Genome-Wide Identification and Expression Analysis of the SUC Gene Family in Peanut (Arachis hypogaea L.) Reveals Its Role in Seed Sucrose Accumulation
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of SUC Genes in Peanut
2.2. Chromosomal Localization and Physicochemical Properties
2.3. Phylogenetic Analysis of the SUC Gene Family
2.4. Gene Structure and Analysis of Conserved Motifs
2.5. Protein Structure Analysis of Peanut AhSUCs
2.6. Promoter Cis-Element Analysis
2.7. Collinearity Analysis
2.8. Gene Expression Analysis
2.9. Plant Materials and Determination of Sucrose Content
2.10. RNA Isolation and Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
2.11. Verification of Sucrose Transport Capacity of Peanut AhSUC
2.12. Statistical Analysis
3. Results
3.1. Identification and Physicochemical Analysis of SUC Genes in Peanut
3.2. Analysis of AhSUC Protein Motifs and Gene Structure
3.3. Phylogenetic Tree Analysis in Peanut
3.4. Prediction and Analysis of Peanut AhSUC Protein Structure
3.5. Cis-Acting Elements in the AhSUC Gene Promoter
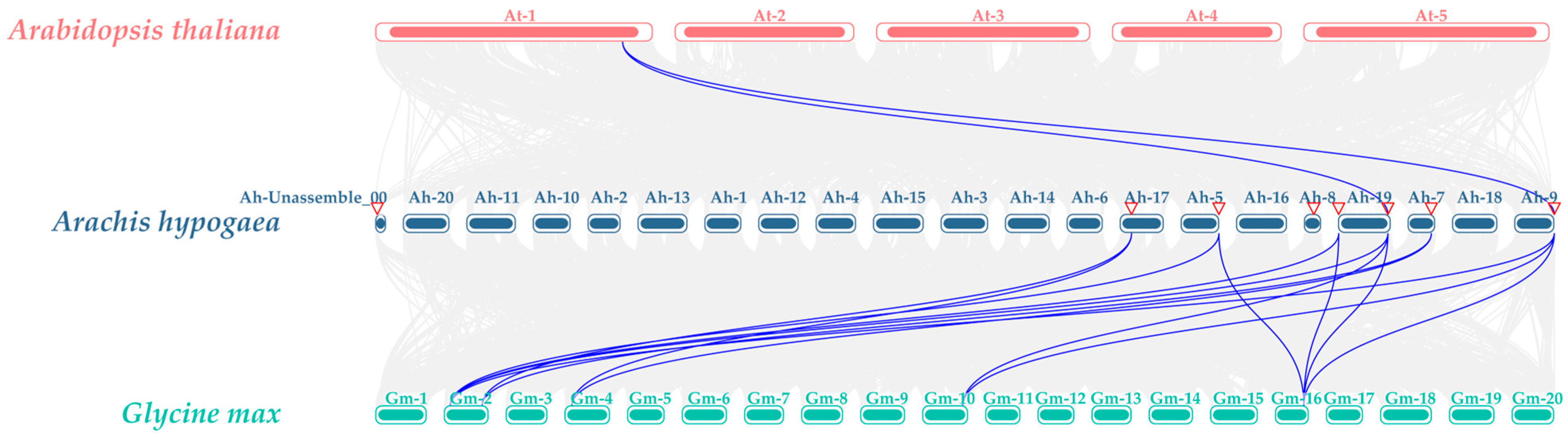
3.6. AhSUC Gene Collinearity Analysis
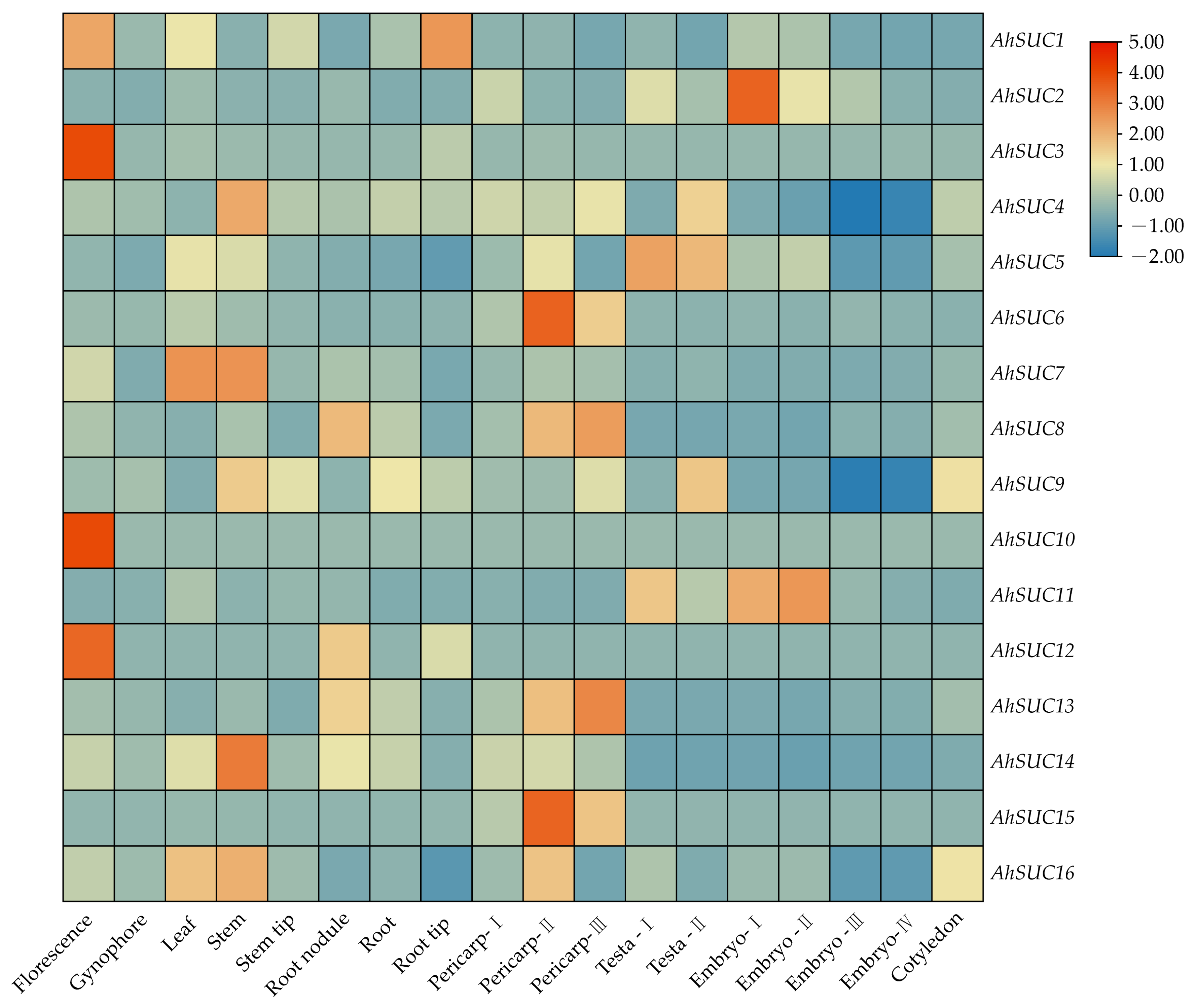
3.7. Analysis of AhSUC Gene Expression Datasets
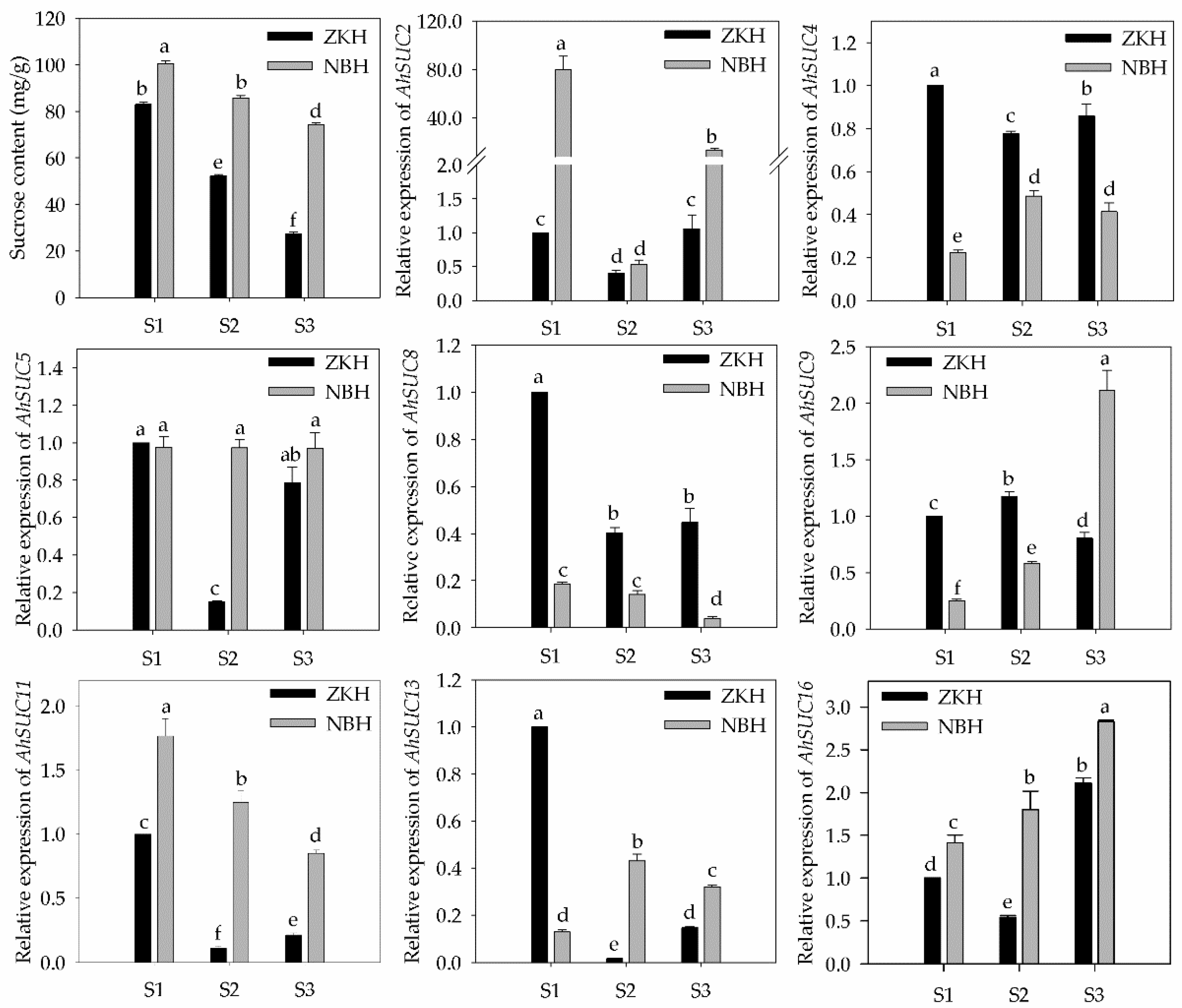
