Integrator Complex Subunit 6 Regulates Biological Nature of Hepatocellular Carcinoma by Modulating Epithelial–Mesenchymal Transition
Abstract
1. Introduction
2. Materials and Methods
2.1. Open Data Analysis
2.2. Cell Lines and Cell Culture
2.3. SaRNA/SiRNA Transfection
2.4. Gene Expression Analysis
2.5. Immunoblot Analysis
2.6. Migration and Invasion Assay
2.7. Scratch Assay
2.8. Cell-Cycle Analysis Using Flow Cytometry
2.9. Cell Proliferation Assay
2.10. Brdu Incorporation Assay
2.11. Assessment of Sorafenib Sensitivity in Huh7 Cells
2.12. Statistical Analysis
3. Results
3.1. Clinical Characteristics of INTS6 Expression in HCC
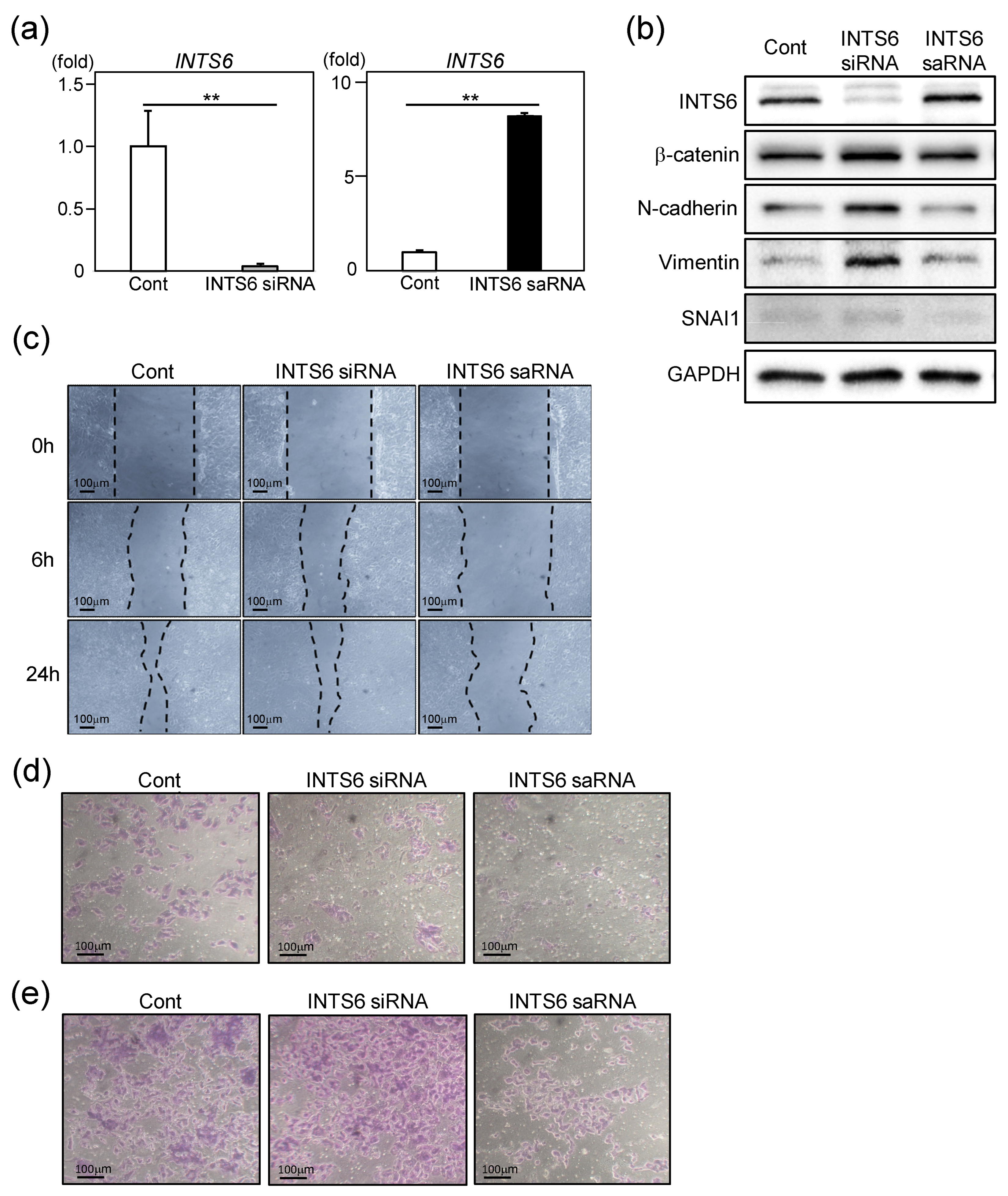
3.2. INTS6 Regulates EMT in HCC Cells
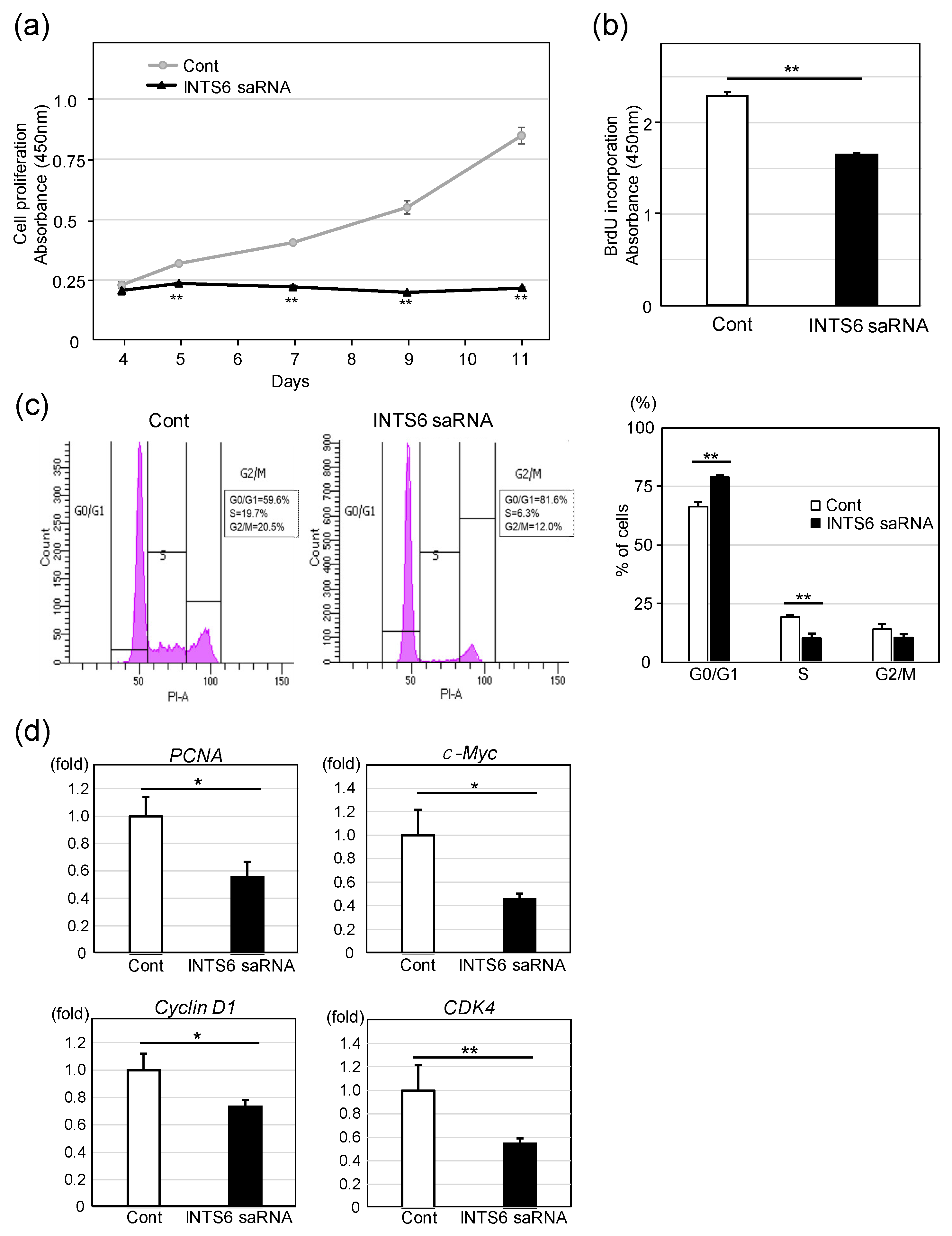
3.3. Upregulation of INTS6 Reduces HCC Cell Proliferation
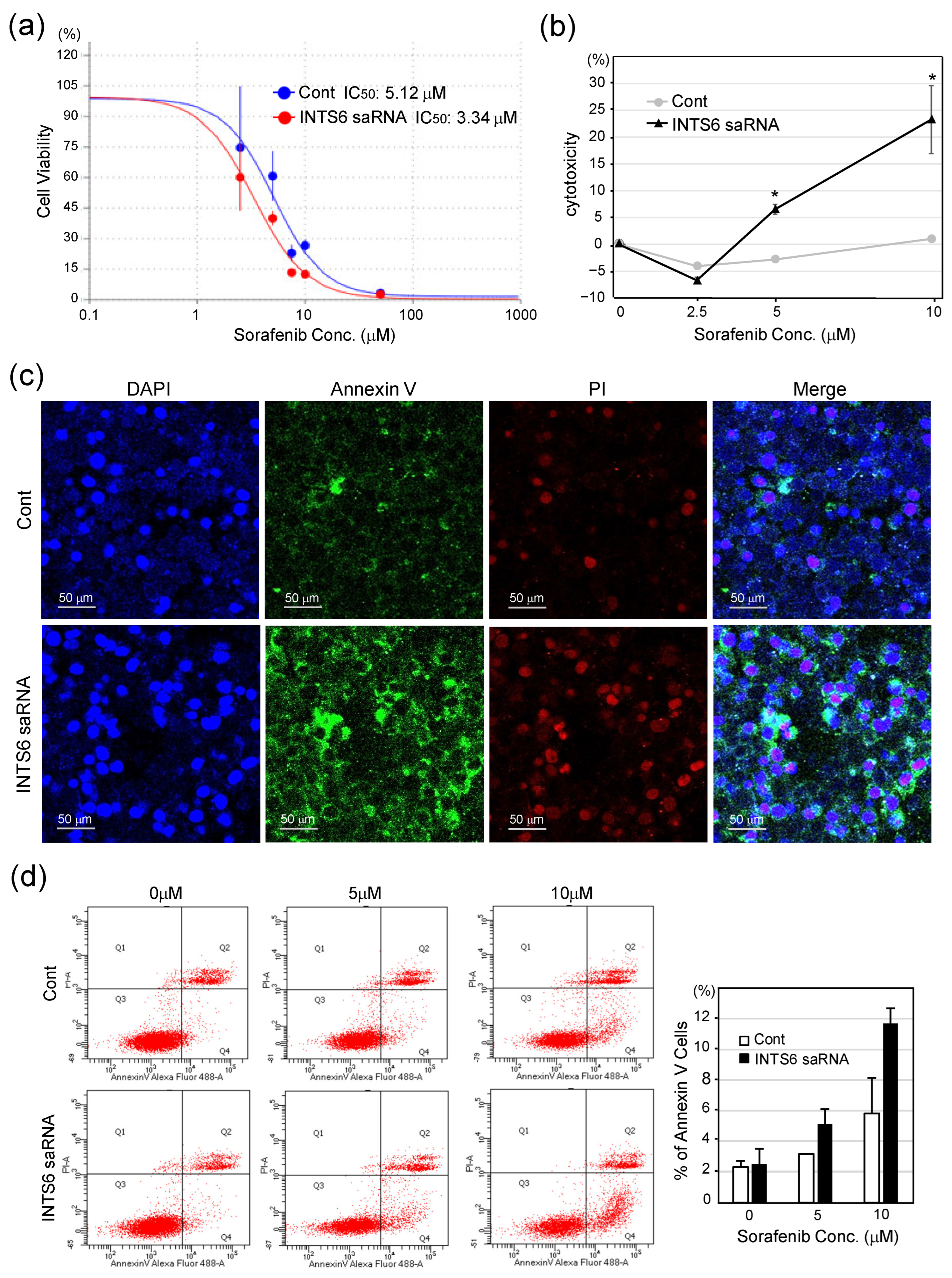
3.4. INTS6 Induction Enhances the Sensitivity of HCC Cells to Sorafenib
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CAR | chimeric antigen receptor |
| CCK-8 | cell-counting Kit-8 |
| CDK9 | cyclin-dependent kinase 9 |
| DFS | disease-free survival |
| EMT | epithelial–mesenchymal transition |
| GAPDH | glyceraldehyde-3-phosphate dehydrogenase |
| HCC | hepatocellular carcinoma |
| IC50 | half-maximal inhibitory concentration |
| INTS6 | integrator complex subunit 6 |
| LDH | lactate dehydrogenase |
| lncRNA | long non-coding RNAs |
| OS | overall survival |
| PI | propidium iodide |
| PP2A | protein phosphatase 2A |
| RNAPII | RNA polymerase II |
| saRNA | small activating RNA |
| SD | standard deviation |
| siRNA | small interfering RNA |
| TCGA | The Cancer Genome Atlas |
References
- Chakraborty, E.; Sarkar, D. Emerging Therapies for Hepatocellular Carcinoma (HCC). Cancers 2022, 14, 2798. [Google Scholar] [CrossRef]
- Llovet, J.M.; Kelley, R.K.; Villanueva, A.; Singal, A.G.; Pikarsky, E.; Roayaie, S.; Lencioni, R.; Koike, K.; Zucman-Rossi, J.; Finn, R.S. Hepatocellular carcinoma. Nat. Rev. Dis. Primers. 2021, 7, 6. [Google Scholar] [CrossRef]
