Transcriptomic Analysis Reveals the Molecular Relationship Between Common Respiratory Infections and Parkinson’s Disease
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Retrieval
- (1)
- The datasets contain control samples to compare with.
- (2)
- Only blood samples were used because circulating mRNA molecules can originate from various tissues and organs, providing a systemic profile of gene expression.
- (3)
- Due to the limited accuracy of DNA microarray, we only included RNA-seq datasets.
- (4)
- We excluded datasets derived from samples treated with special clinical interventions.
2.2. Data Quality Control
2.3. Differential Gene Expression (DGE)
2.4. Weighted Correlation Network Analysis (WGCNA)
2.5. Protein-Protein Interactions (PPIs) and Hub Genes
2.6. Enrichment Analysis
2.7. Machine Learning Model (Random Forest)
3. Results
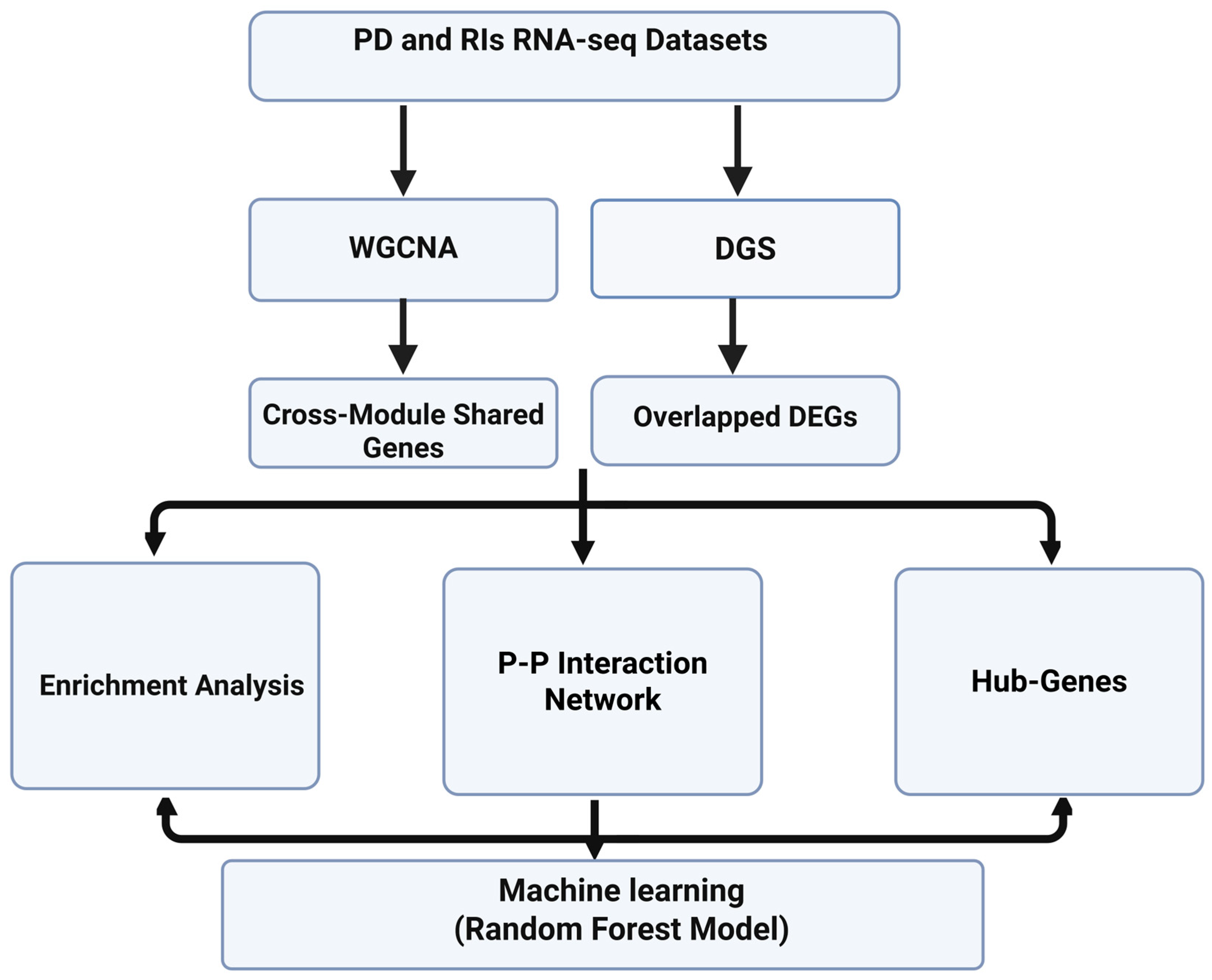
3.1. Study Workflow
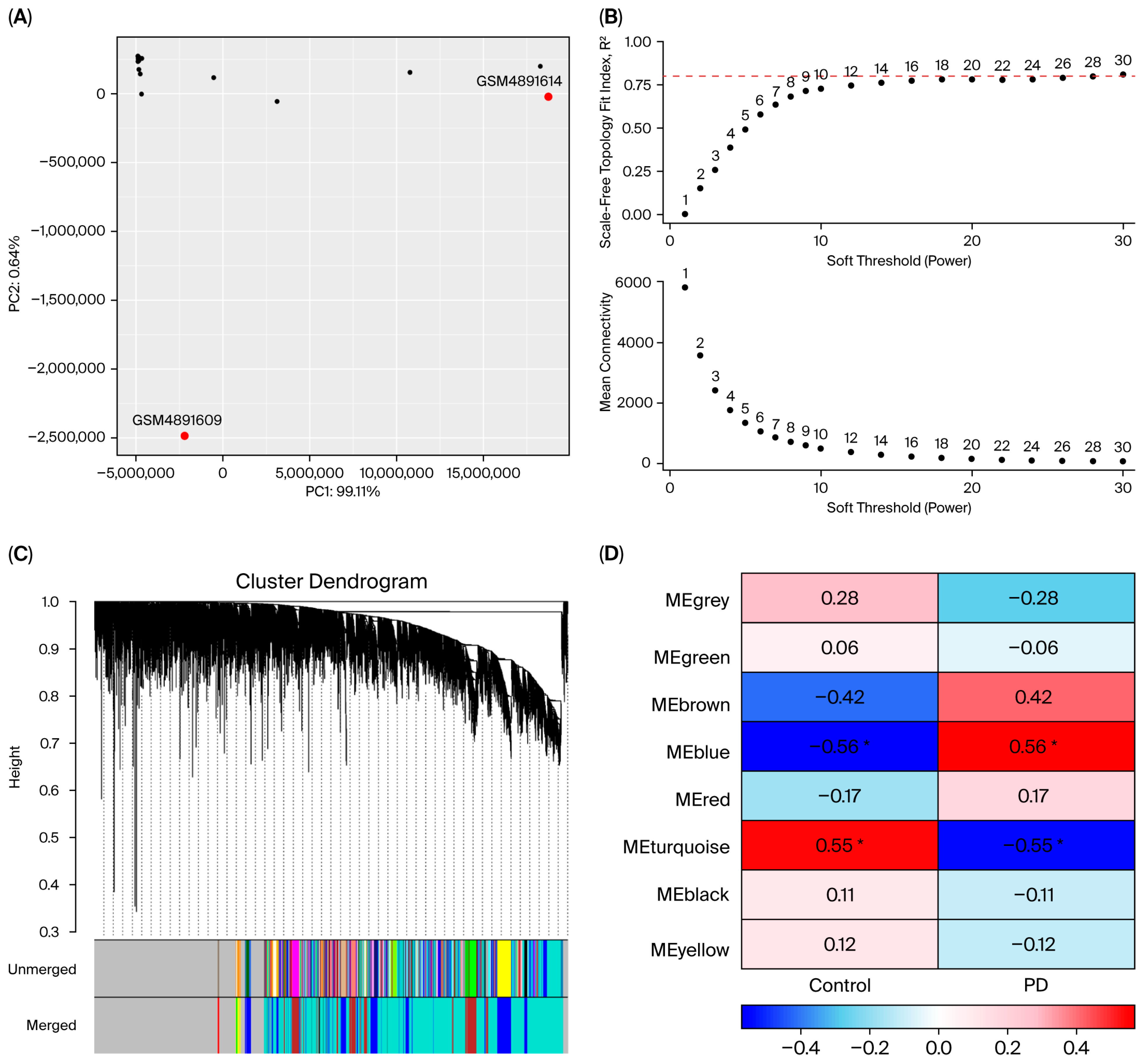
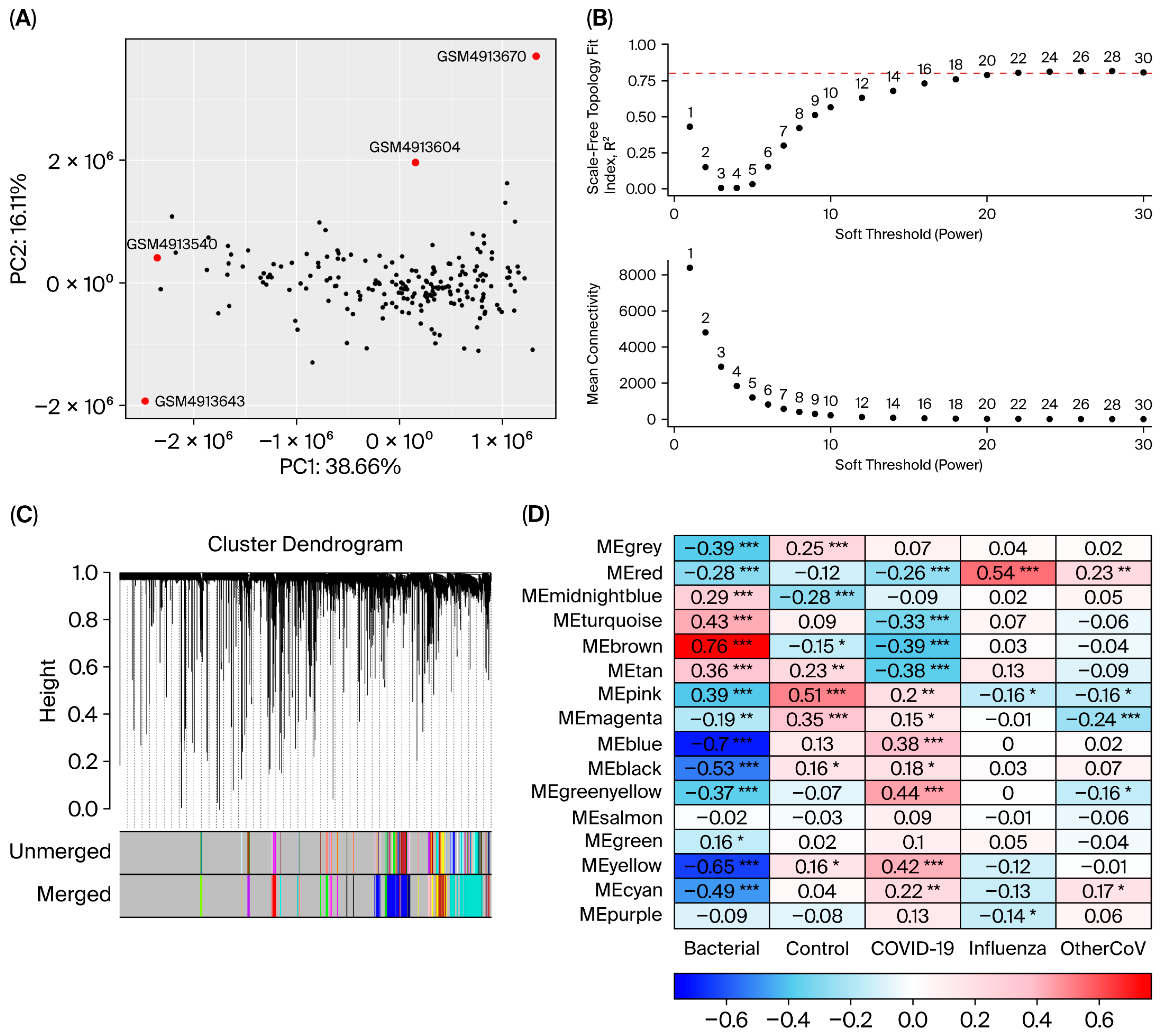
3.2. Quality Control
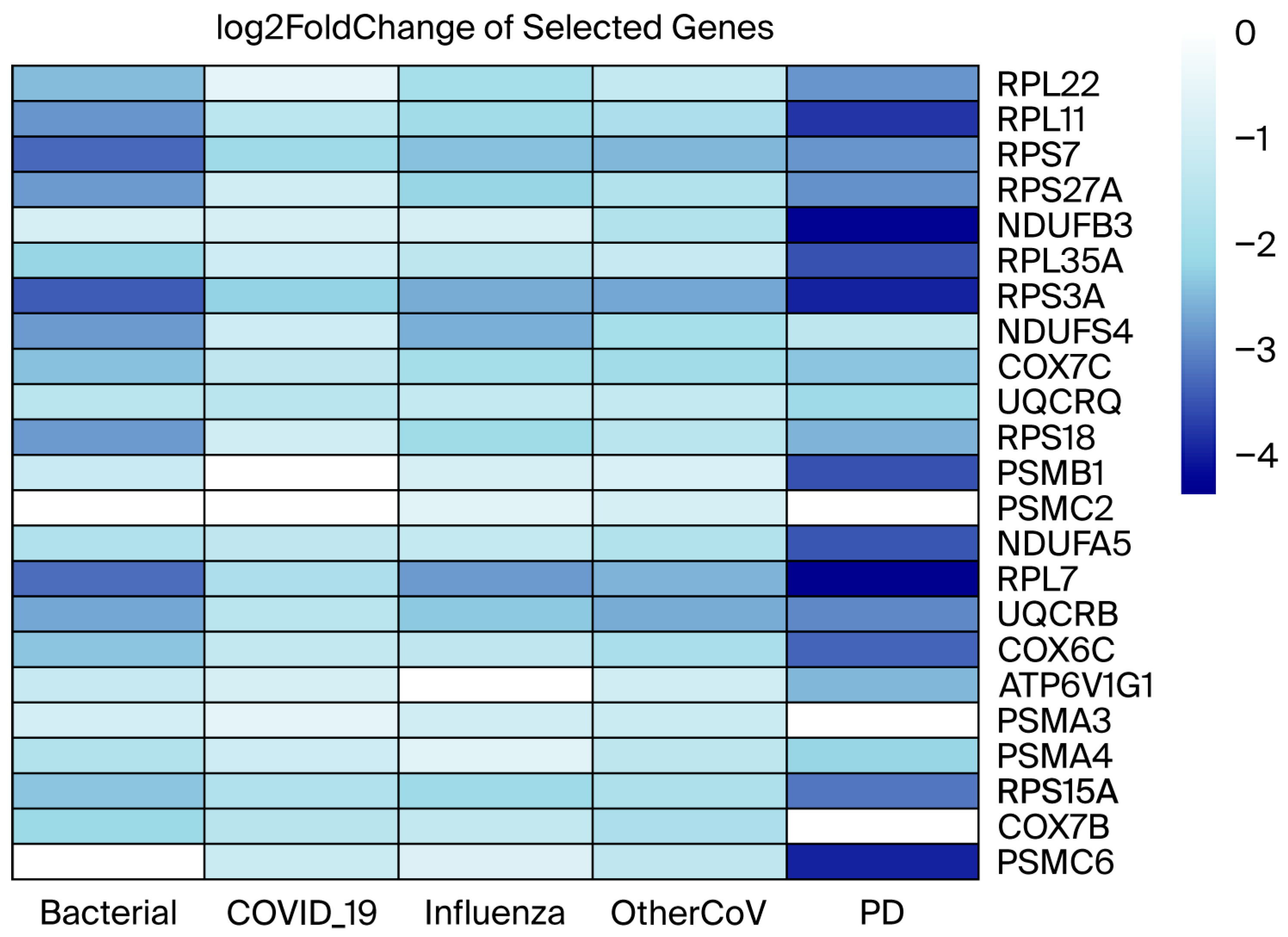
3.3. Differential Gene Expression and Overlapped DEGs
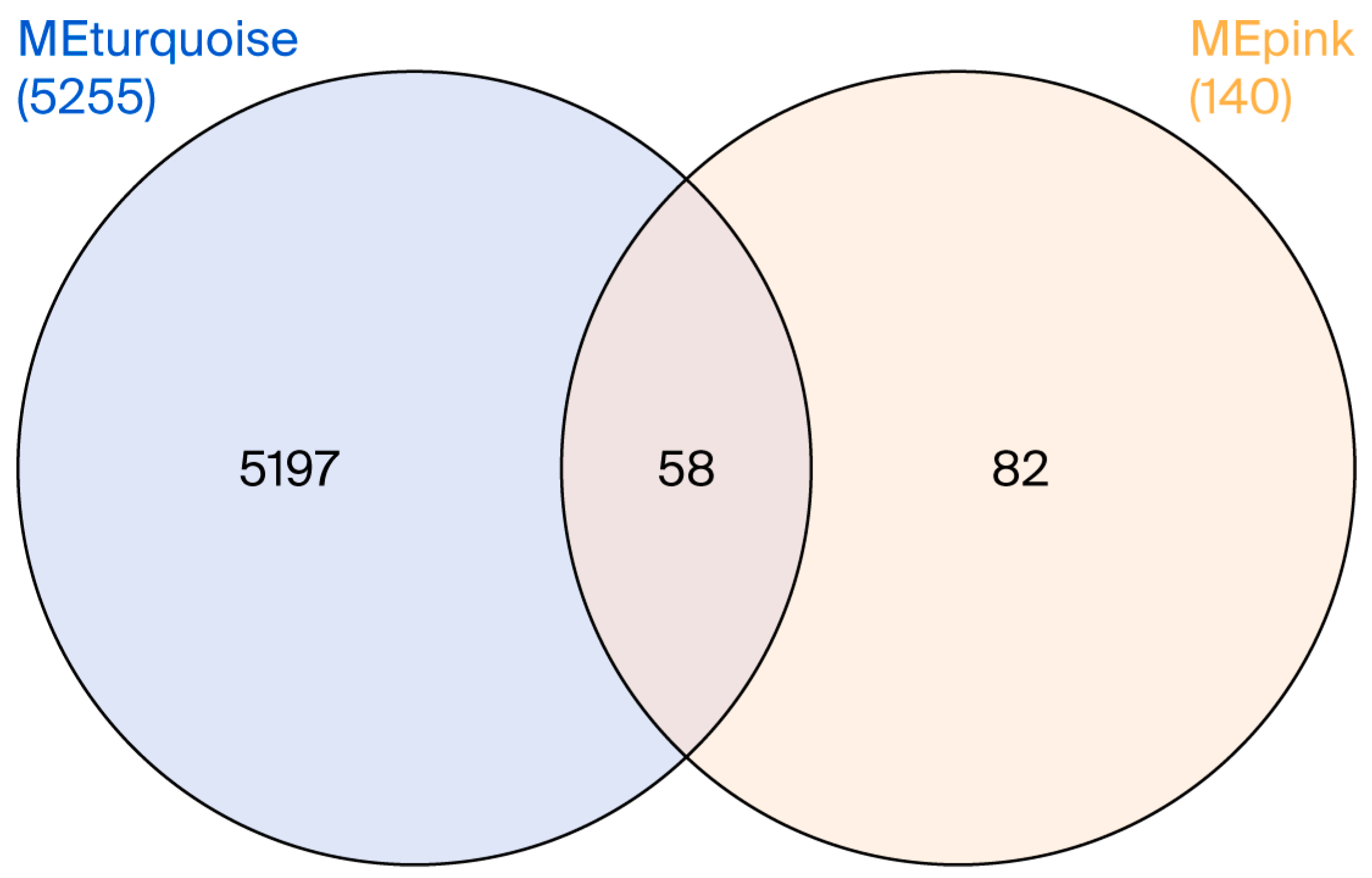
3.4. Co-Expression Modules
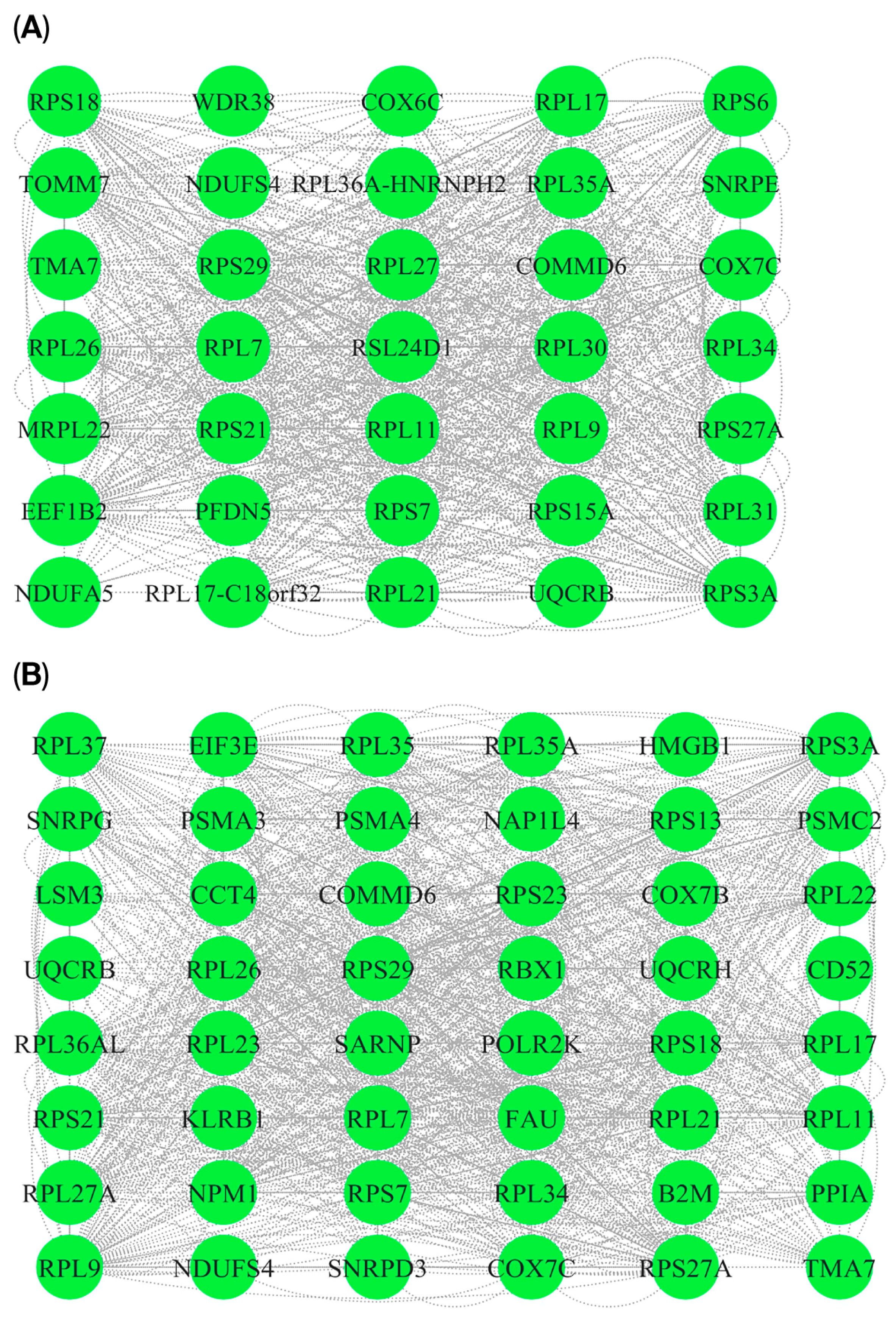
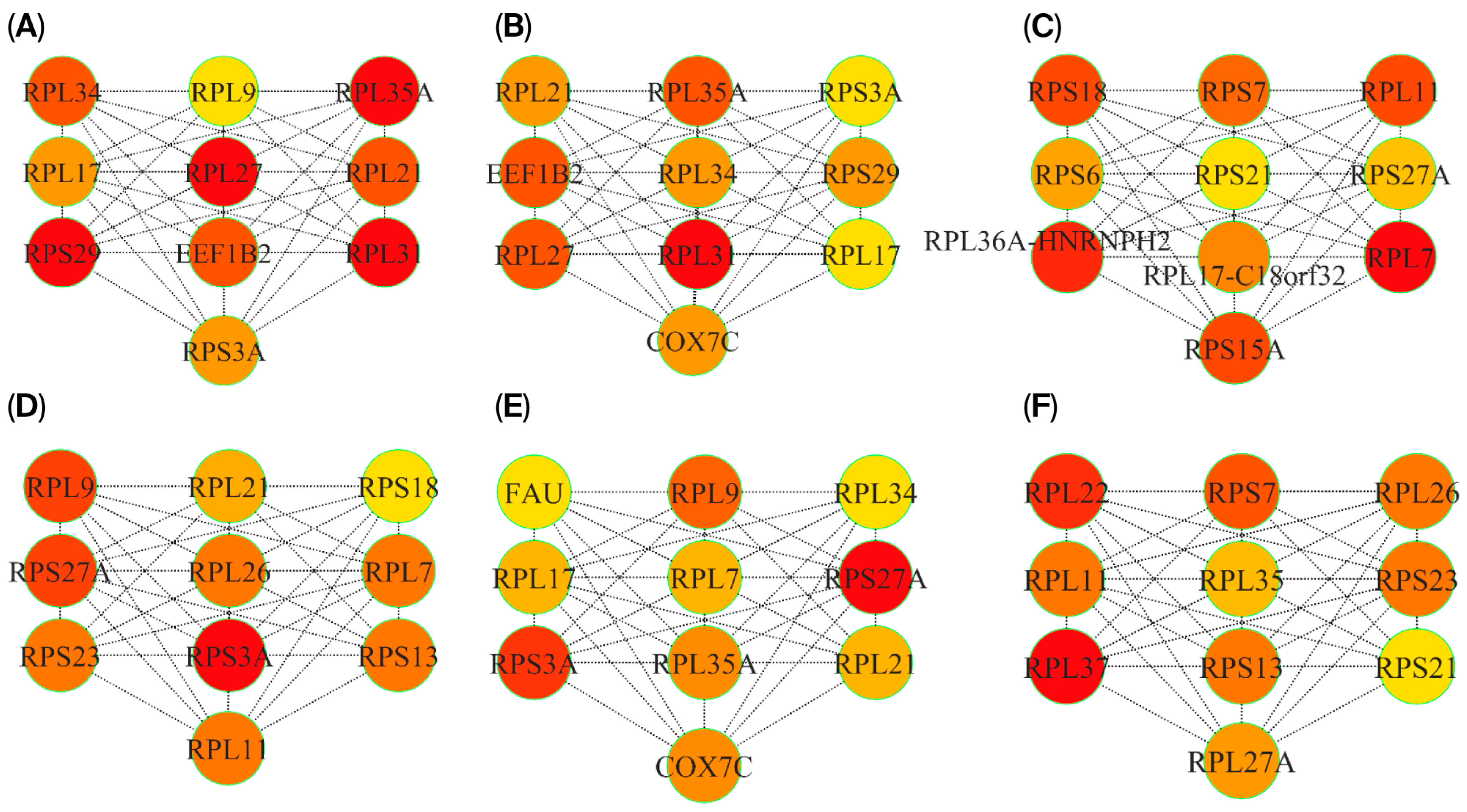
3.5. P-P Networks and HUB Genes
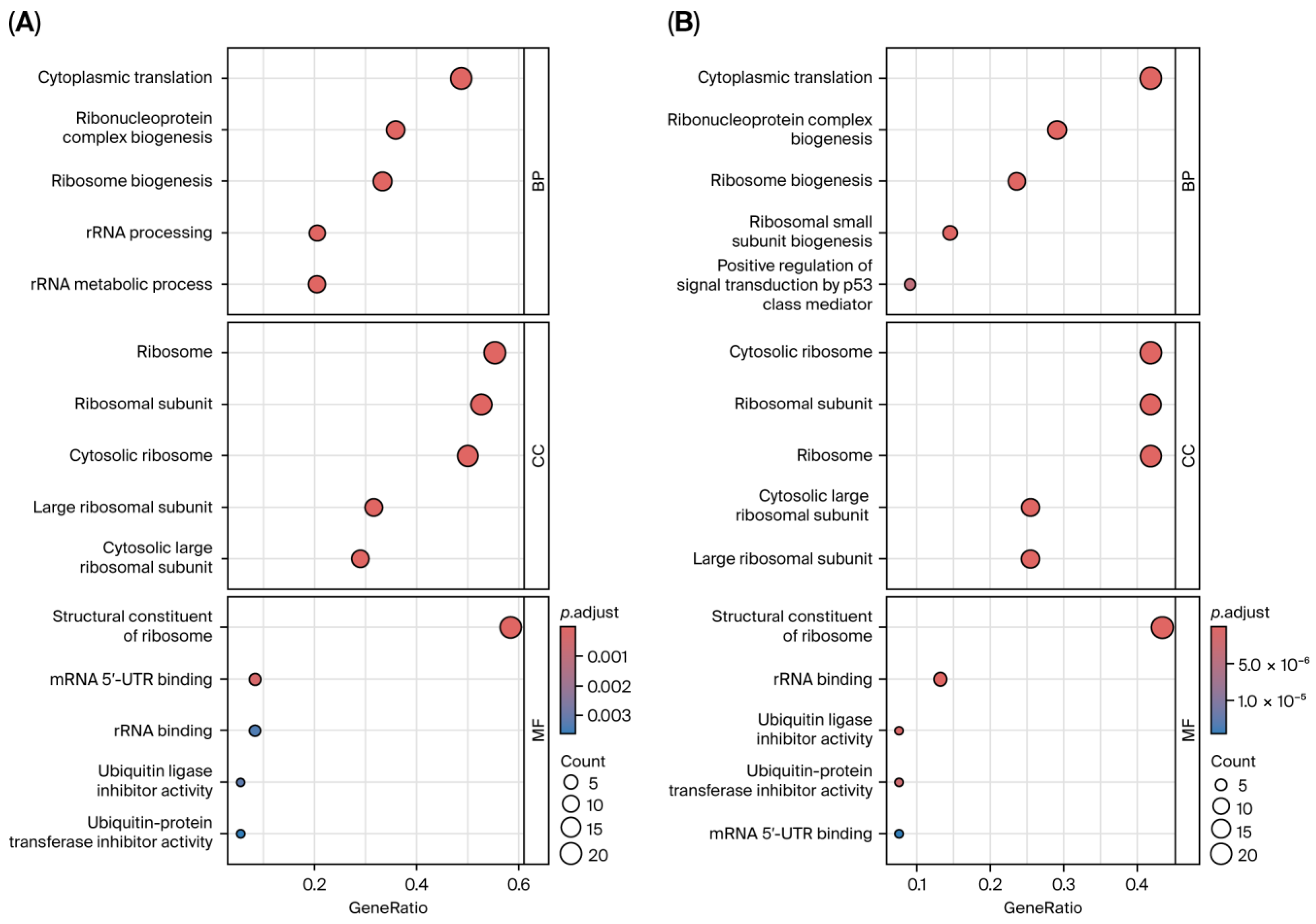
3.6. Functional and Pathway Enrichment Analysis
3.7. Random Forest Model
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PD | Parkinson’s disease |
| RIs | Respiratory infections |
| DEGs | Differentially expressed genes |
| DGE | Differential gene expression |
| PCA | Principle component analysis |
| GO | Gene ontology |
| RF | Random forest |
| CNS | Central nervus system |
| WGCNA | Weighted gene co-expression network analysis |
| PPI | Protein-protein interaction |
References
- Khan, A.U.; Akram, M.; Daniyal, M.; Zainab, R. Awareness and Current Knowledge of Parkinson’s Disease: A Neurodegenerative Disorder. Int. J. Neurosci. 2019, 129, 55–93. [Google Scholar] [CrossRef]