3.8. Sucrose Content and AhSUC Expression During Peanut Development
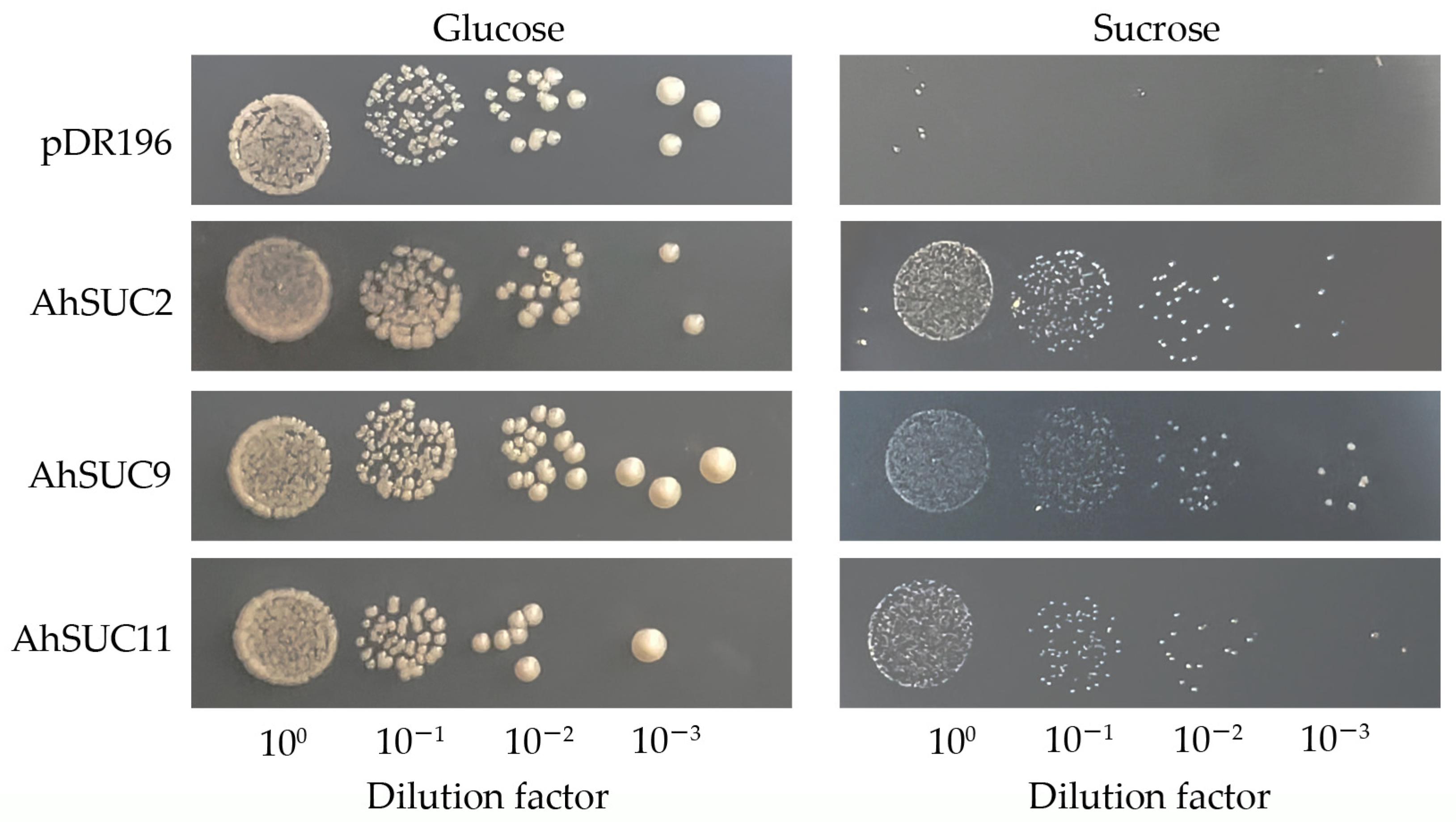
3.9. Sugar Transport Capacity of Peanut AhSUC9
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| SUT | Sucrose transporters |
| SUC | Sucrose/H+ co-transporters |
| SWEET | Sugars Will Eventually Be Exported Transporters |
| MST | Monosaccharide Transporters |
| TM | Transmembrane |
| MFS | Major facilitator superfamily |
| NJ | Neighbor-joining |
| MCScanX | Multiple collinearity scanning tool |
| qRT-PCR | Quantitative Reverse Transcription Polymerase Chain Reaction |
| GPH_sucrose | Conserved sucrose/proton co-transporter |
| GRAVY | Grand Average of Hydropathy |
| PI | Isoelectric Point |
| NO. of TM | Number of Transmembrane Domains. |
| ABRE | Abscisic acid response element |
References
- Bertioli, D.J.; Jenkins, J.; Clevenger, J.; Dudchenko, O.; Gao, D.; Seijo, G.; Leal-Bertioli, S.C.M.; Ren, L.; Farmer, A.D.; Pandey, M.K.; et al. The genome sequence of segmental allotetraploid peanut Arachis hypogaea. Nat. Genet. 2019, 51, 877–884. [Google Scholar] [CrossRef]
- Kassie, F.C.; Nguepjop, J.R.; Ngalle, H.B.; Assaha, D.V.M.; Gessese, M.K.; Abtew, W.G.; Tossim, H.A.; Sambou, A.; Seye, M.; Rami, J.F.; et al. An Overview of Mapping Quantitative Trait Loci in Peanut (Arachis hypogaea L.). Genes 2023, 14, 1176. [Google Scholar] [CrossRef]
