PSMB9 Orchestrates Tumor Immune Landscape and Serves as a Potent Biomarker for Prognosis and T Cell-Based Immunotherapy Response
Abstract
1. Introduction
2. Materials and Methods
2.1. Expression Profiles
2.2. Prognosis Analysis
2.3. Genomic Alterations and Modification Analysis
2.4. Analysis of Tumor Stemness Correlations
2.5. Gene Set Enrichment Analyses
2.6. Immune Cell Infiltration Analysis
2.7. Immunotherapy Analysis
2.8. CAR-T Cell Generation
2.9. CRISPR/Cas9 Screening
2.10. Establishment of PSMB9KO Cell Line
2.11. Western Blot
2.12. Statistical Analysis
3. Results
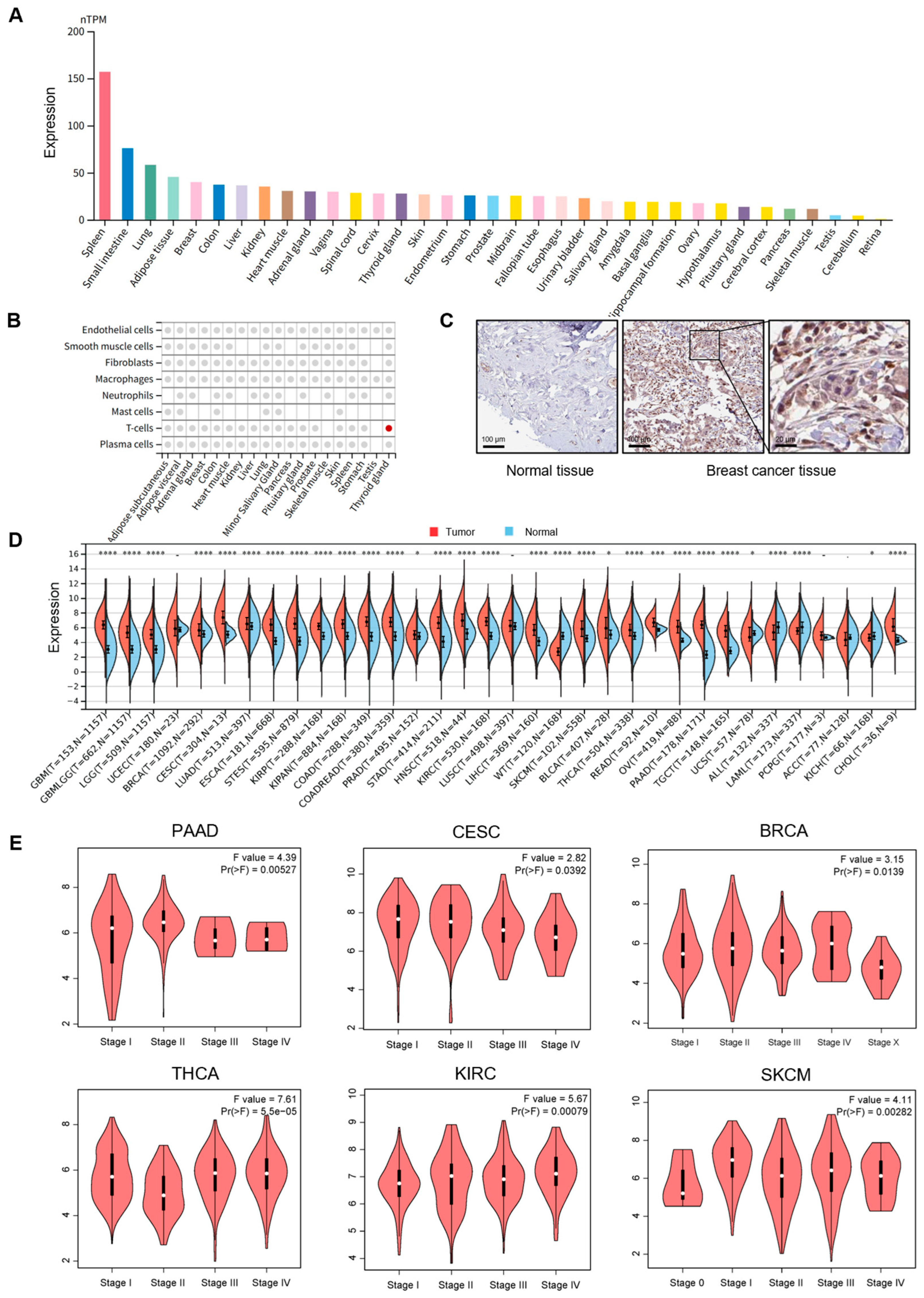
3.1. PSMB9 Displays Dysregulated Expression Patterns Across Diverse Human Cancers
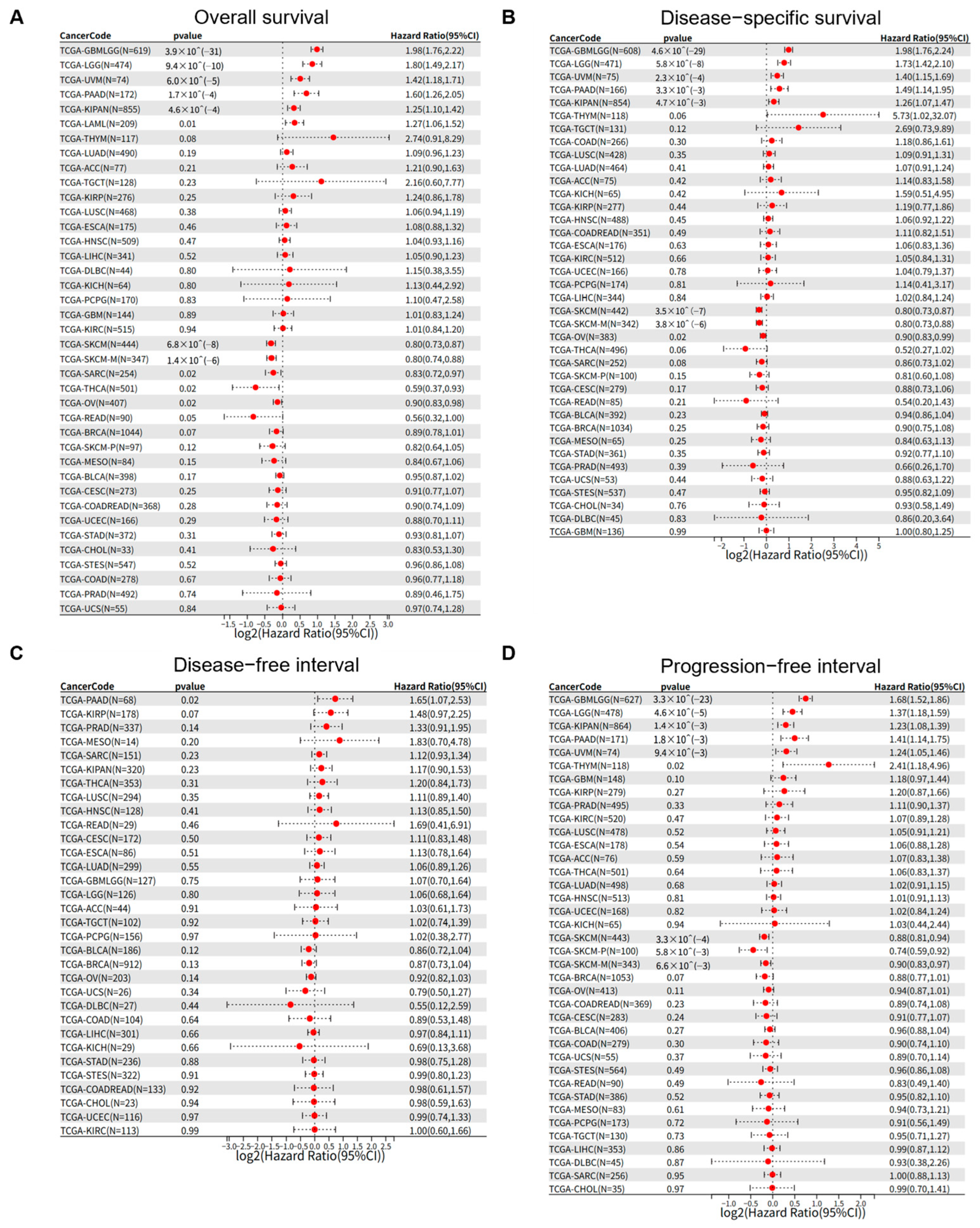
3.2. PSMB9 Expression Levels Exert Distinct Prognostic Impacts Across Different Tumor Types
3.3. PSMB9 Alteration Profiles and Their Role in Tumor Stemness
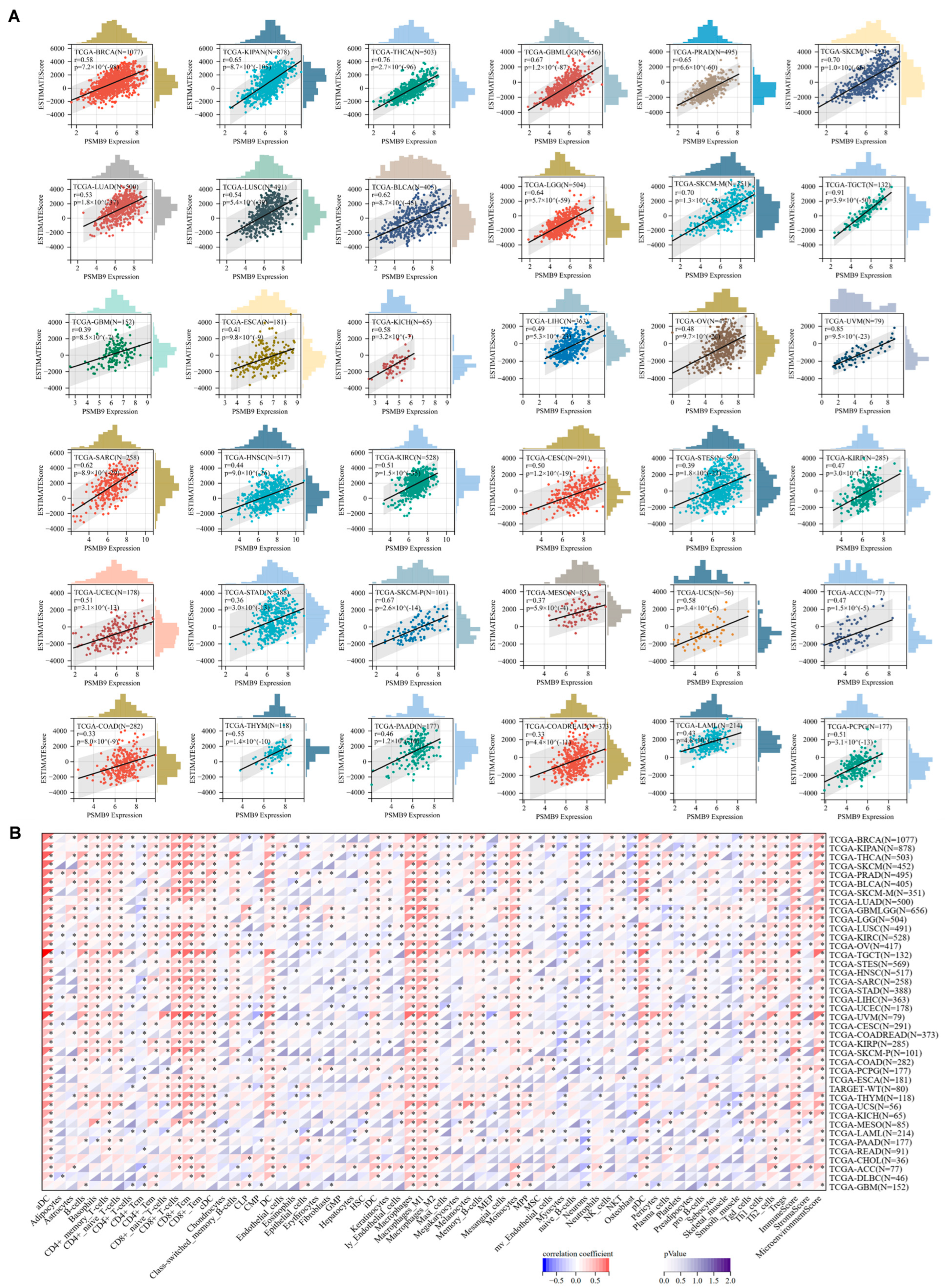
3.4. PSMB9 Associates with Tumor Immune Responses
3.5. Higher PSMB9 Reveals Superior Tumor Immune Cell Infiltration
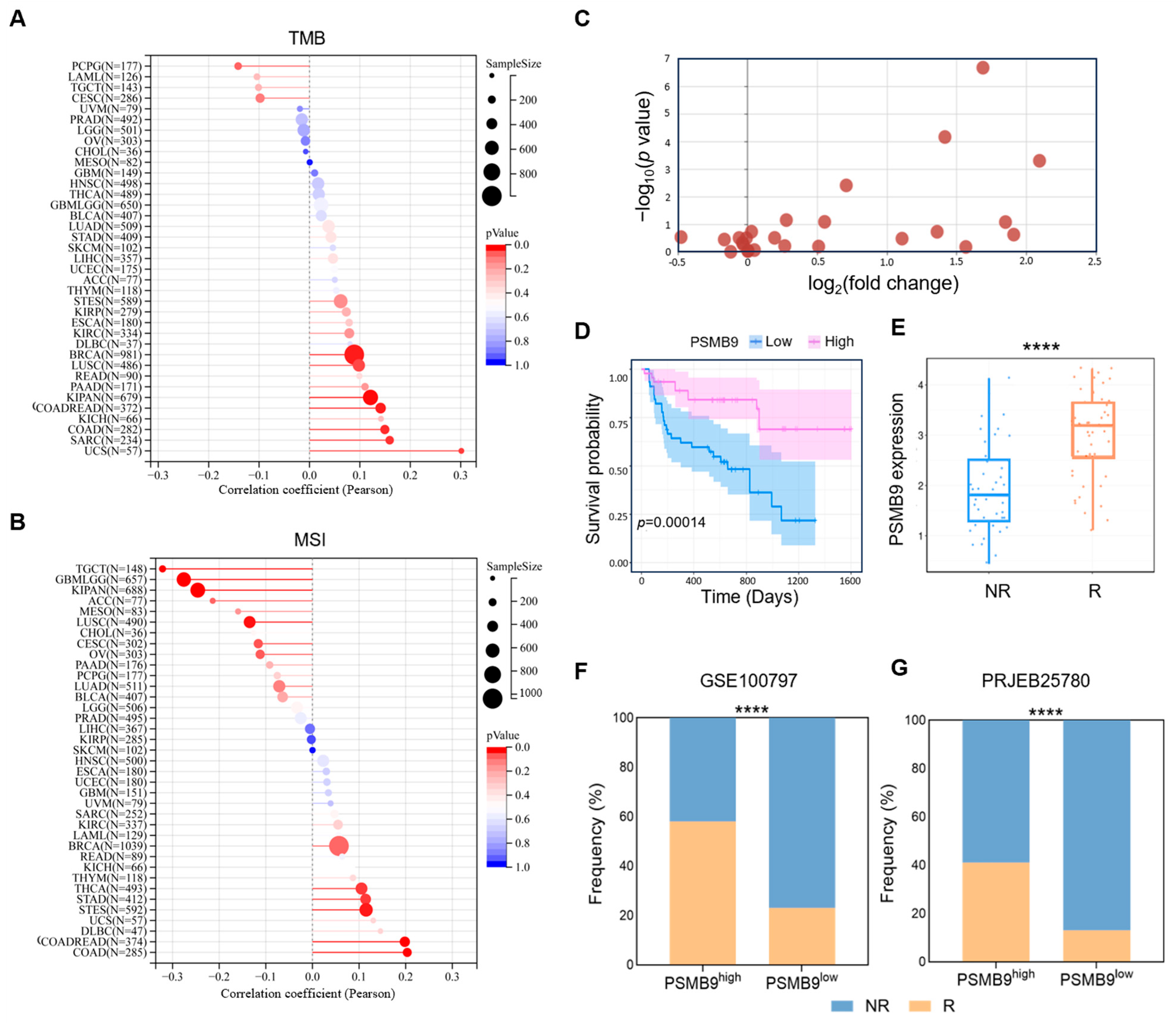
3.6. PSMB9 Modulates Responses to ICIs
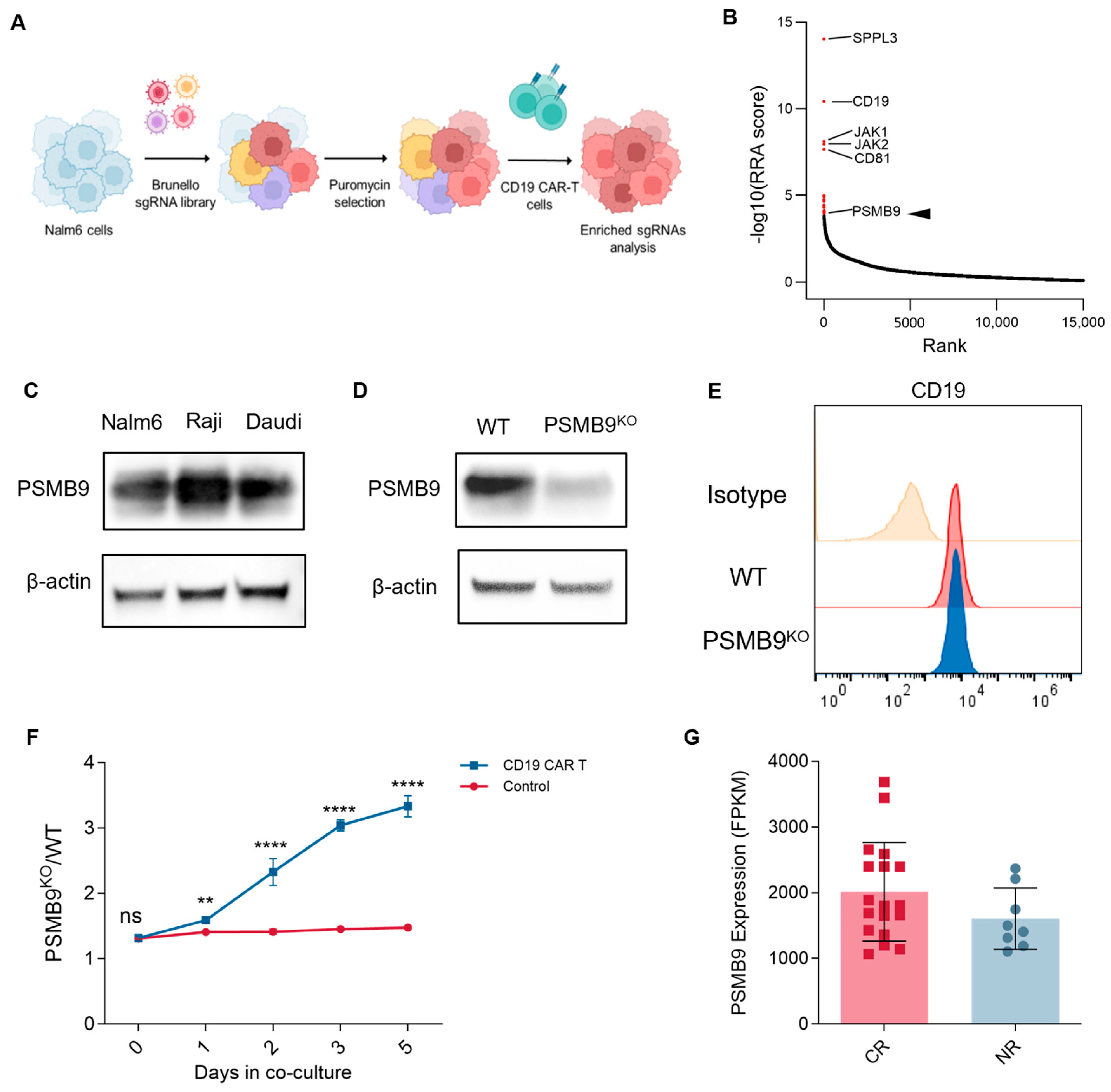
3.7. PSMB9 Plays a Potential Role in CAR-T Cell Therapy
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PSMB9 | Proteasome subunit beta type-9 |
| MHC | Major histocompatibility complex |
| CAR T | Chimeric antigen receptor T |
| NSCLC | Non-small-cell lung cancer |
| GTEx | Genotype-tissue expression |
| IHC | Immunohistochemical |
| HPA | Human Protein Atlas |
| TCGA | The Cancer Genome Atlas |
| GBM | Glioblastoma multiforme |
| GBMLGG | Glioma (including GBM and LGG) |
| LGG | Brain lower-grade glioma |
| BRCA | Breast invasive carcinoma |
| CESC | Endocervical adenocarcinoma |
| LUAD | Lung adenocarcinoma |
| ESCA | Esophageal carcinoma |
| STES | Stomach and esophageal carcinoma |
| KIRP | Kidney renal papillary cell carcinoma |
| COAD | Colon adenocarcinoma |
| PRAD | Prostate adenocarcinoma |
| STAD | Stomach adenocarcinoma |
| HNSC | Head and neck squamous cell carcinoma |