- Jankovic, J.; Tan, E.K. Parkinson’s Disease: Etiopathogenesis and Treatment. J. Neurol. Neurosurg. Psychiatry 2020, 91, 795–808. [Google Scholar] [CrossRef]
- Dorsey, E.R.; Sherer, T.; Okun, M.S.; Bloem, B.R. The Emerging Evidence of the Parkinson Pandemic. J. Park. Dis. 2018, 8 (Suppl. 1), S3–S8. [Google Scholar] [CrossRef]
- Chia, S.J.; Tan, E.-K.; Chao, Y.-X. Historical Perspective: Models of Parkinson’s Disease. Int. J. Mol. Sci. 2020, 21, 2464. [Google Scholar] [CrossRef] [PubMed]
- Mei, J.; Desrosiers, C.; Frasnelli, J. Machine Learning for the Diagnosis of Parkinson’s Disease: A Review of Literature. Front. Aging Neurosci. 2021, 13, 633752. [Google Scholar] [CrossRef]
- Limphaibool, N.; Iwanowski, P.; Holstad, M.J.V.; Kobylarek, D.; Kozubski, W. Infectious Etiologies of Parkinsonism: Pathomechanisms and Clinical Implications. Front. Neurol. 2019, 10, 652. [Google Scholar] [CrossRef]
- Tran, V.T.A.; Lee, L.P.; Cho, H. Neuroinflammation in Neurodegeneration via Microbial Infections. Front. Immunol. 2022, 13, 907804. [Google Scholar] [CrossRef]
- Klein, R.S.; Garber, C.; Howard, N. Infectious Immunity in the Central Nervous System and Brain Function. Nat. Immunol. 2017, 18, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Valerio, F.; Whitehouse, D.P.; Menon, D.K.; Newcombe, V.F.J. The Neurological Sequelae of Pandemics and Epidemics. J. Neurol. 2021, 268, 2629–2655. [Google Scholar] [CrossRef] [PubMed]
- DiSabato, D.J.; Quan, N.; Godbout, J.P. Neuroinflammation: The Devil Is in the Details. J. Neurochem. 2016, 139, 136–153. [Google Scholar] [CrossRef]
- McGeer, P.L.; Itagaki, S.; Boyes, B.E.; McGeer, E.G. Reactive Microglia Are Positive for HLA-DR in the Substantia Nigra of Parkinson’s and Alzheimer’s Disease Brains. Neurology 1988, 38, 1285–1291. [Google Scholar] [CrossRef]
- Nair, S.; Diamond, M.S. Innate Immune Interactions Within the Central Nervous System Modulate Pathogenesis of Viral Infections. Curr. Opin. Immunol. 2015, 36, 47–53. [Google Scholar] [CrossRef]