- Pan, Y.; Zhuang, Y.; Liu, T.; Chen, H.; Wang, L.; Varshney, R.K.; Zhuang, W.; Wang, X. Deciphering peanut complex genomes paves a way to understand its origin and domestication. Plant Biotechnol. J. 2023, 21, 2173–2181. [Google Scholar] [CrossRef]
- Liu, Y.; Shao, L.; Zhou, J.; Li, R.; Pandey, M.K.; Han, Y.; Cui, F.; Zhang, J.; Guo, F.; Chen, J.; et al. Genomic insights into the genetic signatures of selection and seed trait loci in cultivated peanut. J. Adv. Res. 2022, 42, 237–248. [Google Scholar] [CrossRef]
- Mingrou, L.; Guo, S.; Ho, C.T.; Bai, N. Review on chemical compositions and biological activities of peanut (Arachis hypogeae L.). J. Food Biochem. 2022, 46, e14119. [Google Scholar] [CrossRef]
- Nunes, Y.C.; de Oliveira Santos, G.; Machado, N.M.; Otoboni, A.M.; Laurindo, L.F.; Bishayee, A.; Fimognari, C.; Bishayee, A.; Barbalho, S.M. Corrigendum to Peanut (Arachis hypogaea L.) seeds and by-products in metabolic syndrome and cardiovascular disorders: A systematic review of clinical studies Phytomedicine 123 (2024) 155170. Phytomedicine 2025, 136, 155870. [Google Scholar] [CrossRef] [PubMed]
- FAOSTAT. Available online: https://www.fao.org/faostat/zh/#data (accessed on 17 September 2025).
