Exploring the Complete Chloroplast Genome of Pyrola decorata Andres: Structure, Variability, Phylogenetic Relationship
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Morphological Observations
2.2. DNA Extraction, Genome Sequencing, and Assembly
2.3. Codon Usage Frequency Analysis
2.4. Repeat Sequence Analysis
2.5. Analysis of IR Region Boundaries and Global Comparison of CpDNA
2.6. Phylogenetic Analysis
3. Results
3.1. Morphological Observations
3.2. Nuclear rDNA ITS Sequence Comparisons and Phylogenetic Analyses
3.3. CpDNA Assembly and Gene Annotation
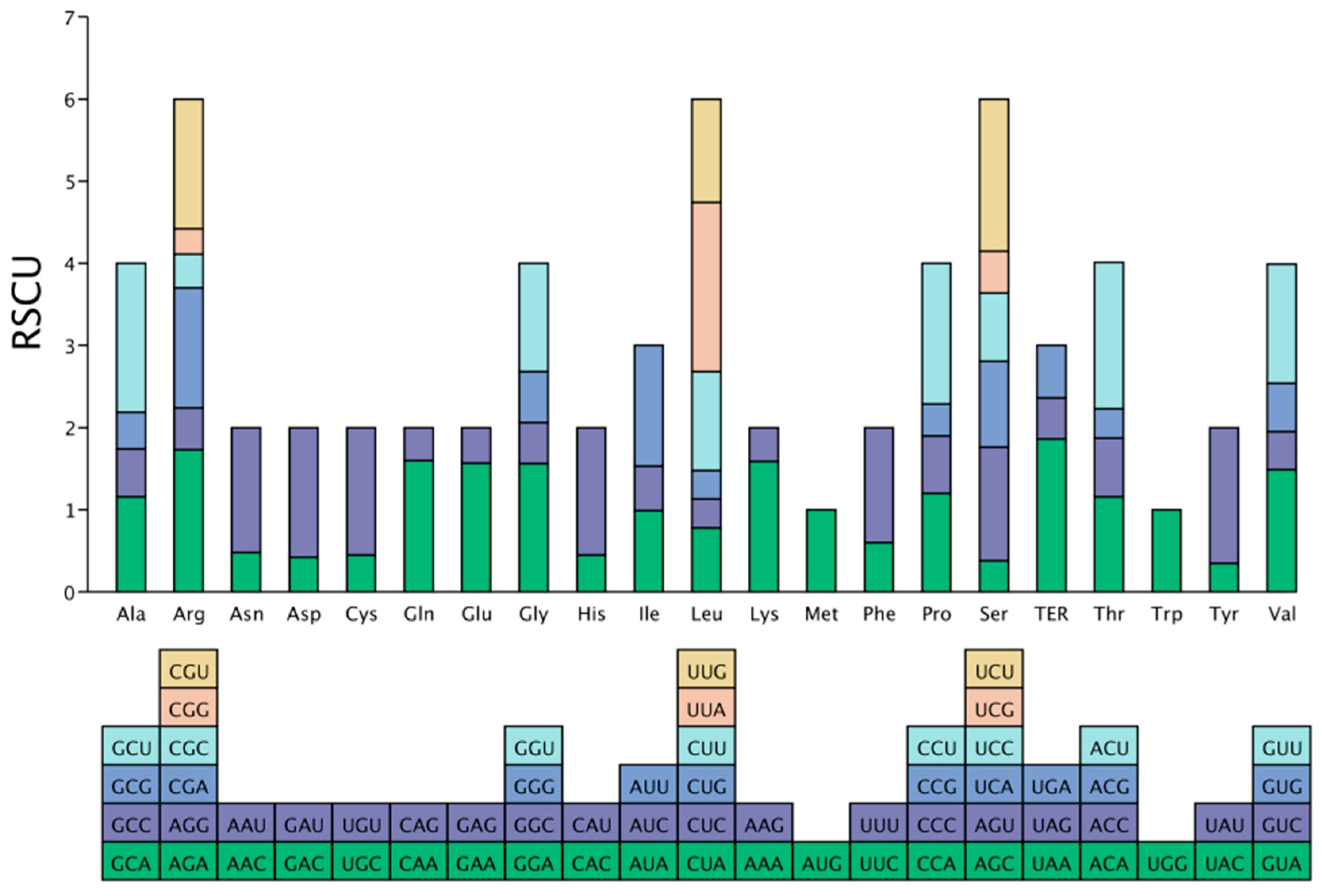
3.4. Analysis of Relative Synonymous Codon Usage
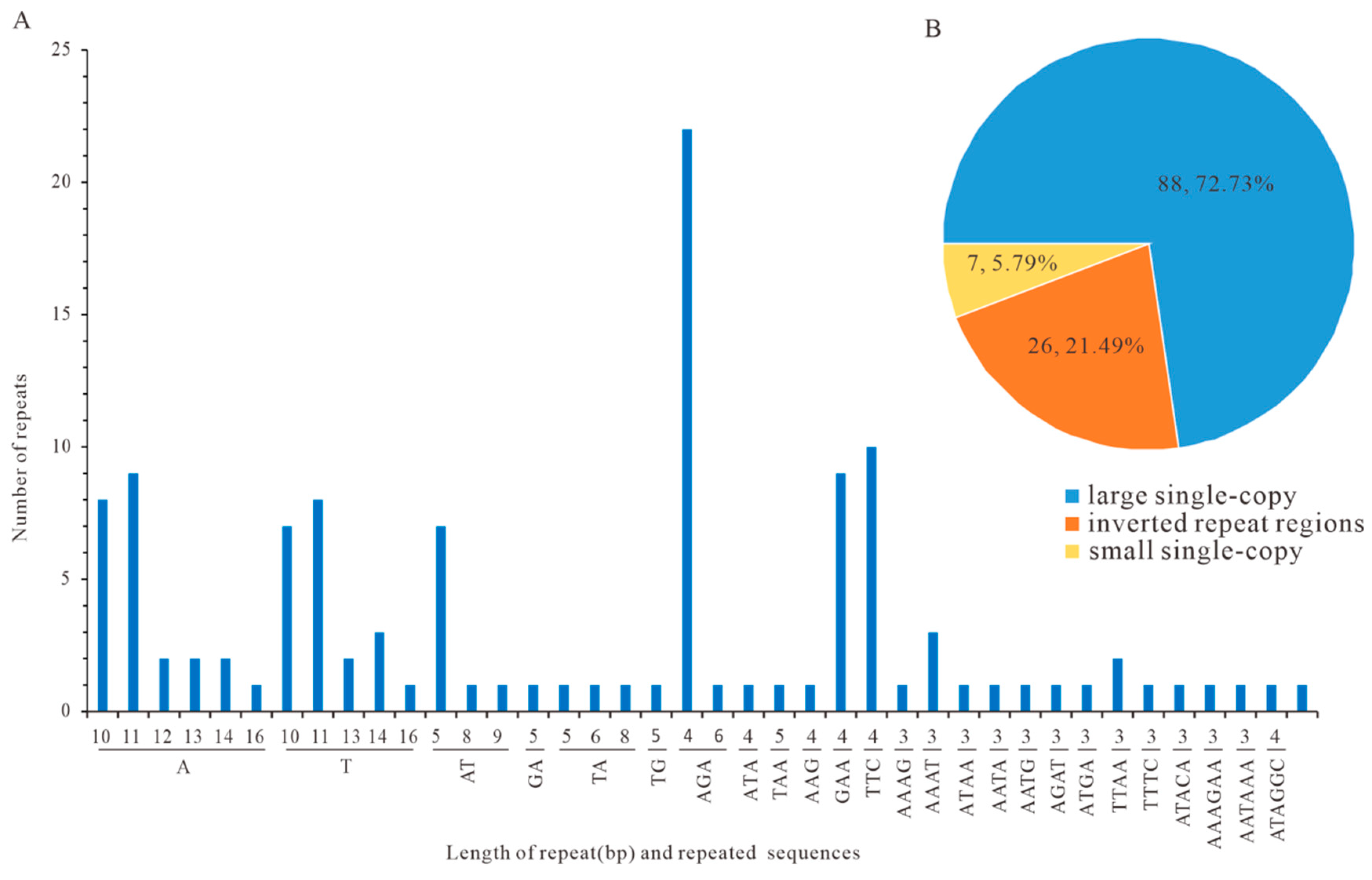
3.5. Repetitive Sequences in Analysis
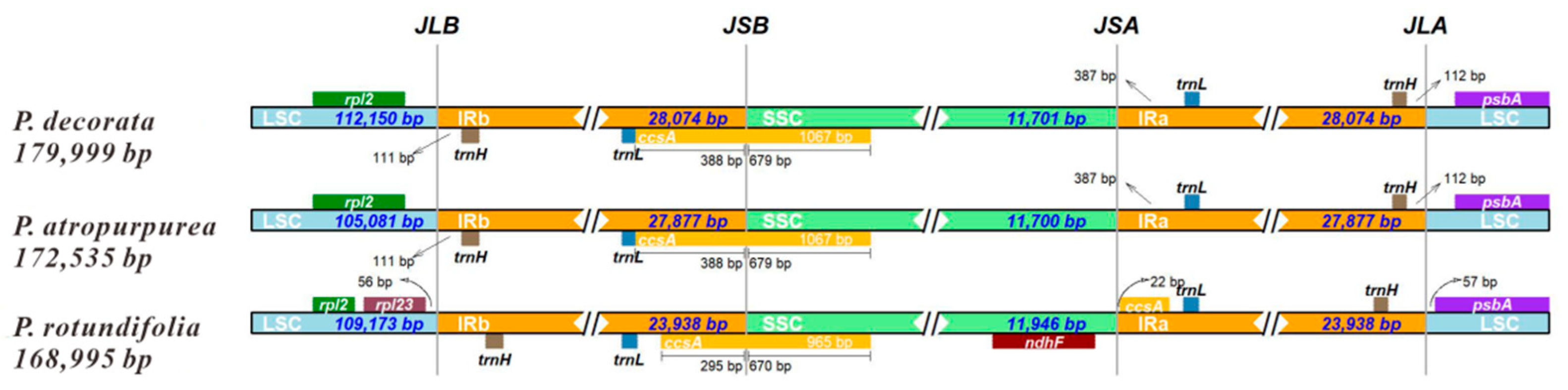
3.6. CpDNA Boundary Analysis of Three Pyrola Species
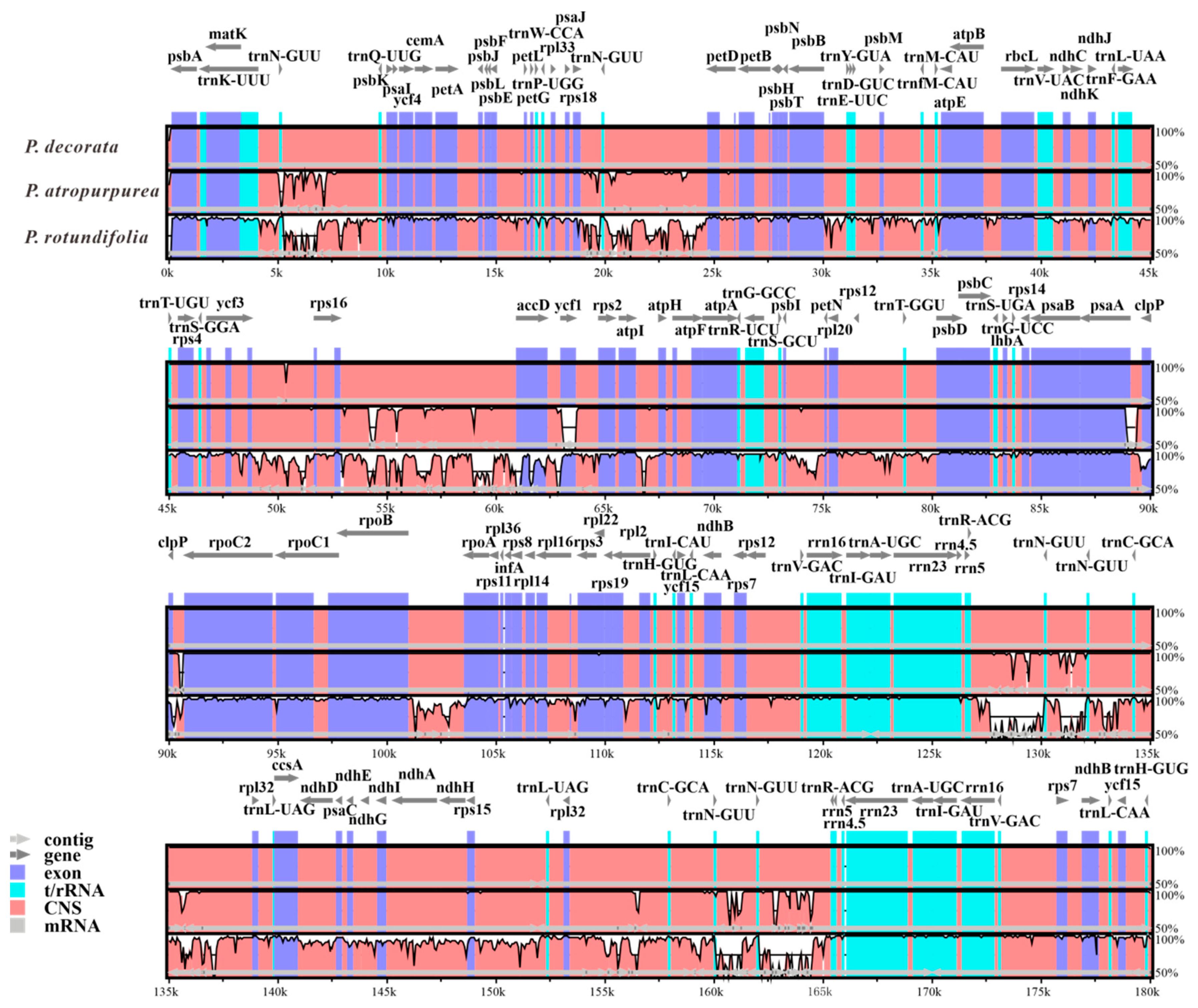
3.7. Comparative Divergence Analysis of Three Pyrola CpDNAs
3.8. Phylogenetic Inferences of Ericaceae Family Based on cpDNA Comparative Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dong, H.; Liu, Z.; Peng, H. Geographical distribution and floristic significance of Pyrola in China. Plant Sci. J. 2024, 42, 43–47. [Google Scholar] [CrossRef]
- Chinese Pharmacopoeia Commission. Pharmacopoeia of the People’s Republic of China, 2020 ed.; China Medical Science and Technology Press: Beijing, China, 2020; Volume 1, pp. 337–338.
- Deng, Q.; Peng, Z.; Liu, N.; Peng, J. Exploration of the mechanism of action of Pyrola calliantha in the treatment of rheumatoid arthritis based on weighted gene co-expression network analysis combined with network pharmacology. Asian J. Surg. 2024, 48, 720–721. [Google Scholar] [CrossRef]
- He, C.; Liu, J.; Ke, T.; Luo, Y.; Zhang, S.; Mao, T.; Li, Z.; Qin, X.; Jin, S. Pyrolae herba: A review on its botany, traditional uses, phytochemistry, pharmacology and quality control. J. Ethnopharmacol. 2022, 298, 115584. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, Y.; Bollas, A.; Wang, Y.; Au, K.F. Nanopore sequencing technology, bioinformatics and applications. Nat. Biotechnol. 2021, 39, 1348–1365. [Google Scholar] [CrossRef]