- Win, K.K.; Parmasivam, P.A. Immune Responses in Infections of the Central Nervous System. In Viral and Fungal Infections of the Central Nervous System: A Microbiological Perspective; Sami, H., Firoze, S., Khan, P.A., Eds.; Springer Nature: Singapore, 2023; pp. 59–71. ISBN 978-981-99-6444-4. [Google Scholar]
- Dowd, E.; McKernan, D.P. Back to the Future: Lessons from Past Viral Infections and the Link with Parkinson’s Disease. Neuronal Signal. 2021, 5, NS20200051. [Google Scholar] [CrossRef]
- Mori, I.; Nishiyama, Y.; Yokochi, T.; Kimura, Y. Olfactory Transmission of Neurotropic Viruses. J. Neurovirol. 2005, 11, 129–137. [Google Scholar] [CrossRef] [PubMed]
- Bang, Y.; Lim, J.; Choi, H.J. Recent Advances in the Pathology of Prodromal Non-Motor Symptoms Olfactory Deficit and Depression in Parkinson’s Disease: Clues to Early Diagnosis and Effective Treatment. Arch. Pharm. Res. 2021, 44, 588–604. [Google Scholar] [CrossRef]
- Braak, H.; Rüb, U.; Gai, W.P.; Del Tredici, K. Idiopathic Parkinson’s Disease: Possible Routes by Which Vulnerable Neuronal Types May Be Subject to Neuroinvasion by an Unknown Pathogen. J. Neural Transm. 2003, 110, 517–536. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Shrestha, S.; Huang, X.; Jain, S.; Guo, X.; Tranah, G.J.; Garcia, M.E.; Satterfield, S.; Phillips, C.; Harris, T.B.; et al. Olfaction and Incident Parkinson Disease in US White and Black Older Adults. Neurology 2017, 89, 1441–1447. [Google Scholar] [CrossRef]
- Doty, R.L.; Hawkes, C.H.; Good, K.P.; Duda, J.E. Odor Perception and Neuropathology in Neurodegenerative Diseases and Schizophrenia. In Handbook of Olfaction and Gustation; Doty, R.L., Ed.; Wiley: New York, NY, USA, 2015; pp. 403–452. ISBN 978-1-118-13922-6. [Google Scholar]
- Jafek, B.W.; Hartman, D.; Eller, P.M.; Johnson, E.W.; Strahan, R.C.; Moran, D.T. Postviral Olfactory Dysfunction. Am. J. Rhinol. 1990, 4, 91–100. [Google Scholar] [CrossRef]