- Pattee, H.E.; Isleib, T.G.; Giesbrecht, F.G.; McFeeters, R.F. Investigations into genotypic variations of peanut carbohydrates. J. Agric. Food Chem. 2000, 48, 750–756. [Google Scholar] [CrossRef] [PubMed]
- Lei, Y.; Wang, Z.; Huai, D.; Gao, H.; Yan, L.; Li, J.; Li, W.; Chen, Y.; Kang, Y.; Liu, H.; et al. Development and application of a near infrared spectroscopy model for predicting high sucrose content of peanut seed. Acta Agron. Sin. 2020, 47, 332–341. (In Chinese) [Google Scholar] [CrossRef]
- Li, W.; Huang, L.; Liu, N.; Chen, Y.; Guo, J.; Yu, B.; Luo, H.; Zhou, X.; Huai, D.; Chen, W.; et al. Identification of a stable major sucrose-related QTL and diagnostic marker for flavor improvement in peanut. Theor. Appl. Genet. 2023, 136, 78. [Google Scholar] [CrossRef]
- Ma, Q.J.; Sun, M.H.; Liu, Y.J.; Lu, J.; Hu, D.G.; Hao, Y.J. Molecular cloning and functional characterization of the apple sucrose transporter gene MdSUT2. Plant Physiol. Biochem. 2016, 109, 442–451. [Google Scholar] [CrossRef]
- Pommerrenig, B.; Müdsam, C.; Kischka, D.; Neuhaus, H.E. Treat and trick: Common regulation and manipulation of sugar transporters during sink establishment by the plant and the pathogen. J. Exp. Bot. 2020, 71, 3930–3940. [Google Scholar] [CrossRef]
- Keller, I.; Rodrigues, C.M.; Neuhaus, H.E.; Pommerrenig, B. Improved resource allocation and stabilization of yield under abiotic stress. J. Plant Physiol. 2021, 257, 153336. [Google Scholar] [CrossRef]
- Baker, R.F.; Leach, K.A.; Boyer, N.R.; Swyers, M.J.; Benitez-Alfonso, Y.; Skopelitis, T.; Luo, A.; Sylvester, A.; Jackson, D.; Braun, D.M. Sucrose Transporter ZmSut1 Expression and Localization Uncover New Insights into Sucrose Phloem Loading. Plant Physiol. 2016, 172, 1876–1898. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Fang, W.; Peng, W.; Jiang, M.; Chen, G.; Xiong, F. Sucrose transporter in rice. Plant Signal Behav. 2021, 16, 1952373. [Google Scholar] [CrossRef]
- Dhungana, S.R.; Braun, D.M. Sugar transporters in grasses: Function and modulation in source and storage tissues. J. Plant Physiol. 2021, 266, 153541. [Google Scholar] [CrossRef]
- Julius, B.T.; Leach, K.A.; Tran, T.M.; Mertz, R.A.; Braun, D.M. Sugar Transporters in Plants: New Insights and Discoveries. Plant Cell Physiol. 2017, 58, 1442–1460. [Google Scholar] [CrossRef]
- Salvi, P.; Agarrwal, R.; Kajal; Gandass, N.; Manna, M.; Kaur, H.; Deshmukh, R. Sugar transporters and their molecular tradeoffs during abiotic stress responses in plants. Physiol. Plant 2022, 174, e13652. [Google Scholar] [CrossRef]
- Liang, Y.; Bai, J.; Xie, Z.; Lian, Z.; Guo, J.; Zhao, F.; Liang, Y.; Huo, H.; Gong, H. Tomato sucrose transporter SlSUT4 participates in flowering regulation by modulating gibberellin biosynthesis. Plant Physiol. 2023, 192, 1080–1098. [Google Scholar] [CrossRef]
- Sun, W.T.; Cheng, S.C.; Chao, Y.T.; Lin, S.Y.; Yang, T.T.; Ho, Y.P.; Shih, M.C.; Ko, S.S. Sugars and sucrose transporters in pollinia of Phalaenopsis aphrodite (Orchidaceae). J. Exp. Bot. 2023, 74, 2556–2571. [Google Scholar] [CrossRef] [PubMed]