- Hu, T.; Chitnis, N.; Monos, D.; Dinh, A. Next-generation sequencing technologies: An overview. Hum. Immunol. 2021, 82, 801–811. [Google Scholar] [CrossRef]
- Zhang, T.; Li, H.; Jiang, M.; Hou, H.; Gao, Y.; Li, Y.; Wang, F.; Wang, J.; Peng, K.; Liu, Y.-X. Nanopore sequencing: Flourishing in its teenage years. J. Genet. Genom. 2024, 51, 1361–1374. [Google Scholar] [CrossRef]
- Tang, B.; Chen, Z.; Sun, Y.; Jin, Z. Identification of Luxiancao (Pyrolae Herba) and Its Pseudo Species Honghua Luticao (Pyrola Incamata Fisch. ex DC.). J. Shandong Univ. TCM 2019, 43, 519–522. [Google Scholar] [CrossRef]
- Su, Y.; Kang, Y.; Zhang, S.; Yu, B.; Xue, L. Distribution Pattern of Pyrola decorata and Pyrola calliantha Population in Taibai Mountain. J. Northeast. For. Univ. 2013, 41, 89–93. [Google Scholar] [CrossRef]
- Liu, Z.W.; Zhou, J.; Liu, E.D.; Peng, H. A molecular phylogeny and a new classification of Pyrola (Pyroleae, Ericaceae). Taxon 2010, 59, 1690–1700. [Google Scholar] [CrossRef]
- Yu, J.; Wang, C.; Gong, X. Degeneration of photosynthetic capacity in mixotrophic plants, Chimaphila japonica and Pyrola decorata (Ericaceae). Plant Divers. 2017, 39, 80–88. [Google Scholar] [CrossRef] [PubMed]
- Quiñones, K.J.O.; Gentallan, R.P.; Timog, E.B.S.; Cruz, J.R.A.V.; Macabecha, C.G.A.; Papa, I.A.; Coronado, N.B.; Bartolome, M.C.B.; Ceribo, D.B.; Madayag, R.E.; et al. Liquid-nitrogen-free CTAB DNA extraction method from silica-dried specimens for next-generation sequencing and assembly. MethodsX 2024, 12, 102758. [Google Scholar] [CrossRef]
- De Coster, W.; D’Hert, S.; Schultz, D.T.; Cruts, M.; Van Broeckhoven, C. NanoPack: Visualizing and processing long-read sequencing data. Bioinformatics 2018, 34, 2666–2669. [Google Scholar] [CrossRef] [PubMed]