- Doty, R.L. Olfactory Dysfunction in COVID-19: Pathology and Long-Term Implications for Brain Health. Trends Mol. Med. 2022, 28, 781–794. [Google Scholar] [CrossRef]
- Falsey, A.R. Neurologic Complications of Influenza and Potential Protective Vaccine Effects. Influenza Other Respir. Viruses 2025, 19, e70071. [Google Scholar] [CrossRef]
- Levine, K.S.; Leonard, H.L.; Blauwendraat, C.; Iwaki, H.; Johnson, N.; Bandres-Ciga, S.; Ferrucci, L.; Faghri, F.; Singleton, A.B.; Nalls, M.A. Virus Exposure and Neurodegenerative Disease Risk across National Biobanks. Neuron 2023, 111, 1086–1093.e2. [Google Scholar] [CrossRef]
- Marreiros, R.; Müller-Schiffmann, A.; Trossbach, S.V.; Prikulis, I.; Hänsch, S.; Weidtkamp-Peters, S.; Moreira, A.R.; Sahu, S.; Soloviev, I.; Selvarajah, S.; et al. Disruption of Cellular Proteostasis by H1N1 Influenza A Virus Causes α-Synuclein Aggregation. Proc. Natl. Acad. Sci. USA 2020, 117, 6741–6751. [Google Scholar] [CrossRef]
- Wichmann, D.; Sperhake, J.-P.; Lütgehetmann, M.; Steurer, S.; Edler, C.; Heinemann, A.; Heinrich, F.; Mushumba, H.; Kniep, I.; Schröder, A.S.; et al. Autopsy Findings and Venous Thromboembolism in Patients with COVID-19: A Prospective Cohort Study. Ann. Intern. Med. 2020, 173, 268–277. [Google Scholar] [CrossRef]
- Puelles, V.G.; Lütgehetmann, M.; Lindenmeyer, M.T.; Sperhake, J.P.; Wong, M.N.; Allweiss, L.; Chilla, S.; Heinemann, A.; Wanner, N.; Liu, S.; et al. Multiorgan and Renal Tropism of SARS-CoV-2. N. Engl. J. Med. 2020, 383, 590–592. [Google Scholar] [CrossRef] [PubMed]
- Calculli, A.; Bocci, T.; Porcino, M.; Avenali, M.; Casellato, C.; Arceri, S.; Regalbuto, S.; Priori, A.; Pisani, A. Parkinson Disease Following COVID-19: Report of Six Cases. Eur. J. Neurol. 2023, 30, 1272–1280. [Google Scholar] [CrossRef]
- Makhoul, K.; Jankovic, J. Parkinson’s Disease After COVID-19. J. Neurol. Sci. 2021, 422, 117331. [Google Scholar] [CrossRef] [PubMed]