- Aoki, N.; Hirose, T.; Scofield, G.N.; Whitfeld, P.R.; Furbank, R.T. The sucrose transporter gene family in rice. Plant Cell Physiol. 2003, 44, 223–232. [Google Scholar] [CrossRef] [PubMed]
- Reinders, A.; Sivitz, A.B.; Ward, J.M. Evolution of plant sucrose uptake transporters. Front. Plant Sci. 2012, 3, 22. [Google Scholar] [CrossRef]
- Lasin, P.; Weise, A.; Reinders, A.; Ward, J.M. Arabidopsis Sucrose Transporter AtSuc1 introns act as strong enhancers of expression. Plant Cell Physiol. 2020, 61, 1054–1063. [Google Scholar] [CrossRef]
- Wang, L.F.; Qi, X.X.; Huang, X.S.; Xu, L.L.; Jin, C.; Wu, J.; Zhang, S.L. Overexpression of sucrose transporter gene PbSUT2 from Pyrus bretschneideri, enhances sucrose content in Solanum lycopersicum fruit. Plant Physiol. Biochem. 2016, 105, 150–161. [Google Scholar] [CrossRef] [PubMed]
- Durand, M.; Mainson, D.; Porcheron, B.; Maurousset, L.; Lemoine, R.; Pourtau, N. Carbon source-sink relationship in Arabidopsis thaliana: The role of sucrose transporters. Planta 2018, 247, 587–611. [Google Scholar] [CrossRef]
- Rottmann, T.M.; Fritz, C.; Lauter, A.; Schneider, S.; Fischer, C.; Danzberger, N.; Dietrich, P.; Sauer, N.; Stadler, R. Protoplast-Esculin Assay as a New Method to Assay Plant Sucrose Transporters: Characterization of AtSUC6 and AtSUC7 Sucrose Uptake Activity in Arabidopsis Col-0 Ecotype. Front. Plant Sci. 2018, 9, 430. [Google Scholar] [CrossRef] [PubMed]
- Seitz, J.; Reimann, T.M.; Fritz, C.; Schröder, C.; Knab, J.; Weber, W.; Stadler, R. How pollen tubes fight for food: The impact of sucrose carriers and invertases of Arabidopsis thaliana on pollen development and pollen tube growth. Front. Plant Sci. 2023, 14, 1063765. [Google Scholar] [CrossRef] [PubMed]
- Leach, K.A.; Tran, T.M.; Slewinski, T.L.; Meeley, R.B.; Braun, D.M. Sucrose transporter2 contributes to maize growth, development, and crop yield. J. Integr. Plant Biol. 2017, 59, 390–408. [Google Scholar] [CrossRef]
- Usha, B.; Bordoloi, D.; Parida, A. Diverse expression of sucrose transporter gene family in Zea mays. J. Genet. 2015, 94, 151–154. [Google Scholar] [CrossRef]
- Bihmidine, S.; Baker, R.F.; Hoffner, C.; Braun, D.M. Sucrose accumulation in sweet sorghum stems occurs by apoplasmic phloem unloading and does not involve differential Sucrose transporter expression. BMC Plant Biol. 2015, 15, 186. [Google Scholar] [CrossRef]
- Milne, R.J.; Reinders, A.; Ward, J.M.; Offler, C.E.; Patrick, J.W.; Grof, C.P.L. Contribution of sucrose transporters to phloem unloading within Sorghum bicolor stem internodes. Plant Signal Behav. 2017, 12, e1319030. [Google Scholar] [CrossRef]
- Hu, Z.; Tang, Z.; Zhang, Y.; Niu, L.; Yang, F.; Zhang, D.; Hu, Y. Rice SUT and SWEET Transporters. Int. J. Mol. Sci. 2021, 22, 11198. [Google Scholar] [CrossRef]