| KIRC | Kidney renal clear cell carcinoma |
| LIHC | Liver hepatocellular carcinoma |
| SKCM | Skin cutaneous melanoma |
| BLCA | Bladder urothelial carcinoma |
| THCA | Thyroid carcinoma |
| READ | Rectum adenocarcinoma |
| OV | Ovarian serous cystadenocarcinoma |
| PAAD | Pancreatic adenocarcinoma |
| TGCT | Testicular germ cell tumors |
| LAML | Acute myeloid leukemia |
| KICH | Kidney chromophobe |
| CHOL | Cholangiocarcinoma |
| UCS | Uterine carcinosarcoma |
| ALL | Acute lymphoblastic leukemia |
| WT | Wilms tumor |
| OS | Overall survival |
| UVM | Uveal melanoma |
| KIPAN | Pan-kidney cancer cohort (including KIRC, KIRP, KICH) |
| SKCM-M | Skin cutaneous melanoma-metastatic |
| SARC | Sarcoma |
| DSS | Disease-specific survival |
| DFI | Disease-free interval |
| PFI | Progression-free interval |
| THYM | Thymoma |
| SKCM-P | Skin cutaneous melanoma-primary |
| DNAss | DNA stemness score |
| EREG-METHss | Epigenetic regulation of stemness-methylation score |
| ENHss | Enhancer stemness score |
| DEGs | Differential expression genes |
| GSEA | Gene set enrichment analysis |
| NFKB | Nuclear factor kappa-B |
| TMB | Tumor mutational burden |
| MSI | Microsatellite instability |
| ICIs | Immune checkpoint inhibitors |
| TIGER | Tumor Immunotherapy Gene Expression Resource |
| ACT | Adoptive cell transfer |
| SNV | Simple nucleotide variation |
| CRISPR | Clustered regularly interspaced short palindromic repeat |
| sgRNA | Single-guide RNA |
| KO | Knockout |
| PBMCs | Peripheral blood mononuclear cells |
| ECL | Enhanced chemiluminescence |
References
- Oliveira, G.; Wu, C.J. Dynamics and specificities of T cells in cancer immunotherapy. Nat. Rev. Cancer 2023, 23, 295–316. [Google Scholar] [CrossRef]
- O’Donnell, J.S.; Teng, M.W.L.; Smyth, M.J. Cancer immunoediting and resistance to T cell-based immunotherapy. Nat. Rev. Clin. Oncol. 2019, 16, 151–167. [Google Scholar] [CrossRef]
