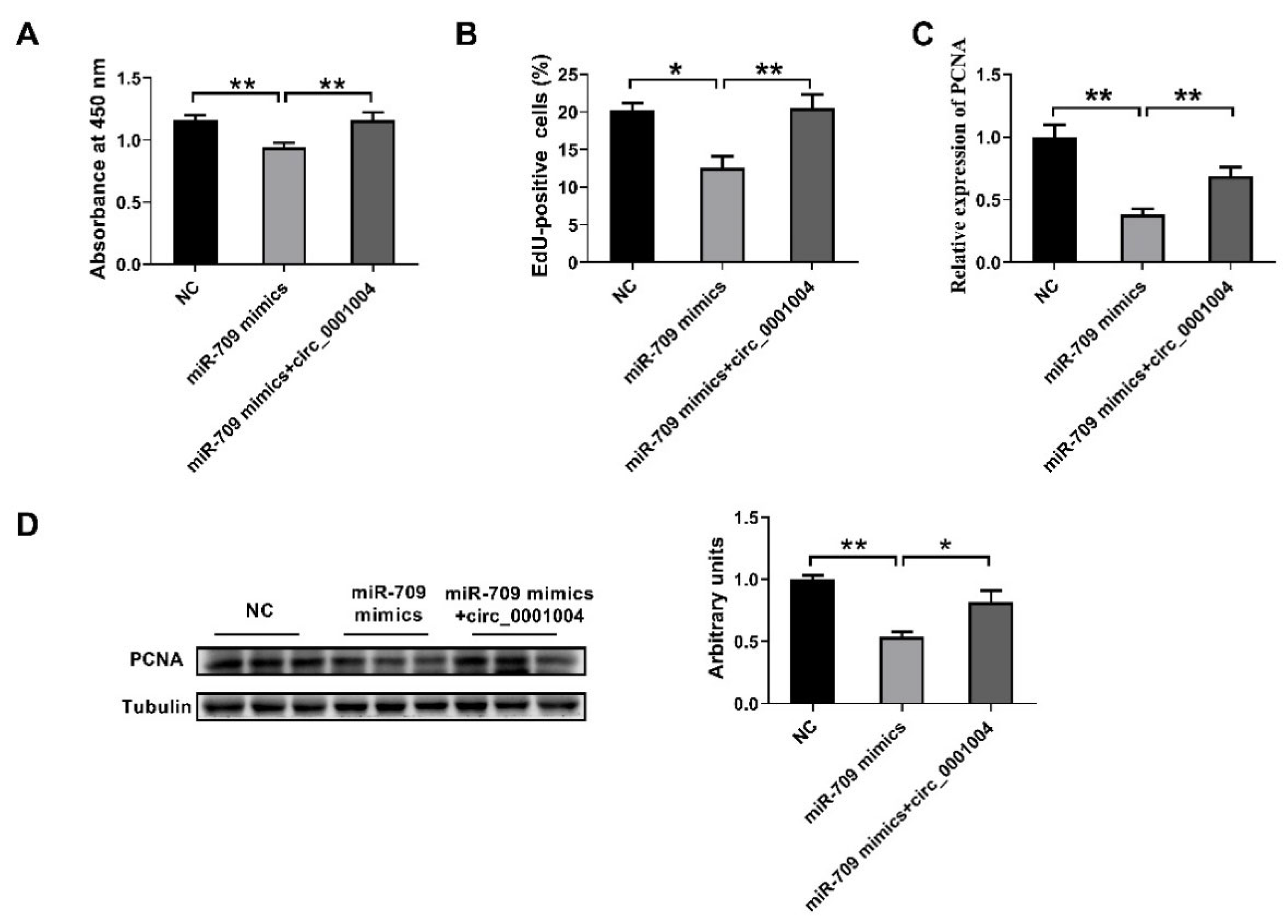
2.5. Rno_circ_0001004 Reversed the Inhibition of Cell Proliferation by miR-709
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Transfection
4.2. RAN Isolation, cDNA Synthesis, RT-PCR and Sanger Sequencing
4.3. Vector Construction
4.4. Dual-Luciferase Reporter Assay
4.5. Evaluation of GH3 Proliferation
4.6. Western Blot Analysis
4.7. Quantification of Secretory GH by ELISA
4.8. Statistics Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nyberg, F. Growth Hormone in the Brain: Characteristics of Specific Brain Targets for the Hormone and Their Functional Significance. Front. Neuroendocrinol. 2000, 21, 330–348. [Google Scholar] [CrossRef] [PubMed]
- Mao, Z.G.; He, D.S.; Zhou, J.; Yao, B.; Xiao, W.W.; Chen, C.H.; Zhu, Y.H.; Wang, H.J. Differential expression of microRNAs in GH-secreting pituitary adenomas. Diagn. Pathol. 2010, 5, 79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z. Florez Sergio Gutierrez-Hartmann Arthur Martin James, F.; Amendt Brad, A. MicroRNAs Regulate Pituitary Development, and MicroRNA 26b Specifically Targets Lymphoid Enhancer Factor 1 (Lef-1), Which Modulates Pituitary Transcription Factor 1 (Pit-1) Expression. J. Biol. Chem. 2010, 285, 34718–34728. [Google Scholar] [PubMed] [Green Version]
- Yu, Z.W.; Gao, W.; Feng, X.Y.; Zhang, J.Y.; Yuan, B. Roles of differential expression of miR-543-5p in GH regulation in rat anterior pituitary cells and GH3 cells. PLoS ONE 2019, 14, e0222340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.W. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [Google Scholar]
- Gee, H.E.; Camps, C.; Buffa, F.M.; Colella, S.; Sheldon, H.; Gleadle, J.M.; Ragoussis, J.; Harris, A.L. MicroRNA-10b and breast cancer metastasis. Nature 2008, 455, E8. [Google Scholar] [CrossRef]
- Kota, J.; Chivukula, R.R.; O’Donnell, K.A.; Wentzel, E.A.; Montgomery, C.L.; Hwang, H.W.; Chang, T.C.; Vivekanandan, P.; Torbenson, M.; Clark, K.R. Therapeutic delivery of miR-26a inhibits cancer cell proliferation and induces tumor-specific apoptosis. Cell 2009, 137, 1005. [Google Scholar] [CrossRef] [Green Version]
- Ma, L.; Teruya-Feldstein, J.; Weinberg, R.A. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 2007, 449, 682–688. [Google Scholar] [CrossRef]
- Png, K.J.; Halberg, N.; Yoshida, M.; Tavazoie, S.F. A microRNA regulon that mediates endothelial recruitment and metastasis by cancer cells. Nature 2012, 481, 190–194. [Google Scholar] [CrossRef]