- Patel, R.K.; Jain, M. NGS QC Toolkit: A toolkit for quality control of next generation sequencing data. PLoS ONE 2012, 7, e30619. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Wick, R.R.; Schultz, M.B.; Zobel, J.; Holt, K.E. Bandage: Interactive visualization of de novo genome assemblies. Bioinformatics 2015, 31, 3350–3352. [Google Scholar] [CrossRef]
- Alverson, A.J.; Wei, X.; Rice, D.W.; Stern, D.B.; Barry, K.; Palmer, J.D. Insights into the evolution of mitochondrial genome size from complete sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae). Mol. Biol. Evol. 2010, 27, 1436–1448. [Google Scholar] [CrossRef]
- Qu, X.-J.; Moore, M.J.; Li, D.-Z.; Yi, T.-S. PGA: A software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods 2019, 15, 50. [Google Scholar] [CrossRef]
- Greiner, S.; Lehwark, P.; Bock, R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef]
- Ling, L.; Zhang, S.; Yang, T. Analysis of Codon Usage Bias in Chloroplast Genomes of Dryas octopetala var. asiatica (Rosaceae). Genes 2024, 15, 899. [Google Scholar] [CrossRef]
- Zhao, M.; Wu, Y.; Ren, Y. Complete Chloroplast Genome Sequence Structure and Phylogenetic Analysis of Kohlrabi (Brassica oleracea var. gongylodes L.). Genes 2024, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Fernández-Jiménez, A.; Estefanía-Fresco, R.; García-De-La-Fuente, A.M.; Marichalar-Mendia, X.; Aguirre-Urizar, J.M.; Aguirre-Zorzano, L.A. Comparative study of the modified VISTA technique (m-VISTA) versus the coronally advanced flap (CAF) in the treatment of multiple Miller class III/RT2 recessions: A randomized clinical trial. Clin. Oral. Investig. 2023, 27, 505–517. [Google Scholar] [CrossRef]