- Leone, P.; Shin, E.C.; Perosa, F.; Vacca, A.; Dammacco, F.; Racanelli, V. MHC class I antigen processing and presenting machinery: Organization, function, and defects in tumor cells. J. Natl. Cancer Inst. 2013, 105, 1172–1187. [Google Scholar] [CrossRef]
- Basler, M.; Groettrup, M. On the Role of the Immunoproteasome in Protein Homeostasis. Cells 2021, 10, 3216. [Google Scholar] [CrossRef]
- Maia Falcao, R.; Kokaraki, G.; De Wispelaere, W.; Amant, F.; De Souza, G.A.; de Souza, J.E.S.; Carlson, J.W.; Petta, T.B. The Expression of the Immunoproteasome Subunit PSMB9 Is Related to Distinct Molecular Subtypes of Uterine Leiomyosarcoma. Cancers 2022, 14, 5007. [Google Scholar] [CrossRef]
- Li, S.; He, R.C.; Wu, S.G.; Song, Y.; Zhang, K.L.; Tang, M.L.; Bei, Y.R.; Zhang, T.; Lu, J.B.; Ma, X.; et al. LncRNA PSMB8-AS1 Instigates Vascular Inflammation to Aggravate Atherosclerosis. Circ. Res. 2024, 134, 60–80. [Google Scholar] [CrossRef]
- Yuan, J.; Wang, T.; Wang, L.; Li, P.; Shen, H.; Mo, Y.; Zhang, Q.; Ni, C. Transcriptome-wide association study identifies PSMB9 as a susceptibility gene for coal workers’ pneumoconiosis. Environ. Toxicol. 2022, 37, 2103–2114. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, K.; Jinnin, M.; Kudo, H.; Inoue, K.; Nakayama, W.; Honda, N.; Kajihara, I.; Masuguchi, S.; Fukushima, S.; Ihn, H. The role of PSMB9 upregulated by interferon signature in the pathophysiology of cutaneous lesions of dermatomyositis and systemic lupus erythematosus. Br. J. Dermatol. 2016, 174, 1030–1041. [Google Scholar] [CrossRef]
- Zeng, L.; Chen, K.; Xiao, F.; Zhu, C.Y.; Bai, J.Y.; Tan, S.; Long, L.; Wang, Y.; Zhou, Q. Potential common molecular mechanisms between Sjogren syndrome and inclusion body myositis: A bioinformatic analysis and in vivo validation. Front. Immunol. 2023, 14, 1161476. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Chen, Y.; Zulipikaer, M.; Xu, C.; Fu, J.; Deng, T.; Hao, L.B.; Chen, J.Y. Identification of PSMB9 and CXCL13 as Immune-related Diagnostic Markers for Rheumatoid Arthritis by Machine Learning. Curr. Pharm. Des. 2022, 28, 2842–2854. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.; Kwon, A.Y.; Jeong, J.Y.; Kim, S.; Kang, H.; Park, J.; Kim, J.H.; Han, O.J.; Lim, S.M.; An, H.J. Immune gene signatures for predicting durable clinical benefit of anti-PD-1 immunotherapy in patients with non-small cell lung cancer. Sci. Rep. 2020, 10, 643. [Google Scholar] [CrossRef]