- Cavallieri, F.; Fioravanti, V.; Toschi, G.; Grisanti, S.; Napoli, M.; Moratti, C.; Pascarella, R.; Versari, A.; Fraternali, A.; Casali, M.; et al. COVID-19 and Parkinson’s Disease: A Casual Association or a Possible Second Hit in Neurodegeneration? J. Neurol. 2022, 269, 59–61. [Google Scholar] [CrossRef] [PubMed]
- Cohen, M.E.; Eichel, R.; Steiner-Birmanns, B.; Janah, A.; Ioshpa, M.; Bar-Shalom, R.; Paul, J.J.; Gaber, H.; Skrahina, V.; Bornstein, N.M.; et al. A Case of Probable Parkinson’s Disease After SARS-CoV-2 Infection. Lancet Neurol. 2020, 19, 804–805. [Google Scholar] [CrossRef]
- Méndez-Guerrero, A.; Laespada-García, M.I.; Gómez-Grande, A.; Ruiz-Ortiz, M.; Blanco-Palmero, V.A.; Azcarate-Diaz, F.J.; Rábano-Suárez, P.; Álvarez-Torres, E.; De Fuenmayor-Fernández De La Hoz, C.P.; Vega Pérez, D.; et al. Acute Hypokinetic-Rigid Syndrome Following SARS-CoV-2 Infection. Neurology 2020, 95, e2109–e2118. [Google Scholar] [CrossRef]
- Douaud, G.; Lee, S.; Alfaro-Almagro, F.; Arthofer, C.; Wang, C.; McCarthy, P.; Lange, F.; Andersson, J.L.R.; Griffanti, L.; Duff, E.; et al. SARS-CoV-2 Is Associated with Changes in Brain Structure in UK Biobank. Nature 2022, 604, 697–707. [Google Scholar] [CrossRef]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for Functional Genomics Data Sets—Update. Nucleic Acids Res. 2012, 41, D991–D995. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Stat. Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Heberle, H.; Meirelles, G.V.; Da Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A Web-Based Tool for the Analysis of Sets through Venn Diagrams. BMC Bioinform. 2015, 16, 169. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R Package for Weighted Correlation Network Analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2. WIREs Comput. Stat. 2011, 3, 180–185. [Google Scholar] [CrossRef]
- Blighe, K. CorLevelPlot: Visualise Correlation Results and Test Significancies of These. 2018. Available online: https://rdrr.io/github/kevinblighe/CorLevelPlot/f/README.md (accessed on 17 July 2025).