- Amiryousefi, A.; Hyvönen, J.; Poczai, P. IRscope: An online program to visualize the junction sites of chloroplast genomes. Bioinformatics 2018, 34, 3030–3031. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Xiang, C.-Y.; Gao, F.; Jakovlić, I.; Lei, H.-P.; Hu, Y.; Zhang, H.; Zou, H.; Wang, G.-T.; Zhang, D. Using PhyloSuite for molecular phylogeny and tree-based analyses. Imeta 2023, 2, e87. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Nguyen, M.A.T.; von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Xie, J.; Chen, Y.; Cai, G.; Cai, R.; Hu, Z.; Wang, H. Tree Visualization By One Table (tvBOT): A web application for visualizing, modifying and annotating phylogenetic trees. Nucleic Acids Res. 2023, 51, W587–W592. [Google Scholar] [CrossRef]
- Zhu, J.; Huang, Y.; Chai, W.; Xia, P. Decoding the Chloroplast Genome of Tetrastigma (Vitaceae): Variations and Phylogenetic Selection Insights. Int. J. Mol. Sci. 2024, 25, 8290. [Google Scholar] [CrossRef]
- Yaradua, S.S.; Yessoufou, K. Chloroplast genome of Ecbolium viride (Forssk.) Alston: Plastome evolution and phylogenomics of Justiceae (Acanthaceae, Acanthoideae). Genome 2024, 67, 267–280. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, C.; Guo, X.; Wang, Y.; Chen, Y.; Shen, J.; He, C.; Yu, Y.; Wang, Q. Phylogenomics analysis of Scutellaria (Lamiaceae) of the world. BMC Biol. 2024, 22, 185. [Google Scholar] [CrossRef]
- Wei, X.-P.; Zhang, X.-Y.; Dong, Y.-Q.; Cheng, J.-L.; Bai, Y.-J.; Liu, J.-S.; Qi, Y.-D.; Zhang, B.-G.; Liu, H.-T. Molecular Structure and Phylogenetic Analyses of the Complete Chloroplast Genomes of Three Medicinal Plants Conioselinum vaginatum, Ligusticum sinense, and Ligusticum jeholense. Front. Plant Sci. 2022, 13, 878263. [Google Scholar] [CrossRef]
- Huang, Y.; Jin, X.-J.; Zhang, C.-Y.; Li, P.; Meng, H.-H.; Zhang, Y.-H. Plastome evolution of Engelhardia facilitates phylogeny of Juglandaceae. BMC Plant Biol. 2024, 24, 634. [Google Scholar] [CrossRef]
- Sheng, W.T. Chloroplast Genome Sequence Characterization and Phylogenetic Analysis of Pyrola Atropurpurea Franch. Phyton-Int. J. Exp. Bot. 2025, 94, 331–345. [Google Scholar] [CrossRef]