- Hu, X.; Hu, Z.; Zhang, H.; Zhang, N.; Feng, H.; Jia, X.; Zhang, C.; Cheng, Q. Deciphering the tumor-suppressive role of PSMB9 in melanoma through multi-omics and single-cell transcriptome analyses. Cancer Lett. 2024, 581, 216466. [Google Scholar] [CrossRef]
- Uhlen, M.; Fagerberg, L.; Hallstrom, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, A.; Kampf, C.; Sjostedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Tang, Z.; Zhang, W.; Ye, Z.; Liu, F. GEPIA2021: Integrating multiple deconvolution-based analysis into GEPIA. Nucleic Acids Res. 2021, 49, W242–W246. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]
- de Bruijn, I.; Kundra, R.; Mastrogiacomo, B.; Tran, T.N.; Sikina, L.; Mazor, T.; Li, X.; Ochoa, A.; Zhao, G.; Lai, B.; et al. Analysis and Visualization of Longitudinal Genomic and Clinical Data from the AACR Project GENIE Biopharma Collaborative in cBioPortal. Cancer Res. 2023, 83, 3861–3867. [Google Scholar] [CrossRef] [PubMed]
- Goldman, M.J.; Craft, B.; Hastie, M.; Repecka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.; Rogers, D.; Brooks, A.N.; et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020, 38, 675–678. [Google Scholar] [CrossRef]
- Chen, D.; Xu, L.; Xing, H.; Shen, W.; Song, Z.; Li, H.; Zhu, X.; Li, X.; Wu, L.; Jiao, H.; et al. Sangerbox 2: Enhanced functionalities and update for a comprehensive clinical bioinformatics data analysis platform. iMeta 2024, 3, e238. [Google Scholar] [CrossRef]
- Malta, T.M.; Sokolov, A.; Gentles, A.J.; Burzykowski, T.; Poisson, L.; Weinstein, J.N.; Kaminska, B.; Huelsken, J.; Omberg, L.; Gevaert, O.; et al. Machine Learning Identifies Stemness Features Associated with Oncogenic Dedifferentiation. Cell 2018, 173, 338–354.e315. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdottir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Hanzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef]