- Villanueva, A.; Longo, D.L. Hepatocellular Carcinoma. New Engl. J. Med. 2019, 380, 1450–1462. [Google Scholar] [CrossRef] [PubMed]
- Galle, P.R.; Dufour, J.-F.; Peck-Radosavljevic, M.; Trojan, J.; Vogel, A. Systemic Therapy of Advanced Hepatocellular Carcinoma. Futur. Oncol. 2020, 17, 1237–1251. [Google Scholar] [CrossRef] [PubMed]
- Holohan, C.; Van Schaeybroeck, S.; Longley, D.B.; Johnston, P.G. Cancer drug resistance: An evolving paradigm. Nat. Rev. Cancer 2013, 13, 714–726. [Google Scholar] [CrossRef]
- Shi, D.; Shi, Y.; Kaseb, A.O.; Qi, X.; Zhang, Y.; Chi, J.; Lu, Q.; Gao, H.; Jiang, H.; Wang, H.; et al. Chimeric Antigen Receptor-Glypican-3 T-Cell Therapy for Advanced Hepatocellular Carcinoma: Results of Phase I Trials. Clin. Cancer Res. 2020, 26, 3979–3989. [Google Scholar] [CrossRef] [PubMed]
- Mino, M.; Kanno, K.; Okimoto, K.; Sugiyama, A.; Kishikawa, N.; Kobayashi, T.; Ono, J.; Izuhara, K.; Kobayashi, T.; Ohigashi, T.; et al. Periostin promotes malignant potential by induction of epithelial-mesenchymal transition in intrahepatic cholangiocarcinoma. Hepatol. Commun. 2017, 1, 1099–1109. [Google Scholar] [CrossRef]
- Negri, M.; Amatrudo, F.; Provvisiero, D.P.; Patalano, R.; Trinchese, G.; Cimmino, F.; de Angelis, C.; Simeoli, C.; Auriemma, R.S.; Mollica, M.P.; et al. PER2 expression and cellular localization play a critical role in tumor aggressiveness and drug resistance in an in vitro model of hepatocellular carcinoma. Cancer Drug Resist. 2025, 8, 26. [Google Scholar] [CrossRef]
- Hu, C.; Xin, Z.; Sun, X.; Hu, Y.; Zhang, C.; Yan, R.; Wang, Y.; Lu, M.; Huang, J.; Du, X.; et al. Activation of ACLY by SEC63 deploys metabolic reprogramming to facilitate hepatocellular carcinoma metastasis upon endoplasmic reticulum stress. J. Exp. Clin. Cancer Res. 2023, 42, 108. [Google Scholar] [CrossRef]