- Jolles, D.D. Morphometric analysis of floral variation in the Pyrola picta species complex (Ericaceae): Interpretation and implications for ecological and phylogenetic differentiation. Bot. J. Linn. Soc. 2015, 177, 462–480. [Google Scholar] [CrossRef]
- Flora of China Editorial Committee. Flora of China; Science Press: Beijing, China, 1990; Volume 56. [Google Scholar]
- Liu, Z.W.; Wang, Z.H.; Zhou, J.; Peng, H. Phylogeny of Pyroleae (Ericaceae): Implications for character evolution. J. Plant Res. 2011, 124, 325–337. [Google Scholar] [CrossRef]
- Liu, Z.W.; Jolles, D.D.; Zhou, J.; Peng, H.; Milne, R.I. Multiple origins of circumboreal taxa in Pyrola (Ericaceae), a group with a Tertiary relict distribution. Ann. Bot. 2014, 114, 1701–1709. [Google Scholar] [CrossRef]
- Yang, T.; Aishan, S.; Zhu, J.; Qin, Y.; Liu, J.; Liu, H.; Tie, J.; Wang, J.; Qin, R. Chloroplast Genomes and Phylogenetic Analysis of Three Carthamus (Asteraceae) Species. Int. J. Mol. Sci. 2023, 24, 15634. [Google Scholar] [CrossRef]
- Zhang, P.; Xu, W.; Lu, X.; Wang, L. Analysis of codon usage bias of chloroplast genomes in Gynostemma species. Physiol. Mol. Biol. Plants 2021, 27, 2727–2737. [Google Scholar] [CrossRef]
- Shang, M. Comparative chloroplast genomics and phylogenetic relationships in medicinal plants of genus Dendrobium Sw. In Chinese Traditional and Herbal Drugs; Dali University: Dali, China, 2024; pp. 35–36. [Google Scholar]
- Robbins, E.H.J.; Kelly, S. The Evolutionary Constraints on Angiosperm Chloroplast Adaptation. Genome Biol. Evol. 2023, 15, evad101. [Google Scholar] [CrossRef]
- Parvathy, S.T.; Udayasuriyan, V.; Bhadana, V. Codon usage bias. Mol. Biol. Rep. 2022, 49, 539–565. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Miao, Y.; Qian, J.; Zheng, Y.; Xia, C.; Yang, Q.; Liu, C.; Huang, L.; Duan, B. Comparative analysis of complete chloroplast genome sequences of five endangered species and new insights into phylogenetic relationships of Paris. Gene 2022, 833, 146572. [Google Scholar] [CrossRef]