- Chen, Z.; Luo, Z.; Zhang, D.; Li, H.; Liu, X.; Zhu, K.; Zhang, H.; Wang, Z.; Zhou, P.; Ren, J.; et al. TIGER: A Web Portal of Tumor Immunotherapy Gene Expression Resource. Genom. Proteom. Bioinform. 2023, 21, 337–348. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.T.; Cristescu, R.; Bass, A.J.; Kim, K.M.; Odegaard, J.I.; Kim, K.; Liu, X.Q.; Sher, X.; Jung, H.; Lee, M.; et al. Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nat. Med. 2018, 24, 1449–1458. [Google Scholar] [CrossRef]
- Gide, T.N.; Quek, C.; Menzies, A.M.; Tasker, A.T.; Shang, P.; Holst, J.; Madore, J.; Lim, S.Y.; Velickovic, R.; Wongchenko, M.; et al. Distinct Immune Cell Populations Define Response to Anti-PD-1 Monotherapy and Anti-PD-1/Anti-CTLA-4 Combined Therapy. Cancer Cell 2019, 35, 238–255.e236. [Google Scholar] [CrossRef]
- Wang, Y.; Tong, C.; Dai, H.; Wu, Z.; Han, X.; Guo, Y.; Chen, D.; Wei, J.; Ti, D.; Liu, Z.; et al. Low-dose decitabine priming endows CAR T cells with enhanced and persistent antitumour potential via epigenetic reprogramming. Nat. Commun. 2021, 12, 409. [Google Scholar] [CrossRef]
- Tong, C.; Zhang, Y.; Liu, Y.; Ji, X.; Zhang, W.; Guo, Y.; Han, X.; Ti, D.; Dai, H.; Wang, C.; et al. Optimized tandem CD19/CD20 CAR-engineered T cells in refractory/relapsed B-cell lymphoma. Blood 2020, 136, 1632–1644, Erratum in Blood 2023, 141, 1896. [Google Scholar] [CrossRef]
- Yan, X.; Chen, D.; Ma, X.; Wang, Y.; Guo, Y.; Wei, J.; Tong, C.; Zhu, Q.; Lu, Y.; Yu, Y.; et al. CD58 loss in tumor cells confers functional impairment of CAR T cells. Blood Adv. 2022, 6, 5844–5856. [Google Scholar] [CrossRef]
- Yan, X.; Chen, D.; Wang, Y.; Guo, Y.; Tong, C.; Wei, J.; Zhang, Y.; Wu, Z.; Han, W. Identification of NOXA as a pivotal regulator of resistance to CAR T-cell therapy in B-cell malignancies. Signal Transduct. Target. Ther. 2022, 7, 98. [Google Scholar] [CrossRef]