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING Database in 2023: Protein–Protein Association Networks and Functional Enrichment Analyses for Any Sequenced Genome of Interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Chin, C.-H.; Chen, S.-H.; Wu, H.-H.; Ho, C.-W.; Ko, M.-T.; Lin, C.-Y. cytoHubba: Identifying Hub Objects and Sub-Networks from Complex Interactome. BMC Syst. Biol. 2014, 8 (Suppl. 4), S11. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Carlson, M. org.Hs.eg.db. 2017. Available online: https://bioconductor.org/packages/release/data/annotation/html/org.Hs.eg.db.html (accessed on 17 July 2025).
- Leek, J.T.; Johnson, W.E.; Parker, H.S.; Jaffe, A.E.; Storey, J.D. The Sva Package for Removing Batch Effects and Other Unwanted Variation in High-Throughput Experiments. Bioinformatics 2012, 28, 882–883. [Google Scholar] [CrossRef] [PubMed]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.-C.; Müller, M. pROC: An Open-Source Package for R and S+ to Analyze and Compare ROC Curves. BMC Bioinform. 2011, 12, 77. [Google Scholar] [CrossRef] [PubMed]
- Konopka, T. Umap: Uniform Manifold Approximation and Projection. 2018. Available online: https://cran.r-project.org/web/packages/umap/index.html (accessed on 17 July 2025).
- R Core Team. R: A Language and Environment for Statistical Computing [Computer Software]. R Foundation for Statistical Computing. 2024. Available online: https://www.R-project.org/ (accessed on 17 July 2025).
- Shannon, K.M. Infections and Changes in Commensal Bacteria and the Pathogenesis of Parkinson’s Disease. J. Park. Dis. 2022, 12 (Suppl. 1), S45–S51. [Google Scholar] [CrossRef]
- Ma, Q.; Yao, C.; Wu, Y.; Wang, H.; Fan, Q.; Yang, Q.; Xu, J.; Dai, H.; Zhang, Y.; Xu, F.; et al. Neurological Disorders After Severe Pneumonia Are Associated with Translocation of Endogenous Bacteria from the Lung to the Brain. Sci. Adv. 2023, 9, eadi0699. [Google Scholar] [CrossRef] [PubMed]
- Cannon, T.; Gruenheid, S. Microbes and Parkinson’s Disease: From Associations to Mechanisms. Trends Microbiol. 2022, 30, 749–760. [Google Scholar] [CrossRef] [PubMed]
- Jang, Y.; Pletnikova, O.; Troncoso, J.C.; Pantelyat, A.Y.; Dawson, T.M.; Rosenthal, L.S.; Na, C.H. Mass Spectrometry–Based Proteomics Analysis of Human Substantia Nigra From Parkinson’s Disease Patients Identifies Multiple Pathways Potentially Involved in the Disease. Mol. Cell. Proteomics 2023, 22, 100452, Erratum in Mol. Cell. Proteomics 2023, 22, 100570. [Google Scholar] [CrossRef]
- D’Angiolini, S.; Lui, M.; Mazzon, E.; Calabrò, M. Network Analysis Performed on Transcriptomes of Parkinson’s Disease Patients Reveals Dysfunction in Protein Translation. Int. J. Mol. Sci. 2024, 25, 1299. [Google Scholar] [CrossRef] [PubMed]
- Ishii, K.; Washio, T.; Uechi, T.; Yoshihama, M.; Kenmochi, N.; Tomita, M. Characteristics and Clustering of Human Ribosomal Protein Genes. BMC Genom. 2006, 7, 37. [Google Scholar] [CrossRef]
- Das, A.S.; Basu, A.; Kumar, R.; Borah, P.K.; Bakshi, S.; Sharma, M.; Duary, R.K.; Ray, P.S.; Mukhopadhyay, R. Post-transcriptional Regulation of C-C Motif Chemokine Ligand 2 Expression by Ribosomal Protein L22 during LPS-mediated Inflammation. FEBS J. 2020, 287, 3794–3813. [Google Scholar] [CrossRef]
- Kang, J.; Brajanovski, N.; Chan, K.T.; Xuan, J.; Pearson, R.B.; Sanij, E. Ribosomal Proteins and Human Diseases: Molecular Mechanisms and Targeted Therapy. Signal Transduct. Target. Ther. 2021, 6, 323. [Google Scholar] [CrossRef]
- Das, A.S.; Basu, A.; Mukhopadhyay, R. Ribosomal Proteins: The Missing Piece in the Inflammation Puzzle? Mol. Cell. Biochem. 2025, 480, 785–797. [Google Scholar] [CrossRef]
- Zhou, X.; Liao, W.-J.; Liao, J.-M.; Liao, P.; Lu, H. Ribosomal Proteins: Functions Beyond the Ribosome. J. Mol. Cell Biol. 2015, 7, 92–104. [Google Scholar] [CrossRef]
- Stein, K.C.; Morales-Polanco, F.; Van Der Lienden, J.; Rainbolt, T.K.; Frydman, J. Ageing Exacerbates Ribosome Pausing to Disrupt Cotranslational Proteostasis. Nature 2022, 601, 637–642. [Google Scholar] [CrossRef]
- Zhao, Z.; Ruan, S.; Li, Y.; Qi, T.; Qi, Y.; Huang, Y.; Liu, Z.; Ruan, Q.; Ma, Y. The Influence of Extra-Ribosomal Functions of Eukaryotic Ribosomal Proteins on Viral Infection. Biomolecules 2024, 14, 1565. [Google Scholar] [CrossRef] [PubMed]