- Giannelli, G.; Koudelkova, P.; Dituri, F.; Mikulits, W. Role of epithelial to mesenchymal transition in hepatocellular carcinoma. J. Hepatol. 2016, 65, 798–808. [Google Scholar] [CrossRef]
- Klingenberg, M.; Matsuda, A.; Diederichs, S.; Patel, T. Non-coding RNA in hepatocellular carcinoma: Mechanisms, biomarkers and therapeutic targets. J. Hepatol. 2017, 67, 603–619. [Google Scholar] [CrossRef] [PubMed]
- Wan, T.; Zhang, T.; Si, X.; Zhou, Y. Overexpression of EMT-inducing transcription factors as a potential poor prognostic factor for hepatocellular carcinoma in Asian populations: A meta-analysis. Oncotarget 2017, 8, 59500–59508. [Google Scholar] [CrossRef] [PubMed]
- Wagner, E.J.; Tong, L.; Adelman, K. Integrator is a global promoter-proximal termination complex. Mol. Cell 2023, 83, 416–427. [Google Scholar] [CrossRef] [PubMed]
- Welsh, S.A.; Gardini, A. Genomic regulation of transcription and RNA processing by the multitasking Integrator complex. Nat. Rev. Mol. Cell Biol. 2022, 24, 204–220. [Google Scholar] [CrossRef]
- Vervoort, S.J.; Welsh, S.A.; Devlin, J.R.; Barbieri, E.; Knight, D.A.; Offley, S.; Bjelosevic, S.; Costacurta, M.; Todorovski, I.; Kearney, C.J.; et al. The PP2A-Integrator-CDK9 axis fine-tunes transcription and can be targeted therapeutically in cancer. Cell 2021, 184, 3143–3162.e32. [Google Scholar] [CrossRef]
- Sablina, A.A.; Hahn, W.C. The Role of PP2A A Subunits in Tumor Suppression. Cell Adhes. Migr. 2007, 1, 140–141. [Google Scholar] [CrossRef]
- Seshacharyulu, P.; Pandey, P.; Datta, K.; Batra, S.K. Phosphatase: PP2A structural importance, regulation and its aberrant expression in cancer. Cancer Lett. 2013, 335, 9–18. [Google Scholar] [CrossRef]
- Xu, Y.; Liao, W.; Wang, T.; Zhang, L.; Zhang, H. Comprehensive bioinformatics analysis of integrator complex subunits: Expression patterns, immune infiltration, and prognostic signature, validated through experimental approaches in hepatocellular carcinoma. Discov. Oncol. 2024, 15, 246. [Google Scholar] [CrossRef]