- Vilar, E.; Gruber, S.B. Microsatellite instability in colorectal cancer-the stable evidence. Nat. Rev. Clin. Oncol. 2010, 7, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Palmeri, M.; Mehnert, J.; Silk, A.W.; Jabbour, S.K.; Ganesan, S.; Popli, P.; Riedlinger, G.; Stephenson, R.; de Meritens, A.B.; Leiser, A.; et al. Real-world application of tumor mutational burden-high (TMB-high) and microsatellite instability (MSI) confirms their utility as immunotherapy biomarkers. ESMO Open 2022, 7, 100336. [Google Scholar] [CrossRef] [PubMed]
- Lauss, M.; Donia, M.; Harbst, K.; Andersen, R.; Mitra, S.; Rosengren, F.; Salim, M.; Vallon-Christersson, J.; Torngren, T.; Kvist, A.; et al. Mutational and putative neoantigen load predict clinical benefit of adoptive T cell therapy in melanoma. Nat. Commun. 2017, 8, 1738, Correction in Nat. Commun. 2020, 11, 1714. [Google Scholar] [CrossRef]
- Jiang, P.; Gu, S.; Pan, D.; Fu, J.; Sahu, A.; Hu, X.; Li, Z.; Traugh, N.; Bu, X.; Li, B.; et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 2018, 24, 1550–1558. [Google Scholar] [CrossRef]
- Ayers, M.; Lunceford, J.; Nebozhyn, M.; Murphy, E.; Loboda, A.; Kaufman, D.R.; Albright, A.; Cheng, J.D.; Kang, S.P.; Shankaran, V.; et al. IFN-gamma-related mRNA profile predicts clinical response to PD-1 blockade. J. Clin. Investig. 2017, 127, 2930–2940. [Google Scholar] [CrossRef]
- Neelapu, S.S.; Locke, F.L.; Bartlett, N.L.; Lekakis, L.J.; Miklos, D.B.; Jacobson, C.A.; Braunschweig, I.; Oluwole, O.O.; Siddiqi, T.; Lin, Y.; et al. Axicabtagene Ciloleucel CAR T-Cell Therapy in Refractory Large B-Cell Lymphoma. N. Engl. J. Med. 2017, 377, 2531–2544. [Google Scholar] [CrossRef]
- Feins, S.; Kong, W.; Williams, E.F.; Milone, M.C.; Fraietta, J.A. An introduction to chimeric antigen receptor (CAR) T-cell immunotherapy for human cancer. Am. J. Hematol. 2019, 94, S3–S9. [Google Scholar] [CrossRef]