- Ye, J.; Luo, Q.; Lang, Y.; Ding, N.; Jian, Y.-Q.; Wu, Z.-K.; Wei, S.-H.; Yan, F.-L. Analysis of chloroplast genome structure and phylogeny of the traditional medicinal of Ardisia crispa (Myrsinaceae). Sci. Rep. 2024, 14, 19045. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Huang, H.; Lin, J.; Liu, F.; Wang, X.; Chen, R.; Geng, X. Decoding the Chloroplast Genome of Bitterwood (Picrasma quassioides): Structure, Variability, and Evolutionary Relationships. Ecol. Evol. 2025, 15, e71245. [Google Scholar] [CrossRef]
- Cheng, C.; Fan, S.; Wang, C.; Ye, L.; Wang, Z.; Huang, H. Phylogenetic Analysis of Elaeagnus L. in China: A Basis for Genetic Improvement of a Berry Crop. Front. Plant Sci. 2022, 13, 899079. [Google Scholar] [CrossRef]
- Swarup, S.; Cargill, E.J.; Crosby, K.; Flagel, L.; Kniskern, J.; Glenn, K.C. Genetic diversity is indispensable for plant breeding to improve crops. Crop Sci. 2021, 61, 839–852. [Google Scholar] [CrossRef]




| Character | P. decorata | P. atropurpurea | P. calliantha | P. rotundifolia |
|---|---|---|---|---|
| tall | 15–35 cm | 7–18 cm | 15–30 cm | 15–25 cm |
| leaf morphology/size | leaf blade light green and purplish abaxially, deep green with pale veins adaxially, oblong or obovate-oblong, (3–)5–7 × 2.5–4 cm, thinly leathery, base cuneate | leaf blade reddish purple abaxially, green adaxially, cordate-ovate, (1–)1.5–3 × (1–)1.2–3 cm, papery, base cordate | leaf blade purplish and often glaucous abaxially, green adaxially, elliptic or ovate, (2.5–)3–6 × 2–3.5 cm, leathery, base broadly cuneate or suborbicular | leaf blade slightly green abaxially, green adaxially, slightly shiny, orbicular to ovate, (2–)3–6 × (1.5–)2.5–5.5 cm, leathery, base sometimes subcordate |
| scape bracts | lanceolate, membranous | naked or with 1 or 2 min lanceolate bracts | ligulate, 6–7.5 mm | axillary, imbricate, lanceolate, membranous |