- Lui, K.Y.; Zhao, H.; Qiu, C.; Li, C.; Zhang, Z.; Peng, H.; Fu, R.; Chen, H.-A.; Lu, M.-Q. Integrator complex subunit 6 (INTS6) inhibits hepatocellular carcinoma growth by Wnt pathway and serve as a prognostic marker. BMC Cancer 2017, 17, 644. [Google Scholar] [CrossRef]
- Tong, H.; Liu, X.; Li, T.; Qiu, W.; Peng, C.; Shen, B.; Zhu, Z. INTS8 accelerates the epithelial-to-mesenchymal transition in hepatocellular carcinoma by upregulating the TGF-β signaling pathway. Cancer Manag. Res. 2019, 11, 1869–1879. [Google Scholar] [CrossRef]
- Jia, Y.; Cheng, Z.; Bharath, S.R.; Sun, Q.; Su, N.; Huang, J.; Song, H. Crystal structure of the INTS3/INTS6 complex reveals the functional importance of INTS3 dimerization in DSB repair. Cell Discov. 2021, 7, 66. [Google Scholar] [CrossRef]
- Yoon, S.; Rossi, J.J. Therapeutic Potential of Small Activating RNAs (saRNAs) in Human Cancers. Curr. Pharm. Biotechnol. 2018, 19, 604–610. [Google Scholar] [CrossRef]
- Tan, C.P.; Sinigaglia, L.; Gomez, V.; Nicholls, J.; Habib, N.A. RNA Activation—A Novel Approach to Therapeutically Upregulate Gene Transcription. Molecules 2021, 26, 6530. [Google Scholar] [CrossRef] [PubMed]
- Lachenmayer, A.; Alsinet, C.; Savic, R.; Cabellos, L.; Toffanin, S.; Hoshida, Y.; Villanueva, A.; Minguez, B.; Newell, P.; Tsai, H.-W.; et al. Wnt-Pathway Activation in Two Molecular Classes of Hepatocellular Carcinoma and Experimental Modulation by Sorafenib. Clin. Cancer Res. 2012, 18, 4997–5007. [Google Scholar] [CrossRef]
- Chen, H.; Shen, H.-X.; Lin, Y.-W.; Mao, Y.-Q.; Liu, B.; Xie, L.-P. Small RNA-induced INTS6 gene up-regulation suppresses castration-resistant prostate cancer cells by regulating β-catenin signaling. Cell Cycle 2018, 17, 1602–1613. [Google Scholar] [CrossRef]
- Wang, J.; Place, R.F.; Portnoy, V.; Huang, V.; Kang, M.R.; Kosaka, M.; Ho, M.K.C.; Li, L.-C. Inducing gene expression by targeting promoter sequences using small activating RNAs. J. Biol. Methods 2015, 2, e14. [Google Scholar] [CrossRef]