- Karlas, A.; Machuy, N.; Shin, Y.; Pleissner, K.-P.; Artarini, A.; Heuer, D.; Becker, D.; Khalil, H.; Ogilvie, L.A.; Hess, S.; et al. Genome-Wide RNAi Screen Identifies Human Host Factors Crucial for Influenza Virus Replication. Nature 2010, 463, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Yerlici, V.T.; Astori, A.; Kejiou, N.S.; Jordan, C.A.; Khosraviani, N.; Chan, J.N.Y.; Hakem, R.; Raught, B.; Palazzo, A.F.; Mekhail, K. SARS-CoV-2 Targets Ribosomal RNA Biogenesis. Cell Rep. 2024, 43, 113891. [Google Scholar] [CrossRef] [PubMed]
- Daftuar, L.; Zhu, Y.; Jacq, X.; Prives, C. Ribosomal Proteins RPL37, RPS15 and RPS20 Regulate the Mdm2-P53-MdmX Network. PLoS ONE 2013, 8, e68667. [Google Scholar] [CrossRef] [PubMed]
- Deisenroth, C.; Zhang, Y. Ribosome Biogenesis Surveillance: Probing the Ribosomal Protein-Mdm2-P53 Pathway. Oncogene 2010, 29, 4253–4260. [Google Scholar] [CrossRef]
- Slomnicki, L.P.; Chung, D.-H.; Parker, A.; Hermann, T.; Boyd, N.L.; Hetman, M. Ribosomal Stress and Tp53-Mediated Neuronal Apoptosis in Response to Capsid Protein of the Zika Virus. Sci. Rep. 2017, 7, 16652. [Google Scholar] [CrossRef]
- Friedlander, R.M. Apoptosis and Caspases in Neurodegenerative Diseases. N. Engl. J. Med. 2003, 348, 1365–1375. [Google Scholar] [CrossRef]
- Kuai, X.; Wei, C.; He, X.; Wang, F.; Wang, C.; Ji, J. The Potential Value of RPS27A in Prognosis and Immunotherapy: From Pan-Cancer Analysis to Hepatocellular Carcinoma Validation. ImmunoTargets Ther. 2024, 13, 673–690. [Google Scholar] [CrossRef]
- Nosrati, N.; Kapoor, N.R.; Kumar, V. DNA Damage Stress Induces the Expression of Ribosomal Protein S27a Gene in a P53-Dependent Manner. Gene 2015, 559, 44–51. [Google Scholar] [CrossRef]
- Khayer, N.; Mirzaie, M.; Marashi, S.-A.; Jalessi, M. Rps27a Might Act as a Controller of Microglia Activation in Triggering Neurodegenerative Diseases. PLoS ONE 2020, 15, e0239219. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Qiao, M.; Zhou, Y.; Peng, Y.; Wen, G.; Xie, C.; Zhang, Y. Modulating the RPS27A/PSMD12/NF-κB Pathway to Control Immune Response in Mouse Brain Ischemia-Reperfusion Injury. Mol. Med. 2024, 30, 106. [Google Scholar] [CrossRef]
- Li, Y.; Li, S.; Wu, H. Ubiquitination-Proteasome System (UPS) and Autophagy Two Main Protein Degradation Machineries in Response to Cell Stress. Cells 2022, 11, 851. [Google Scholar] [CrossRef]
- Pohl, C.; Dikic, I. Cellular Quality Control by the Ubiquitin-Proteasome System and Autophagy. Science 2019, 366, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Peters, J.M.; Franke, W.W.; Kleinschmidt, J.A. Distinct 19 S and 20 S Subcomplexes of the 26 S Proteasome and Their Distribution in the Nucleus and the Cytoplasm. J. Biol. Chem. 1994, 269, 7709–7718. [Google Scholar] [CrossRef]
- Morimoto, R.I. Proteotoxic Stress and Inducible Chaperone Networks in Neurodegenerative Disease and Aging. Genes Dev. 2008, 22, 1427–1438. [Google Scholar] [CrossRef]
- Luo, H. Interplay between the Virus and the Ubiquitin–Proteasome System: Molecular Mechanism of Viral Pathogenesis. Curr. Opin. Virol. 2016, 17, 1–10. [Google Scholar] [CrossRef]
- Iovino, F.; Gradstedt, H.; Bijlsma, J.J. The Proteasome-Ubiquitin System Is Required for Efficient Killing of Intracellular Streptococcus Pneumoniae by Brain Endothelial Cells. mBio 2014, 5, e00984-14. [Google Scholar] [CrossRef]
- Grünewald, A.; Kumar, K.R.; Sue, C.M. New Insights into the Complex Role of Mitochondria in Parkinson’s Disease. Prog. Neurobiol. 2019, 177, 73–93. [Google Scholar] [CrossRef] [PubMed]
- Henrich, M.T.; Oertel, W.H.; Surmeier, D.J.; Geibl, F.F. Mitochondrial Dysfunction in Parkinson’s Disease—A Key Disease Hallmark with Therapeutic Potential. Mol. Neurodegener. 2023, 18, 83. [Google Scholar] [CrossRef] [PubMed]
- Purandare, N.; Ghosalkar, E.; Grossman, L.I.; Aras, S. Mitochondrial Oxidative Phosphorylation in Viral Infections. Viruses 2023, 15, 2380. [Google Scholar] [CrossRef]
- Tiku, V.; Tan, M.-W.; Dikic, I. Mitochondrial Functions in Infection and Immunity. Trends Cell Biol. 2020, 30, 263–275. [Google Scholar] [CrossRef]