- Tay, Y.; Zhang, J.; Thomson, A.M.; Bing, L.; Rigoutsos, I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature 2008, 455, 1124. [Google Scholar] [CrossRef]
- Felekkis, K.; Touvana, E.; Stefanou, C.; Deltas, C. MicroRNAs: A newly described class of encoded molecules that play a role in health and disease. Hippokratia 2010, 14, 236–240. [Google Scholar] [PubMed]
- Cheng, Y.; Chen, T.; Song, J.; Qi, Q.; Zhang, Y. miR-709 inhibits GHRP6 induced GH synthesis by targeting PRKCA in pituitary. Mol. Cell. Endocrinol. 2020, 506, 110763. [Google Scholar] [CrossRef] [PubMed]
- Grey, C.L.; Chang, J.P. Differential modulation of ghrelin-induced GH and LH release by PACAP and dopamine in goldfish pituitary cells. Gen. Comp. Endocrinol. 2013, 191, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Pemberton, J.G.; Orr, M.E.; Stafford, J.L.; Chang, J.P. PI3K signalling in GnRH actions on dispersed goldfish pituitary cells: Relationship with PKC-mediated LH and GH release and regulation of long-term effects on secretion and total cellular hormone availability. Gen. Comp. Endocrinol. 2014, 205, 268–278. [Google Scholar] [CrossRef]
- Petiti, J.P.; Gutiérrez, S.; De Paul, A.L. Andreoli: GH3B6 Pituitary Tumor Cell Proliferation is Mediated by PKCα and PKCε via ERK 1/2-dependent Pathway. Cell Physiol. Biochem. 2010, 26, 135–146. [Google Scholar] [CrossRef]
- Meng, S.; Zhou, H.; Feng, Z.; Xu, Z.; Ying, T.; Li, P.; Wu, M. CircRNA: Functions and properties of a novel potential biomarker for cancer. Mol. Cancer 2017, 16, 94. [Google Scholar] [CrossRef]
- Chen, L.L.; Yang, L. Regulation of circRNA biogenesis. RNA Biol. 2015, 12, 381–388. [Google Scholar] [CrossRef]
- Salzman, J.; Gawad, C.; Wang, P.L.; Lacayo, N.; Brown, P.O. Circular RNAs Are the Predominant Transcript Isoform from Hundreds of Human Genes in Diverse Cell Types. PLoS ONE 2012, 7, e30733. [Google Scholar] [CrossRef] [Green Version]
- Ye, C.Y.; Chen, L.; Liu, C.; Zhu, Q.H.; Fan, L. Widespread noncoding circular RNAs in plants. New Phytol. 2015, 208, 88–95. [Google Scholar] [CrossRef]
- Du, W.W.; Fang, L.; Yang, W.; Wu, N.; Awan, F.M.; Yang, Z.; Yang, B.B. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017, 24, 357–370. [Google Scholar] [CrossRef]
- Du, W.W.; Yang, W.; Liu, E.; Yang, Z.; Yang, B.B. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016, 44, gkw027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, Y.; Du, W.W.; Wu, Y.; Yang, Z.; Awan, F.M.; Li, X.; Yang, W.; Zhang, C.; Yang, Q.; Yee, A.; et al. A Circular RNA Binds To and Activates AKT Phosphorylation and Nuclear Localization Reducing Apoptosis and Enhancing Cardiac Repair. Theranostics 2017, 7, 3842–3855. [Google Scholar] [CrossRef] [PubMed]