- Mir, N.; Jayachandran, A.; Dhungel, B.; Shrestha, R.; Steel, J.C. Epithelial-to-Mesenchymal Transition: A Mediator of Sorafenib Resistance in Advanced Hepatocellular Carcinoma. Curr. Cancer Drug Targets 2017, 17, 698–706. [Google Scholar] [CrossRef] [PubMed]
- Kudo, M.; Finn, R.S.; Qin, S.; Han, K.-H.; Ikeda, K.; Piscaglia, F.; Baron, A.; Park, J.-W.; Han, G.; Jassem, J.; et al. Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: A randomised phase 3 non-inferiority trial. Lancet 2018, 391, 1163–1173. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.S.; Ryoo, B.-Y.; Merle, P.; Kudo, M.; Bouattour, M.; Lim, H.Y.; Breder, V.; Edeline, J.; Chao, Y.; Ogasawara, S.; et al. Pembrolizumab As Second-Line Therapy in Patients With Advanced Hepatocellular Carcinoma in KEYNOTE-240: A Randomized, Double-Blind, Phase III Trial. J. Clin. Oncol. 2020, 38, 193–202. [Google Scholar] [CrossRef]
- Finn, R.S.; Qin, S.; Ikeda, M.; Galle, P.R.; Ducreux, M.; Kim, T.-Y.; Kudo, M.; Breder, V.; Merle, P.; Kaseb, A.O.; et al. Atezolizumab plus Bevacizumab in Unresectable Hepatocellular Carcinoma. N. Engl. J. Med. 2020, 382, 1894–1905. [Google Scholar] [CrossRef]
- Li, K.; Lin, S.Y.; Brunicardi, F.C.; Seu, P. Use of RNA interference to target cyclin E-overexpressing hepatocellular carcinoma. Cancer Res. 2003, 63, 3593–3597. [Google Scholar] [PubMed]
- Liu, J.-Y.; Chiang, T.; Liu, C.-H.; Chern, G.-G.; Lin, T.-T.; Gao, D.-Y.; Chen, Y. Delivery of siRNA Using CXCR4-targeted Nanoparticles Modulates Tumor Microenvironment and Achieves a Potent Antitumor Response in Liver Cancer. Mol. Ther. 2015, 23, 1772–1782. [Google Scholar] [CrossRef]
- Bogorad, R.L.; Yin, H.; Zeigerer, A.; Nonaka, H.; Ruda, V.M.; Zerial, M.; Anderson, D.G.; Koteliansky, V. Nanoparticle-formulated siRNA targeting integrins inhibits hepatocellular carcinoma progression in mice. Nat. Commun. 2014, 5, 3869. [Google Scholar] [CrossRef] [PubMed]