- Li, Q.; Zhang, C.; Wen, J.; Chen, L.; Shi, Y.; Yang, Q.; Li, D. Transcriptome Analyses Show Changes in Gene Expression Triggered by a 31-bp InDel within OsSUT3 5′UTR in Rice Panicle. Int. J. Mol. Sci. 2023, 24, 10640. [Google Scholar] [CrossRef]
- Liu, H.T.; Ji, Y.; Liu, Y.; Tian, S.H.; Gao, Q.H.; Zou, X.H.; Yang, J.; Dong, C.; Tan, J.H.; Ni, D.A.; et al. The sugar transporter system of strawberry: Genome-wide identification and expression correlation with fruit soluble sugar-related traits in a Fragaria × ananassa germplasm collection. Hortic. Res. 2020, 7, 132. [Google Scholar] [CrossRef]
- Zhang, B.; Fan, W.; Zhu, Z.; Wang, Y.; Zhao, Z. Functional analysis of MdSUT2. 1, a plasma membrane sucrose transporter from apple. J. Integr. Agric. 2023, 22, 762–775. [Google Scholar] [CrossRef]
- Yang, X.; Yang, W.; Li, J.; Chen, C.; Chen, S.; Wang, H.; Wu, J.; Xue, H.; Liu, Y.; Lu, J.; et al. Major Facilitator Superfamily transporters balance sugar metabolism in peach. Plant Physiol. 2025, 198, kiaf192. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, W.; Chen, H.; Yang, M.; Wang, J.; Pandey, M.K.; Zhang, C.; Chang, W.C.; Zhang, L.; Zhang, X.; Tang, R. The genome of cultivated peanut provides insight into legume karyotypes, polyploid evolution and crop domestication. Nat. Genet. 2019, 51, 865–876. [Google Scholar] [CrossRef] [PubMed]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- He, Q.; He, L.; Feng, Z.; Liu, Y.; Xiao, Y.; Liu, J.; Han, H.; Huang, X. Role of BraSWEET12 in regulating flowering through sucrose transport in flowering Chinese cabbage. Horticulturae 2024, 10, 1037. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Drew, D.; North, R.A.; Nagarathinam, K.; Tanabe, M. Structures and General Transport Mechanisms by the Major Facilitator Superfamily (MFS). Chem. Rev. 2021, 121, 5289–5335. [Google Scholar] [CrossRef] [PubMed]
- Yadav, U.P.; Evers, J.F.; Shaikh, M.A.; Ayre, B.G. Cotton phloem loads from the apoplast using a single member of its nine-member sucrose transporter gene family. J. Exp. Bot. 2022, 73, 848–859. [Google Scholar] [CrossRef]
- Panchy, N.; Lehti-Shiu, M.; Shiu, S.H. Evolution of Gene Duplication in Plants. Plant Physiol. 2016, 171, 2294–2316. [Google Scholar] [CrossRef]
- Zhang, Q.; Hu, W.; Zhu, F.; Wang, L.; Yu, Q.; Ming, R.; Zhang, J. Structure, phylogeny, allelic haplotypes and expression of sucrose transporter gene families in Saccharum. BMC Genom. 2016, 17, 88. [Google Scholar] [CrossRef]
- Chen, W.; Diao, W.; Liu, H.; Guo, Q.; Song, Q.; Guo, G.; Wang, H.; Chen, Y. Molecular characterization of SUT gene family in Solanaceae with emphasis on expression analysis of pepper genes during development and stresses. Bioengineered 2022, 13, 14780–14798. [Google Scholar] [CrossRef]
- Zhang, Z.; Fan, T.; Wu, L.; Ye, Z.; Ye, F. Plant SUT Sucrose Transporters: Structure, Evolution and Biological Functions. Curr. Protein Pept. Sci. 2025, 26, e13892037372125. [Google Scholar] [CrossRef]