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. circRNA Biogenesis Competes with Pre-mRNA Splicing—ScienceDirect. Mol. Cell 2014, 56, 55–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salzman, J. Circular RNA Expression: Its Potential Regulation and Function. Trends Genet. 2016, 32, 309–316. [Google Scholar] [CrossRef] [Green Version]
- Huang, M.; Zhong, Z.; Lv, M.; Shu, J.; Tian, Q.; Chen, J. Comprehensive analysis of differentially expressed profiles of lncRNAs and circRNAs with associated co-expression and ceRNA networks in bladder carcinoma. Oncotarget 2016, 7, 47186. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Lei, P.; Chen, G.; Zhu, Z.; Shen, Z.; Du, C.; Zang, R.; Yang, S.; Hua, X.; Li, H.; Xu, X. Circular RNA ZNF609 functions as a competitive endogenous RNA to regulate AKT3 expression by sponging miR-150-5p in Hirschsprung's disease. Oncotarget 2017, 8, 808. [Google Scholar]
- Li, F.; Zhang, L.; Li, W.; Deng, J.; Zheng, J.; An, M.; Lu, J.; Zhou, Y. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β-catenin pathway. Oncotarget 2015, 6, 6001. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Long, B.; Liu, F.; Wang, J.X.; Li, P.F. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur. Heart J. 2016, 37, ehv713. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Bao, C.; Guo, W.; Li, S.; Chen, J.; Chen, B.; Luo, Y.; Lyu, D.; Li, Y.; Shi, G. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 2016, 7, 11215. [Google Scholar] [CrossRef] [PubMed]
- Du, Q.; Zhang, W.; Feng, Q.; Hao, B.; Cheng, C.; Cheng, Y.; Li, Y.; Fan, X.; Chen, Z. Comprehensive circular RNA profiling reveals that hsa_circ_0001368 is involved in growth hormone-secreting pituitary adenoma development. Brain Res. Bull. 2020, 161, 65–77. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.W.; Ren, W.Z.; Wang, T.; Zhang, W.D.; Yuan, B. CircAgtpbp1 Acts as a Molecular Sponge of miR-543-5p to Regulate the Secretion of GH in Rat Pituitary Cells. Animals 2021, 11, 558. [Google Scholar] [CrossRef] [PubMed]
- Han, D.X.; Sun, X.L.; Fu, Y.; Wang, C.J.; Liu, J.B.; Jiang, H.; Gao, Y.; Chen, C.Z.; Yuan, B.; Zhang, J.B. Identification of long non-coding RNAs in the immature and mature rat anterior pituitary. Sci. Rep. 2017, 7, 17780. [Google Scholar] [CrossRef] [Green Version]