- Reebye, V.; Sætrom, P.; Mintz, P.J.; Huang, K.-W.; Swiderski, P.; Peng, L.; Liu, C.; Liu, X.; Lindkær-Jensen, S.; Zacharoulis, D.; et al. Novel RNA oligonucleotide improves liver function and inhibits liver carcinogenesis in vivo. Hepatology 2013, 59, 216–227. [Google Scholar] [CrossRef]
- Voutila, J.; Reebye, V.; Roberts, T.C.; Protopapa, P.; Andrikakou, P.; Blakey, D.C.; Habib, R.; Huber, H.; Saetrom, P.; Rossi, J.J.; et al. Development and Mechanism of Small Activating RNA Targeting CEBPA, a Novel Therapeutic in Clinical Trials for Liver Cancer. Mol. Ther. 2017, 25, 2705–2714. [Google Scholar] [CrossRef]
- Sarker, D.; Plummer, R.; Meyer, T.; Sodergren, M.H.; Basu, B.; Chee, C.E.; Huang, K.-W.; Palmer, D.H.; Ma, Y.T.; Evans, T.J.; et al. MTL-CEBPA, a Small Activating RNA Therapeutic Upregulating C/EBP-α, in Patients with Advanced Liver Cancer: A First-in-Human, Multicenter, Open-Label, Phase I Trial. Clin. Cancer Res. 2020, 26, 3936–3946. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Ishida, M.; Li, L.; Saito, A.; Kamiya, A.; Hamilton, J.P.; Fu, R.; Olaru, A.V.; An, F.; Popescu, I.; et al. Pseudogene INTS6P1 regulates its cognate gene INTS6 through competitive binding of miR-17-5p in hepatocellular carcinoma. Oncotarget 2015, 6, 5666–5677. [Google Scholar] [CrossRef]
- Suo, J.; Medina, D.; Herrera, S.; Zheng, Z.-Y.; Jin, L.; Chamness, G.C.; Contreras, A.; Gutierrez, C.; Hilsenbeck, S.; Umar, A.; et al. Int6 reduction activates stromal fibroblasts to enhance transforming activity in breast epithelial cells. Cell Biosci. 2015, 5, 10. [Google Scholar] [CrossRef]
- Federico, A.; Rienzo, M.; Abbondanza, C.; Costa, V.; Ciccodicola, A.; Casamassimi, A. Pan-Cancer Mutational and Transcriptional Analysis of the Integrator Complex. Int. J. Mol. Sci. 2017, 18, 936. [Google Scholar] [CrossRef]