- Singh, N.; Lee, Y.G.; Shestova, O.; Ravikumar, P.; Hayer, K.E.; Hong, S.J.; Lu, X.M.; Pajarillo, R.; Agarwal, S.; Kuramitsu, S.; et al. Impaired Death Receptor Signaling in Leukemia Causes Antigen-Independent Resistance by Inducing CAR T-cell Dysfunction. Cancer Discov. 2020, 10, 552–567. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, Z. The history and advances in cancer immunotherapy: Understanding the characteristics of tumor-infiltrating immune cells and their therapeutic implications. Cell Mol. Immunol. 2020, 17, 807–821. [Google Scholar] [CrossRef]
- Chan, T.A.; Yarchoan, M.; Jaffee, E.; Swanton, C.; Quezada, S.A.; Stenzinger, A.; Peters, S. Development of tumor mutation burden as an immunotherapy biomarker: Utility for the oncology clinic. Ann. Oncol. 2019, 30, 44–56. [Google Scholar] [CrossRef] [PubMed]
- Jardim, D.L.; Goodman, A.; de Melo Gagliato, D.; Kurzrock, R. The Challenges of Tumor Mutational Burden as an Immunotherapy Biomarker. Cancer Cell 2021, 39, 154–173. [Google Scholar] [CrossRef] [PubMed]
- Doroshow, D.B.; Bhalla, S.; Beasley, M.B.; Sholl, L.M.; Kerr, K.M.; Gnjatic, S.; Wistuba, I.I.; Rimm, D.L.; Tsao, M.S.; Hirsch, F.R. PD-L1 as a biomarker of response to immune-checkpoint inhibitors. Nat. Rev. Clin. Oncol. 2021, 18, 345–362. [Google Scholar] [CrossRef]
- Gajewski, T.F.; Schreiber, H.; Fu, Y.X. Innate and adaptive immune cells in the tumor microenvironment. Nat. Immunol. 2013, 14, 1014–1022. [Google Scholar] [CrossRef]
- Chapatte, L.; Ayyoub, M.; Morel, S.; Peitrequin, A.L.; Levy, N.; Servis, C.; Van den Eynde, B.J.; Valmori, D.; Levy, F. Processing of tumor-associated antigen by the proteasomes of dendritic cells controls in vivo T-cell responses. Cancer Res. 2006, 66, 5461–5468. [Google Scholar] [CrossRef]
- Demel, U.M.; Boger, M.; Yousefian, S.; Grunert, C.; Zhang, L.; Hotz, P.W.; Gottschlich, A.; Kose, H.; Isaakidis, K.; Vonficht, D.; et al. Activated SUMOylation restricts MHC class I antigen presentation to confer immune evasion in cancer. J. Clin. Investig. 2022, 132, e152383. [Google Scholar] [CrossRef] [PubMed]
- Saw, P.E.; Liu, Q.; Wong, P.P.; Song, E. Cancer stem cell mimicry for immune evasion and therapeutic resistance. Cell Stem Cell 2024, 31, 1101–1112. [Google Scholar] [CrossRef] [PubMed]