| sepal shape | ovate-oblong 3–6 × 2–2.5 mm, apex acute | reddish purple, ovate-triangular, ca. 1.5 × 1.5 mm, apex obtuse | ligulate, (3–)5–7.5 × (1.5–)2–3 mm, margin entire, apex often acute | ovate-lanceolate to lanceolate, 3.5–5.5 mm, reflexed at tip, apex rounded |
| floral coloration | raceme 4–10-flowered, 2.5–4 cm, petals light green to white | raceme 2–4(–5) mm, 2–4-flowered, petals white | raceme 9–13-flowered, 12–16 cm, petals pure white | raceme 8–15-flowered, 6–16 cm, pure white |
| Feature | Pyrola decorata | A | C | G | T | GC |
|---|---|---|---|---|---|---|
| Total length | 179,999 bp | 32.91% 59,242 bp | 17.42% 31,363 bp | 17.33% 31,198 bp | 32.33% 58,196 bp | 34.76% |
| Large single-copy (LSC) length | 112,150 bp | 33.73% 37,827 bp | 16.76% 18,792 bp | 16.71% 18,741 bp | 32.80% 36,790 bp | 33.47% |
| Small single-copy (SSC) length | 11,701 bp | 36.05% 4218 bp | 14.48% 1694 bp | 13.50% 1580 bp | 35.97% 4209 bp | 27.98% |
| Inverted repeat region b (IRb) length | 28,074 bp | 29.85% 8380 bp | 20.99% 5894 bp | 17.75% 4983 bp | 31.41% 8817 bp | 38.74% |
| Inverted repeat region a (IRa) length | 28,074 bp | 31.41% 8817 bp | 17.75% 4983 bp | 20.99% 5894 bp | 29.85% 8380 bp | 38.74% |
| Category of Genes | Groups of Genes | Names of Genes | Gene Groups |
|---|---|---|---|
| Photosynthetic | Photosystem I | psaA, psaB, psaC, psaI, psaJ | 5 |
| Photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT | 14 | |
| NADH | ndhB(×2) a, ndhC, ndhE, ndhI, ndhJ | 6 | |
| Cytochrome b/f complex | petA, petB a, petD a, petG, petL, petN | 6 | |
| ATP synthase; large subunit of rubisco | atpA, atpB, atpE, atpF a, atpH, atpI | 6 | |
| Rubisco | rbcL | 1 | |
| Self-replication | Proteins of large ribosomal subunit | rpl2a, rpl14, rpl16 a, rpl20, rpl22, rpl32(×2), rpl33, rpl36 | 9 |
| Proteins of small ribosomal subunit | rps2, rps3, rps4, rps7(×2), rps8, rps11, rps12(×2), rps14, rps15, rps16 a, rps18, rps19 | 14 | |
| RNA polymerase | rpoA, rpoB, rpoC1 a, rpoC2 | 4 | |
| Ribosomal RNAs | rrn23(×2), rrn16(×2), rrn5(×2), rrn4.5(×2) | 8 | |