- Deng, B.; Gu, X.; Chen, S.; Zhang, M.; Hao, S.; Wei, L.; Cao, Y.; Hu, S. Genome-wide analysis and characterization of Dendrocalamus farinosus SUT gene family reveal DfSUT4 involvement in sucrose transportation in plants. Front. Plant Sci. 2023, 13, 1118398. [Google Scholar] [CrossRef]
- Fan, W.; Gao, H.; Zhang, L.; Mao, D.; Li, Y.; Zhang, L.; Li, J.; Zhao, X.; Hou, H. Genome-wide identification and expression profiling of MST, SUT and SWEET transporters in Dendrobium catenatum. BMC Genom. 2024, 25, 1213. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Zhang, Y.; Huo, X.; Xing, L.; Ge, M.; Suo, N. Genome-Wide Identification, Plasma Membrane Localization, and Functional Validation of the SUT Gene Family in Yam (Dioscorea cayennensis subsp. rotundata). Int. J. Mol. Sci. 2025, 26, 5756. [Google Scholar] [CrossRef]
- Jiang, S.; An, P.; Xia, C.; Ma, W.; Zhao, L.; Liang, T.; Liu, Q.; Xu, R.; Huang, D.; Xia, Z.; et al. Genome-wide identification and expression analysis of the SUT family from three species of Sapindaceae revealed their role in the accumulation of sugars in fruits. Plants 2024, 13, 95. [Google Scholar] [CrossRef] [PubMed]
- Rakkammal, K.; Muthuramalingam, P.; Shin, H.; Ramesh, M. Genome-wide identification and evolutionary analysis of SUT genes reveals key regulators of drought stress response in finger millet (Eleusine coracana). J. Genet. Eng. Biotechnol. 2025, 23, 100592. [Google Scholar] [CrossRef]
- Peng, C.C.; Xu, Y.H.; Xi, R.C.; Zhao, X.L. Expression, subcellular localization and phytohormone stimulation of a functional sucrose transporter (MdSUT1) in apple fruit. Sci. Hortic. 2011, 128, 206–212. [Google Scholar] [CrossRef]
- Ma, Q.J.; Sun, M.H.; Kang, H.; Lu, J.; You, C.X.; Hao, Y.J. A CIPK protein kinase targets sucrose transporter MdSUT2.2 at Ser254 for phosphorylation to enhance salt tolerance. Plant Cell Environ. 2019, 42, 918–930. [Google Scholar] [CrossRef]
- Long, J.; Zhou, H.; Huang, H.; Xiao, Y.; Luo, J.; Pu, Y.; Liu, Z.; Qiu, M.; Lu, X.; He, Y.; et al. The high-affinity pineapple sucrose transporter AcSUT1B, regulated by AcCBF1, exhibited enhanced cold tolerance in transgenic Arabidopsis. Int. J. Biol. Macromol. 2024, 283, 137952. [Google Scholar] [CrossRef]
- Chung, P.; Hsiao, H.H.; Chen, H.J.; Chang, C.W.; Wang, S.J. Influence of temperature on the expression of the rice sucrose transporter 4 gene, OsSUT4, in germinating embryos and maturing pollen. Acta Physiol. Plant 2014, 36, 217–229. [Google Scholar] [CrossRef]
- Li, M.; Wang, G.; Wu, Y.; Ren, Y.; Li, G.; Liu, Z.; Ding, Y.; Ceng, L. Function analysis of sucrose transporter OsSUT4 in sucrose transport in rice. Chin. J. Rice Sci. 2020, 34, 491–498. (In Chinese) [Google Scholar] [CrossRef]
- Baud, S.; Wuilleme, S.; Lemoine, R.; Kronenberger, J.; Caboche, M.; Lepiniec, L.; Rochat, C. The AtSUC5 sucrose transporter specifically expressed in the endosperm is involved in early seed development in Arabidopsis. Plant J. 2005, 43, 824–836. [Google Scholar] [CrossRef] [PubMed]