- Scully, K.M.; Rosenfeld, M.G. Pituitary Development: Regulatory Codes in Mammalian Organogenesis. Science 2002, 295, 2231–2235. [Google Scholar] [CrossRef] [Green Version]
- Haggerjohnson, G. An Introduction to Behavioral Endocrinology. Q. Rev. Biol. 2011, 2, 273–282. [Google Scholar]
- Tissier, P.; Hodson, D.J.; Lafont, C.; Fontanaud, P.; Mollard, P. Anterior pituitary cell networks. Front. Neuroendocrinol. 2012, 33, 252–266. [Google Scholar] [CrossRef] [Green Version]
- Isaksson, O.; Lindahl, A.; Nilsson, A.; Isgaard, J. Mechanism of the stimulatory effect of growth hormone on longitudinal bone growth. Endocr. Rev. 1987, 8, 426–438. [Google Scholar] [CrossRef] [PubMed]
- Salomon, F.; Cuneo, R.C. The effects of treatment with recombinant human growth hormone on body composition and metabolism. N. Engl. J. Med. 1989, 321, 1797. [Google Scholar] [CrossRef]
- Carthew, R.W.; Sontheimer, E.J. Origins and mechanisms of miRNAs and siRNAs. Cell 2009, 136, 642–655. [Google Scholar] [CrossRef] [Green Version]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Miska, E.A. How microRNAs control cell division, differentiation and death. Curr. Opin. Genet. Dev. 2005, 15, 563–568. [Google Scholar] [CrossRef]
- Zamore, P.; Haley, B. Ribo-gnome: The Big World of Small RNAs. Science 2005, 309, 1519–1524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daniela, D.A.; Dario, P.; Paula, M.; Magali, R.; Anne, W.; Gerald, R.; Monica, F.; Maria, C.C.; Jacqueline, T.; Alfredo, F. Altered microRNA expression profile in human pituitary GH adenomas: Down-regulation of miRNA targeting HMGA1, HMGA2, and E2F1. J. Clin. Endocrinol. Metab. 2012, 97, E1128–E1138. [Google Scholar]
- Xiang, L.; Li, Y.; Chen, L.L. The Biogenesis, Functions, and Challenges of Circular RNAs. Mol. Cell 2018, 71, 428–442. [Google Scholar]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. Rna 2013, 19, 141–157. [Google Scholar] [CrossRef] [Green Version]
- Qu, S.; Zhong, Y.; Shang, R.; Zhang, X.; Song, W.; Kjems, J.; Li, H. The emerging landscape of circular RNA in life processes. RNA Biol. 2017, 14, 992–999. [Google Scholar] [CrossRef]
- Li, H.; Wei, X.; Yang, J.; Dong, D.; Hao, D.; Huang, Y.; Lan, X.; Plath, M.; Lei, C.; Ma, Y. circFGFR4 Promotes Differentiation of Myoblasts via Binding miR-107 to Relieve Its Inhibition of Wnt3a. Mol. Ther. Nucleic Acids 2018, 11, 272–283. [Google Scholar] [CrossRef] [Green Version]