| Signature Name | Description | AUC (Melanoma) | AUC (STAD) |
|---|---|---|---|
| PSMB9 | PSMB9 | 0.8164 | 0.7143 |
| T cell-inflamed GEP | CCL5, CD27, CD274, CD276, CD8A, CMKLR1, CXCL9, CXCR6, HLA-DQA1, HLA-DRB1, HLA-E, IDO1, LAG3, NKG7, PDCD1LG2, PSMB10, STAT1 | 0.8229 | 0.7043 |
| CAF | Cancer-associated fibroblasts | 0.5829 | 0.5731 |
| TAM M2 | Tumor-associated macrophages | 0.8229 | 0.6174 |
| IFNG | CXCL10, CXCL9, HLA-DRA, IDO1, IFNG, STAT1 | 0.818 | 0.6901 |
| CD8 | CD8A, CD8B | 0.8438 | 0.7519 |
| CD274 | CD274 | 0.8647 | 0.6817 |
| TLS | CCL19, CCL21, CXCL13, CCR7, SELL, LAMP3, CXCR4, CD86, BCL6 | 0.7327 | 0.6383 |
| TLS-melanoma | CD79B, CD1D, CCR6, LAT, SKAP1, CETP, EIF1AY, RBP5, PTGDS | 0.7279 | 0.4662 |
| T cell dysfunction | Genes regulating dysfunction of T cells in TME | 0.6844 | 0.5656 |
| T cell exclusion | Genes regulating T cell exclusion in TME | 0.7746 | 0.5622 |
| MDSC | Myeloid-derived suppressor cells | 0.6924 | 0.4353 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, X.; Zhu, Q.; Wu, Z.; Han, W. PSMB9 Orchestrates Tumor Immune Landscape and Serves as a Potent Biomarker for Prognosis and T Cell-Based Immunotherapy Response. Curr. Issues Mol. Biol. 2025, 47, 712. https://doi.org/10.3390/cimb47090712
Ma X, Zhu Q, Wu Z, Han W. PSMB9 Orchestrates Tumor Immune Landscape and Serves as a Potent Biomarker for Prognosis and T Cell-Based Immunotherapy Response. Current Issues in Molecular Biology. 2025; 47(9):712. https://doi.org/10.3390/cimb47090712
Chicago/Turabian StyleMa, Xinran, Qi Zhu, Zhiqiang Wu, and Weidong Han. 2025. "PSMB9 Orchestrates Tumor Immune Landscape and Serves as a Potent Biomarker for Prognosis and T Cell-Based Immunotherapy Response" Current Issues in Molecular Biology 47, no. 9: 712. https://doi.org/10.3390/cimb47090712
APA StyleMa, X., Zhu, Q., Wu, Z., & Han, W. (2025). PSMB9 Orchestrates Tumor Immune Landscape and Serves as a Potent Biomarker for Prognosis and T Cell-Based Immunotherapy Response. Current Issues in Molecular Biology, 47(9), 712. https://doi.org/10.3390/cimb47090712