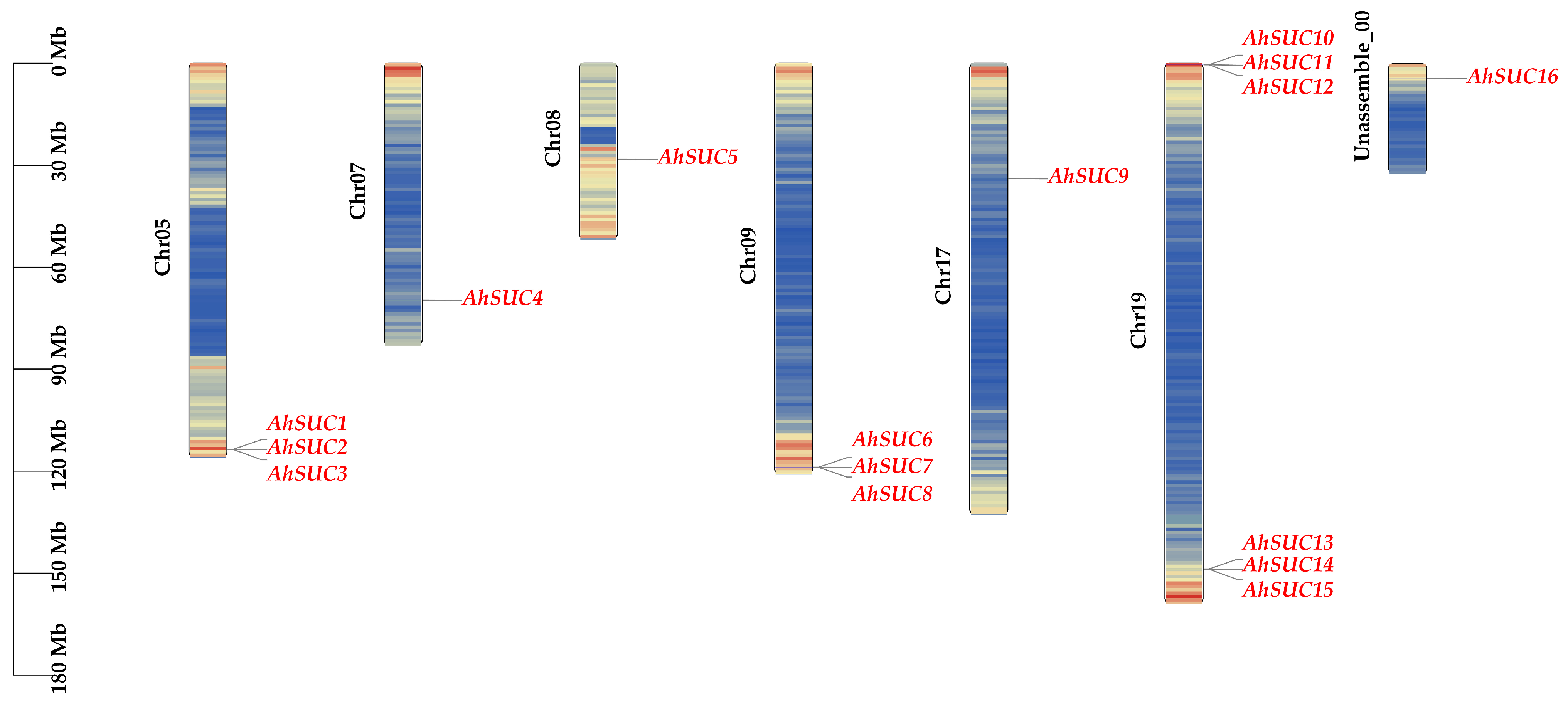
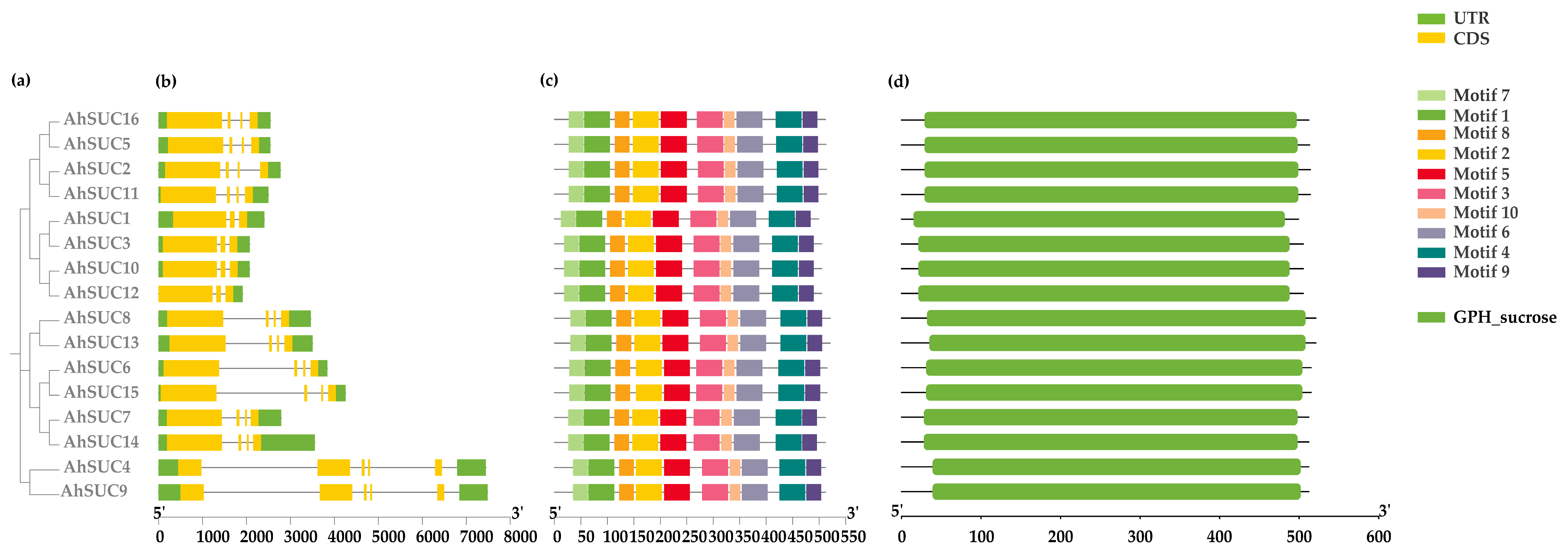
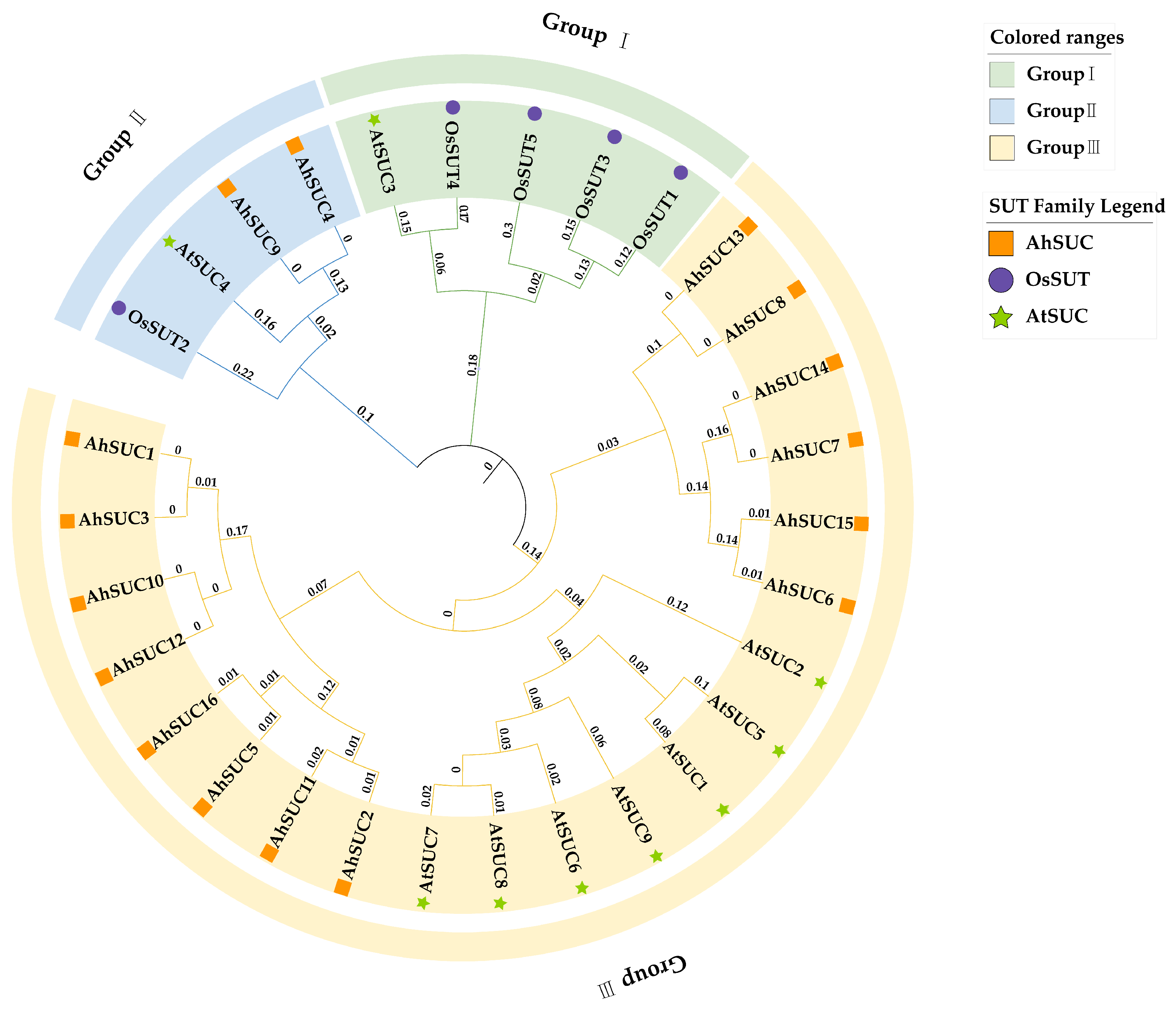


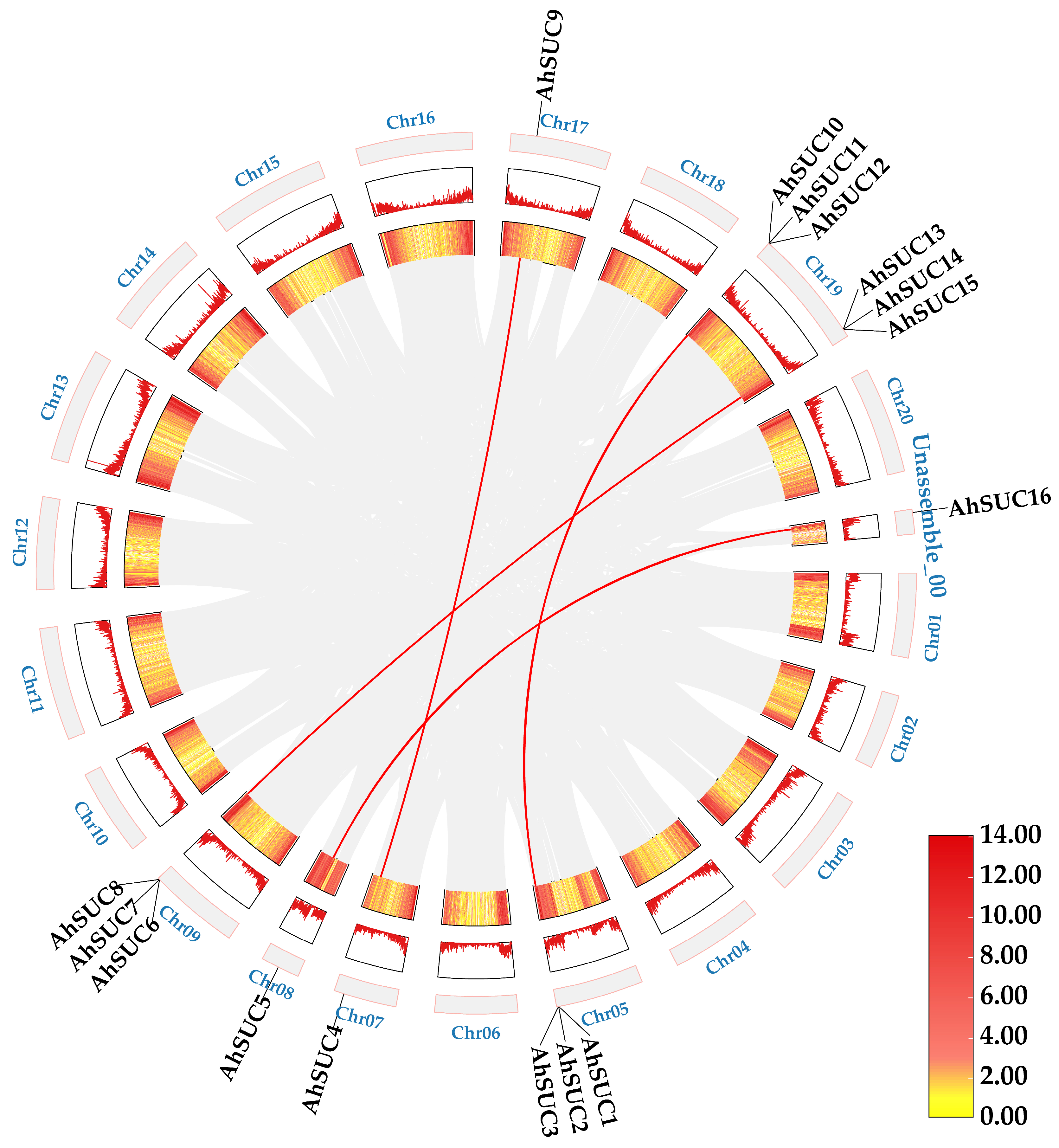
| Name | Cotton Identifier | Chromosome | Protein (aa) | PI | Molecular Weight (Da) | GRAVY | Subcellular Localization | No. of TM |
|---|---|---|---|---|---|---|---|---|
| AhSUC1 | AH05G37290.1 | chr5 | 499 | 8.37 | 52,994.20 | 0.596 | PM | 12 |
| AhSUC2 | AH05G37300.1 | chr5 | 514 | 9.24 | 54,698.10 | 0.494 | PM | 12 |
| AhSUC3 | AH05G37310.1 | chr5 | 505 | 8.07 | 53,622.84 | 0.576 | PM | 12 |
| AhSUC4 | AH07G21450.1 | chr7 | 512 | 9.53 | 54,806.05 | 0.525 | PM | 12 |
| AhSUC5 | AH08G13850.1 | chr8 | 513 | 9.11 | 54,251.44 | 0.514 | PM | 12 |
| AhSUC6 | AH09G33530.1 | chr9 | 515 | 9.13 | 54,319.01 | 0.550 | PM | 12 |
| AhSUC7 | AH09G33540.1 | chr9 | 512 | 8.99 | 54,441.84 | 0.580 | PM | 12 |
| AhSUC8 | AH09G33550.1 | chr9 | 521 | 9.07 | 54,985.48 | 0.496 | PM | 12 |
| AhSUC9 | AH17G15810.1 | chr17 | 512 | 9.53 | 54,806.05 | 0.525 | PM | 12 |
| AhSUC10 | AH19G00570.1 | chr19 | 505 | 7.58 | 53,636.82 | 0.580 | PM | 12 |
| AhSUC11 | AH19G00580.1 | chr19 | 514 | 9.08 | 54,606.95 | 0.509 | PM | 12 |
| AhSUC12 | AH19G00590.1 | chr19 | 505 | 7.58 | 53,593.80 | 0.590 | PM | 12 |
| AhSUC13 | AH19G33930.1 | chr19 | 521 | 9.07 | 55,019.54 | 0.494 | PM | 12 |
| AhSUC14 | AH19G33940.1 | chr19 | 512 | 9.08 | 54,596.05 | 0.572 | PM | 12 |
| AhSUC15 | AH19G33970.1 | chr19 | 515 | 9.13 | 54,304.06 | 0.555 | PM | 12 |
| AhSUC16 | AH00G03340.1 | Unassemble-00 | 512 | 9.12 | 54,136.45 | 0.531 | PM | 12 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Feng, Z.; He, Q.; Zheng, Y.; Zhang, Y.; Chen, X.; Liu, J.; Huang, X. Genome-Wide Identification and Expression Analysis of the SUC Gene Family in Peanut (Arachis hypogaea L.) Reveals Its Role in Seed Sucrose Accumulation. Curr. Issues Mol. Biol. 2026, 48, 29. https://doi.org/10.3390/cimb48010029
Feng Z, He Q, Zheng Y, Zhang Y, Chen X, Liu J, Huang X. Genome-Wide Identification and Expression Analysis of the SUC Gene Family in Peanut (Arachis hypogaea L.) Reveals Its Role in Seed Sucrose Accumulation. Current Issues in Molecular Biology. 2026; 48(1):29. https://doi.org/10.3390/cimb48010029
Chicago/Turabian StyleFeng, Zongqin, Qinqin He, Yixiong Zheng, Yu Zhang, Xiaolin Chen, Jiping Liu, and Xinmin Huang. 2026. "Genome-Wide Identification and Expression Analysis of the SUC Gene Family in Peanut (Arachis hypogaea L.) Reveals Its Role in Seed Sucrose Accumulation" Current Issues in Molecular Biology 48, no. 1: 29. https://doi.org/10.3390/cimb48010029
APA StyleFeng, Z., He, Q., Zheng, Y., Zhang, Y., Chen, X., Liu, J., & Huang, X. (2026). Genome-Wide Identification and Expression Analysis of the SUC Gene Family in Peanut (Arachis hypogaea L.) Reveals Its Role in Seed Sucrose Accumulation. Current Issues in Molecular Biology, 48(1), 29. https://doi.org/10.3390/cimb48010029