- Galle, E.; Thienpont, B.; Cappuyns, S.; Venken, T.; Busschaert, P.; Van Haele, M.; Van Cutsem, E.; Roskams, T.; van Pelt, J.; Verslype, C.; et al. DNA methylation-driven EMT is a common mechanism of resistance to various therapeutic agents in cancer. Clin. Epigenetics 2020, 12, 27. [Google Scholar] [CrossRef]
- Filleur, S.; Hirsch, J.; Wille, A.; Schön, M.; Sell, C.; Shearer, M.H.; Nelius, T.; Wieland, I. INTS6/DICE1 inhibits growth of human androgen-independent prostate cancer cells by altering the cell cycle profile and Wnt signaling. Cancer Cell Int. 2009, 9, 28. [Google Scholar] [CrossRef] [PubMed]
- Oversoe, S.K.; Clement, M.S.; Weber, B.; Grønbæk, H.; Hamilton-Dutoit, S.J.; Sorensen, B.S.; Kelsen, J. Combining tissue and circulating tumor DNA increases the detection rate of a CTNNB1 mutation in hepatocellular carcinoma. BMC Cancer 2021, 21, 376. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yonezawa, S.; Kanno, K.; Shiozaki, M.; Sugiyama, M.; Ito, M. Integrator Complex Subunit 6 Regulates Biological Nature of Hepatocellular Carcinoma by Modulating Epithelial–Mesenchymal Transition. Curr. Issues Mol. Biol. 2025, 47, 733. https://doi.org/10.3390/cimb47090733
Yonezawa S, Kanno K, Shiozaki M, Sugiyama M, Ito M. Integrator Complex Subunit 6 Regulates Biological Nature of Hepatocellular Carcinoma by Modulating Epithelial–Mesenchymal Transition. Current Issues in Molecular Biology. 2025; 47(9):733. https://doi.org/10.3390/cimb47090733
Chicago/Turabian StyleYonezawa, Sayaka, Keishi Kanno, Minami Shiozaki, Masanori Sugiyama, and Masanori Ito. 2025. "Integrator Complex Subunit 6 Regulates Biological Nature of Hepatocellular Carcinoma by Modulating Epithelial–Mesenchymal Transition" Current Issues in Molecular Biology 47, no. 9: 733. https://doi.org/10.3390/cimb47090733
APA StyleYonezawa, S., Kanno, K., Shiozaki, M., Sugiyama, M., & Ito, M. (2025). Integrator Complex Subunit 6 Regulates Biological Nature of Hepatocellular Carcinoma by Modulating Epithelial–Mesenchymal Transition. Current Issues in Molecular Biology, 47(9), 733. https://doi.org/10.3390/cimb47090733