| Transfer RNAs | trnA-UGC(×2) a, trnC-GCA(×2), trnD-GUC, trnE-UUC, trnF-GAA, trnG-GCC a, trnG-UCC, trnH-GUG(×2), trnI-CAU, trnI-GAU(×2) a, trnK-UUUa, trnL-CAA(×2), trnL-UAAa, trnL-UAG(×2), trnM-CAU, trnN-GUU(×6) a,trnP-UGG, trnQ-UUG, trnR-ACG, trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC(×2), trnV-UAC a, trnW-CCA, trnY-GUA, trnfM-CAU, trnL-UAG | 43 | |
| Other | Maturase | matK | 1 |
| Protease | clpP a | 1 | |
| Envelope membrane protein | cemA | 1 | |
| Acetyl-CoA carboxylase | accD | 1 | |
| C-type cytochrome synthesis gene | ccsA | 1 | |
| Translation initiation factor | infA | 1 | |
| LhbA | lhbA | 1 | |
| Unknown function | Conserved open reading frames | ycf1, ycf3 b, ycf4, ycf15(×2) | 5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, R.; Kang, S.; Yu, K.; Jiang, Y.; Qin, Z.; Hu, Y.; Cheng, X.; Wei, F. Exploring the Complete Chloroplast Genome of Pyrola decorata Andres: Structure, Variability, Phylogenetic Relationship. Curr. Issues Mol. Biol. 2025, 47, 688. https://doi.org/10.3390/cimb47090688
Kang R, Kang S, Yu K, Jiang Y, Qin Z, Hu Y, Cheng X, Wei F. Exploring the Complete Chloroplast Genome of Pyrola decorata Andres: Structure, Variability, Phylogenetic Relationship. Current Issues in Molecular Biology. 2025; 47(9):688. https://doi.org/10.3390/cimb47090688
Chicago/Turabian StyleKang, Rong, Shuai Kang, Kunzi Yu, Yuan Jiang, Zeliang Qin, Yuying Hu, Xianlong Cheng, and Feng Wei. 2025. "Exploring the Complete Chloroplast Genome of Pyrola decorata Andres: Structure, Variability, Phylogenetic Relationship" Current Issues in Molecular Biology 47, no. 9: 688. https://doi.org/10.3390/cimb47090688
APA StyleKang, R., Kang, S., Yu, K., Jiang, Y., Qin, Z., Hu, Y., Cheng, X., & Wei, F. (2025). Exploring the Complete Chloroplast Genome of Pyrola decorata Andres: Structure, Variability, Phylogenetic Relationship. Current Issues in Molecular Biology, 47(9), 688. https://doi.org/10.3390/cimb47090688