- Feng, Y.; Yang, Y.; Zhao, X.; Fan, Y.; Zhou, L.; Rong, J.; Yu, Y. Circular RNA circ0005276 promotes the proliferation and migration of prostate cancer cells by interacting with FUS to transcriptionally activate XIAP. Cell Death Dis. 2019, 10, 792. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Li, Y.; Yu, M. CircRNA ZNF609 Knockdown Suppresses Cell Growth via Modulating miR-188/ELF2 Axis in Nasopharyngeal Carcinoma. OncoTargets Ther. 2020, 13, 2399–2409. [Google Scholar] [CrossRef] [Green Version]
- Guo, H.X.; Yuan, B.; Su, M.T.; Zheng, Y.; Zhang, J.B. Identification of Circular RNAs in the Anterior Pituitary in Rats Treated with GnRH. Animals 2021, 11, 2557. [Google Scholar] [CrossRef] [PubMed]
- Han, D.X.; Wang, C.J.; Sun, X.L.; Liu, J.B.; Zhang, J.B. Identification of circular RNAs in the immature and mature rat anterior pituitary. J. Endocrinol. 2019, 240, 393–402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.; Li, X.; Ma, Q.; Zhang, X.; Cao, Y.; Yao, Y.; You, S.; Wang, D.; Quan, R.; Hou, X. Genome-wide analysis of circular RNAs in prenatal and postnatal pituitary glands of sheep. Sci. Rep. 2017, 7, 16143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Hu, B.; Xiong, J.; Chen, T.; Zhang, Y. Genomewide analysis of circular RNA in pituitaries of normal and heat-stressed sows. BMC Genom. 2019, 20, 1013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiong, J.; Zhang, H.; Wang, Y.; Cheng, Y.; Luo, J.; Chen, T.; Xi, Q.; Sun, J.; Zhang, Y. Rno_circ_0001004 Acts as a miR-709 Molecular Sponge to Regulate the Growth Hormone Synthesis and Cell Proliferation. Int. J. Mol. Sci. 2022, 23, 1413. https://doi.org/10.3390/ijms23031413
Xiong J, Zhang H, Wang Y, Cheng Y, Luo J, Chen T, Xi Q, Sun J, Zhang Y. Rno_circ_0001004 Acts as a miR-709 Molecular Sponge to Regulate the Growth Hormone Synthesis and Cell Proliferation. International Journal of Molecular Sciences. 2022; 23(3):1413. https://doi.org/10.3390/ijms23031413
Chicago/Turabian StyleXiong, Jiali, Haojie Zhang, Yuxuan Wang, Yunyun Cheng, Junyi Luo, Ting Chen, Qianyun Xi, Jiajie Sun, and Yongliang Zhang. 2022. "Rno_circ_0001004 Acts as a miR-709 Molecular Sponge to Regulate the Growth Hormone Synthesis and Cell Proliferation" International Journal of Molecular Sciences 23, no. 3: 1413. https://doi.org/10.3390/ijms23031413
APA StyleXiong, J., Zhang, H., Wang, Y., Cheng, Y., Luo, J., Chen, T., Xi, Q., Sun, J., & Zhang, Y. (2022). Rno_circ_0001004 Acts as a miR-709 Molecular Sponge to Regulate the Growth Hormone Synthesis and Cell Proliferation. International Journal of Molecular Sciences, 23(3), 1413. https://doi.org/10.3390/ijms23031413