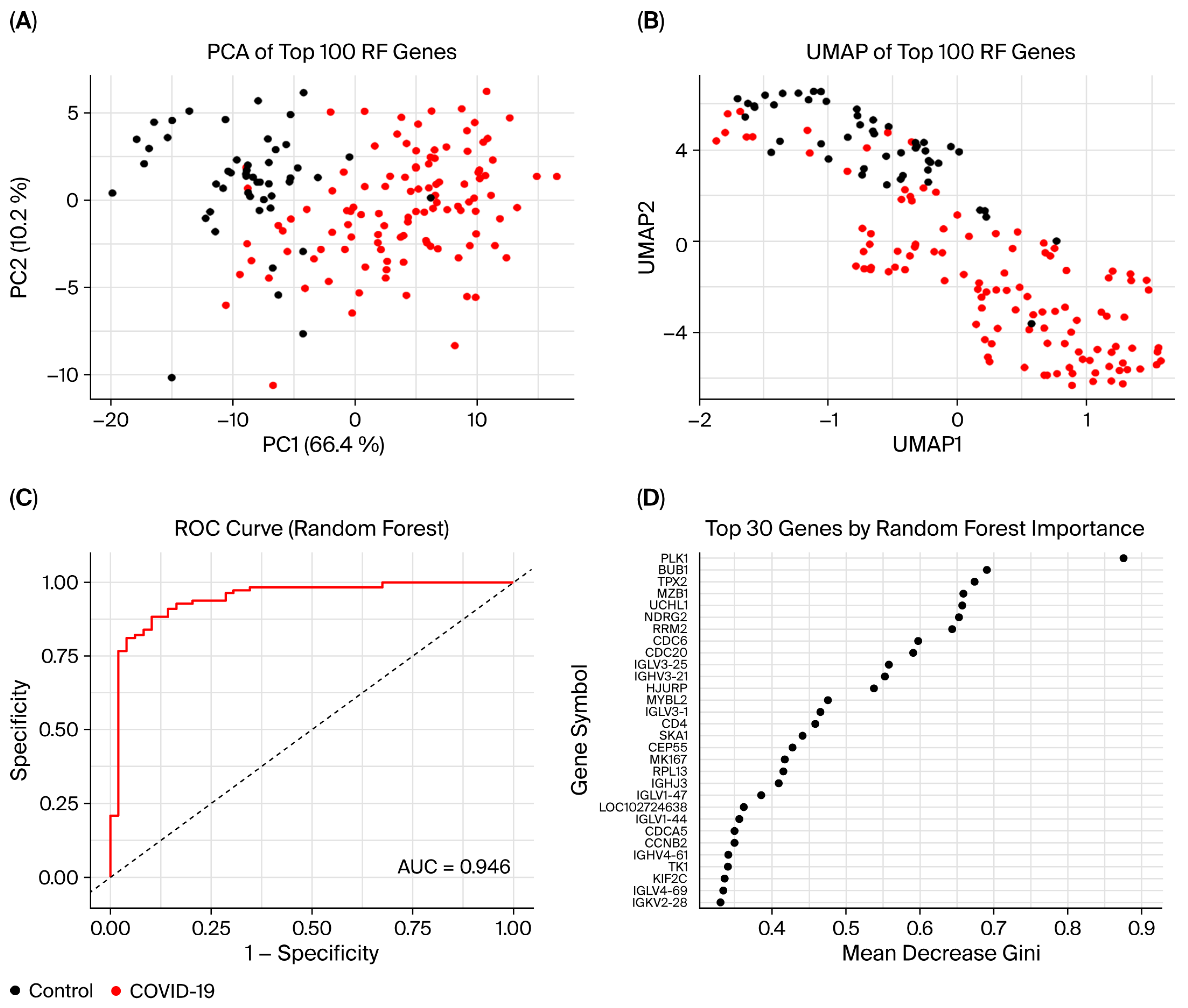
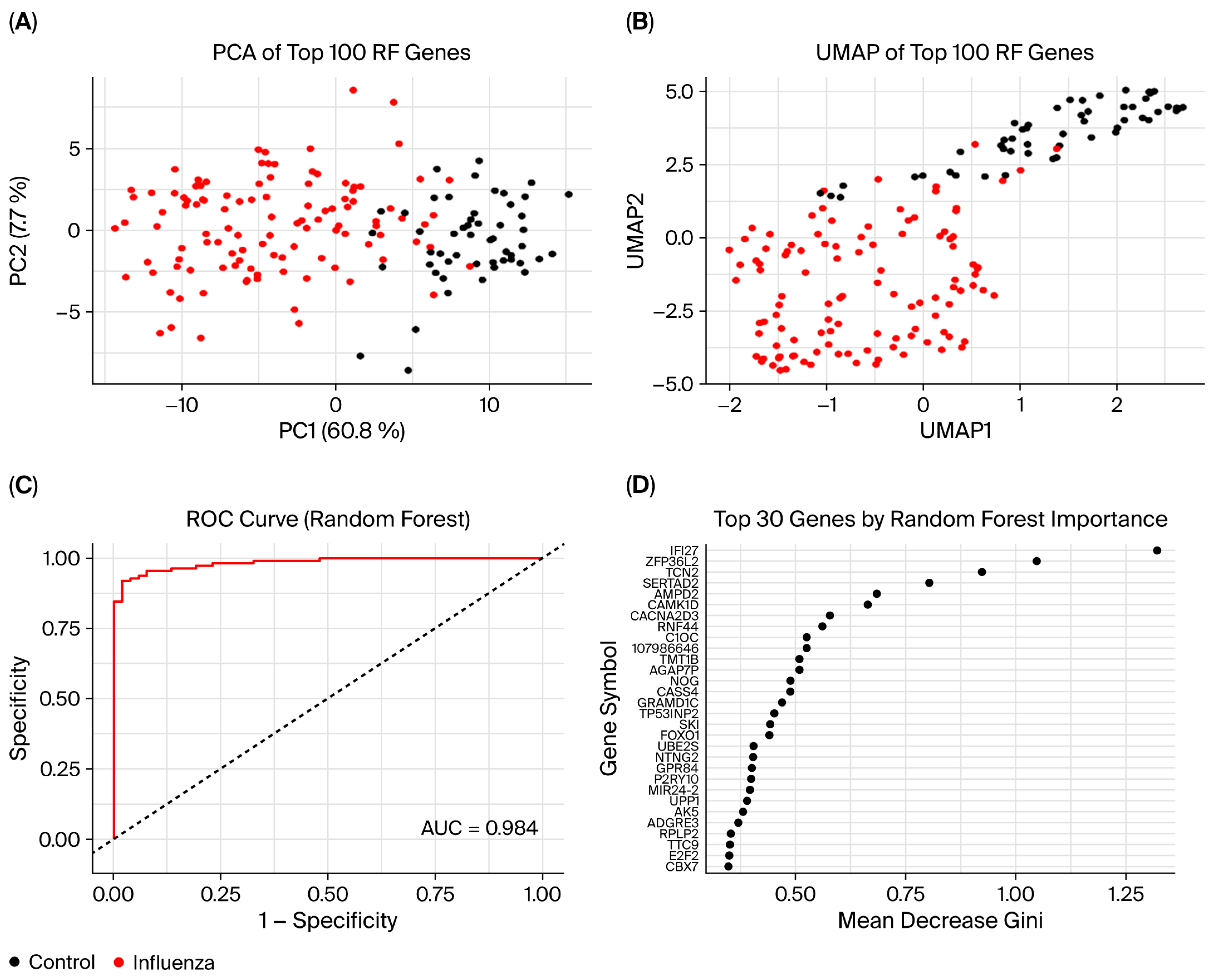
| GEO Accession | Platform | Source | Samples | Analysis | Publishing Year |
|---|---|---|---|---|---|
| GSE161731 | Illumina NovaSeq 6000 | Blood | COVID-19 (n = 77) | DGE and WGCNA | 2021 |
| B. pneumonia (n = 24) | |||||
| Seasonal coronaviruses (n = 61) | |||||
| Influenza (n = 17) | |||||
| Control (n = 19) | |||||
| GSE161199 | Illumina NextSeq 500 | Blood | PD (n = 5) | DGE and WGCNA | 2020 |
| Control (n = 11) | |||||
| GSE196350 | Illumina HiSeq 4000 | Blood | Influenza (n = 33) | RF | 2024 |
| Control (n = 16) | |||||
| GSE155635 | Illumina HiSeq 4000 | Blood | Influenza (n = 60) | RF | 2024 |
| Control (n = 16) | |||||
| GSE213168 | Illumina NovaSeq 6000 | Blood | Influenza (n = 18) | RF | 2024 |
| Control (n = 20) | |||||
| GSE171110 | Illumina HiSeq 2500 | Blood | COVID-19 (n = 44) | RF | 2021 |
| Control (n = 10) | |||||
| GSE167000 | Illumina NovaSeq 6000 | Blood | COVID-19 (n = 51) | RF | 2021 |
| Control (n = 22) | |||||
| GSE152418 | Illumina NovaSeq 6000 | Blood | COVID-19 (n = 16) | RF | 2020 |
| Control (n = 17) |
| Condition | Database | DEGs | Upregulated | Downregulated |
|---|---|---|---|---|
| COVID-19 | GSE161731 | 812 | 327 | 485 |
| Influenza | 1395 | 812 | 583 | |
| OtherCoV | 934 | 497 | 437 | |
| B. pneumonia | 6769 | 2465 | 4304 | |
| PD | GSE161199 | 2479 | 304 | 2175 |
| Shared RIs DEGs | HES4, RPL11, CD52, MAN1A2, RPS27, GAS5-AS1, GAS5, SNORA103, MIR4426, SNRPE, RPL21P28, RPS7, RPS27A, SNRPG, RPL31, DBI, LINC01934, ANKRD44-AS1, PPIL3, EEF1B2, LSM3, TMA7, MIX23, NMRAL2P, RPL35A, RPL9, BDH2, RPL34, SNHG8, RPS3A, NDUFS4, COX7C, HINT1, MRPL22, RPL26L1, RPS18, RPS10-NUDT3, RPS10, NDUFA4, RPL23P8, TOMM7, SEM1, NDUFA5, RPL7, RBIS, UQCRB, RPL30, COX6C, LY6E, RPS6, TOMM5, GKAP1, WDR38, RPS24, KIF20B, IFITM3, KLRB1, LOC107987174, KLRF1, KLRC4, KLRC2, PFDN5, MYG1-AS1, SARNP, RPL41, RPL6, RPL21, CCNA1, TPT1, COMMD6, RPS29, ATP5MJ, RSL24D1, RPS17, CPEB1, RPS15A, RPL26, SNORD3A, LOC107984974, RPL23, RPL27, RPL17-C18orf32, RPL17, SNORD58B, ZNF85, ZNF302, HPN, TRAPPC2B, RPS21, FAM247A, RBFOX2, RBX1, COX7B, RPL36A-HNRNPH2, RPL36A, RPL39, SH2D1A, FGF13 |
| Overlapping DEGs between RIs and PD | HES4, RPL11, GAS5, SNORA103, MIR4426, SNRPE, RPL21P28, RPS7, RPS27A, RPL31, EEF1B2, TMA7, RPL35A, RPL9, RPL34, SNHG8, RPS3A, NDUFS4, COX7C, MRPL22, RPS18, RPL23P8, TOMM7, NDUFA5, RPL7, RBIS, UQCRB, RPL30, COX6C, RPS6, WDR38, KIF20B, KLRB1, PFDN5, MYG1-AS1, RPL21, COMMD6, RPS29, RSL24D1, RPS15A, RPL26, SNORD3A, RPL27, RPL17-C18orf32, RPL17, SNORD58B, ZNF302, RPS21, RPL36A-HNRNPH2 |
| Overlapped genes between MEturquoise (PD) and MEpink (RIs) modules | RPL22, RPL11, UBXN11, CD52, UQCRH, UFC1, RPS7, RPS27A, CCT4, SNRPG, DBI, LSM3, TMA7, CSTA, RPL35A, RPL9, RPL34, RPS3A, RPL37, NDUFS4, RPS23, COX7C, NPM1, RPS18, PPIA, PSMC2, RPL7, UQCRB, POLR2K, EIF3E, ATP6V1G1, RPL35, NAP1L4, RPL27A, RPS13, FAU, KLRB1, LDHB, SARNP, RPL21, HMGB1, COMMD6, RPS29, RPL36AL, PSMA3, ATP5MJ, B2M, PSMA4, RPL26, SNHG29, RPL23, RPL17, RPS21, SNRPD3, RBX1, COX7B, MIR10393, MYG1-AS1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Albeshri, A.; Bahieldin, A.; Ali, H.M. Transcriptomic Analysis Reveals the Molecular Relationship Between Common Respiratory Infections and Parkinson’s Disease. Curr. Issues Mol. Biol. 2025, 47, 727. https://doi.org/10.3390/cimb47090727
Albeshri A, Bahieldin A, Ali HM. Transcriptomic Analysis Reveals the Molecular Relationship Between Common Respiratory Infections and Parkinson’s Disease. Current Issues in Molecular Biology. 2025; 47(9):727. https://doi.org/10.3390/cimb47090727
Chicago/Turabian StyleAlbeshri, Abdulaziz, Ahmed Bahieldin, and Hani Mohammed Ali. 2025. "Transcriptomic Analysis Reveals the Molecular Relationship Between Common Respiratory Infections and Parkinson’s Disease" Current Issues in Molecular Biology 47, no. 9: 727. https://doi.org/10.3390/cimb47090727
APA StyleAlbeshri, A., Bahieldin, A., & Ali, H. M. (2025). Transcriptomic Analysis Reveals the Molecular Relationship Between Common Respiratory Infections and Parkinson’s Disease. Current Issues in Molecular Biology, 47(9), 727. https://doi.org/10.3390/cimb47090727






