Bioinformatic Analysis of the Value of Mitophagy and Immune Responses in Corneal Endothelial Dysfunction
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Extraction
2.2. DEGs Related to CED
2.3. GO and KEGG Analyses of DEGs
2.4. GSEA and GSVA
2.5. PPI Network Analysis
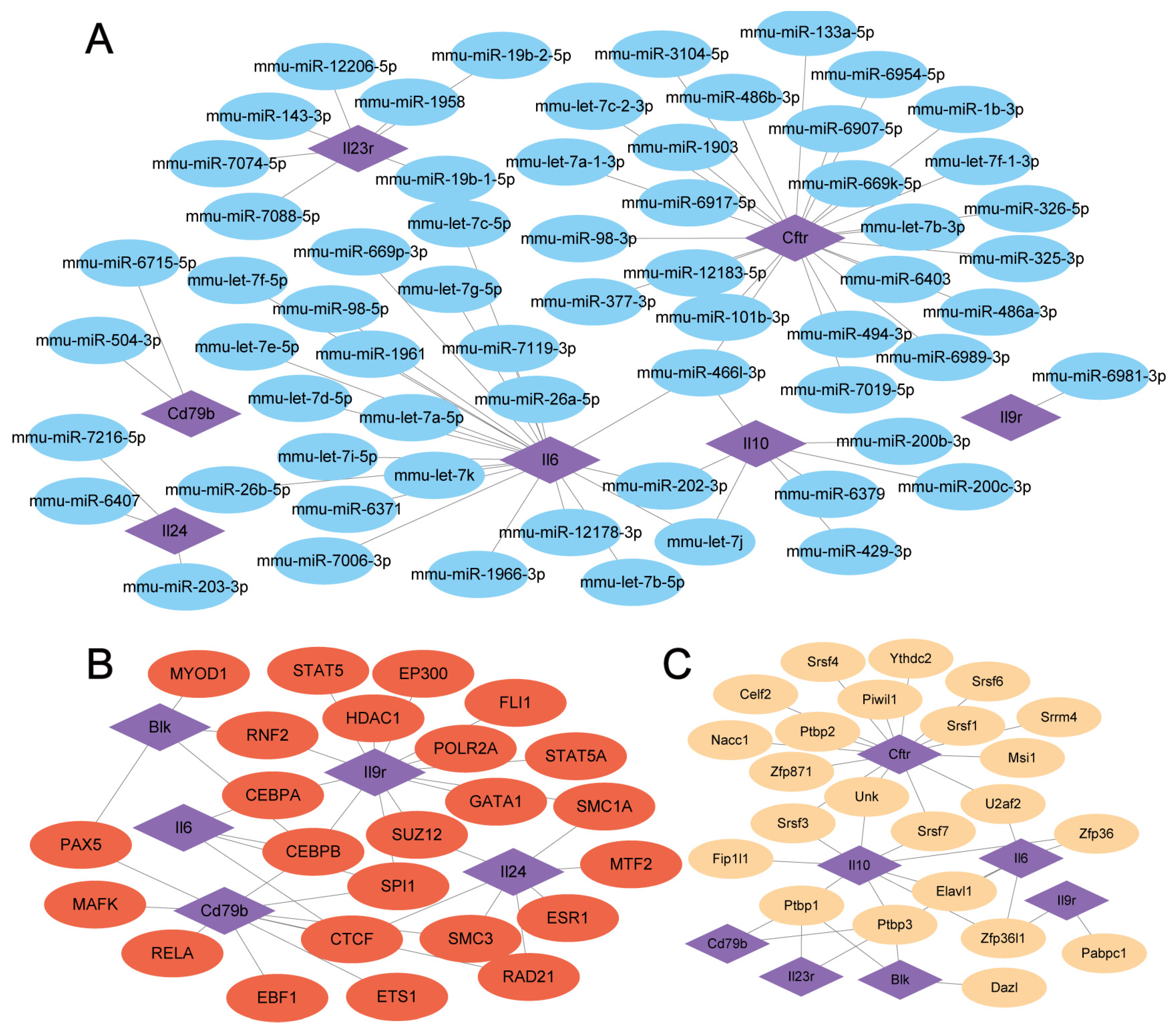
2.6. Construction of the mRNA-miRNA, mRNA-TF, and mRNA-RBP Interaction Networks
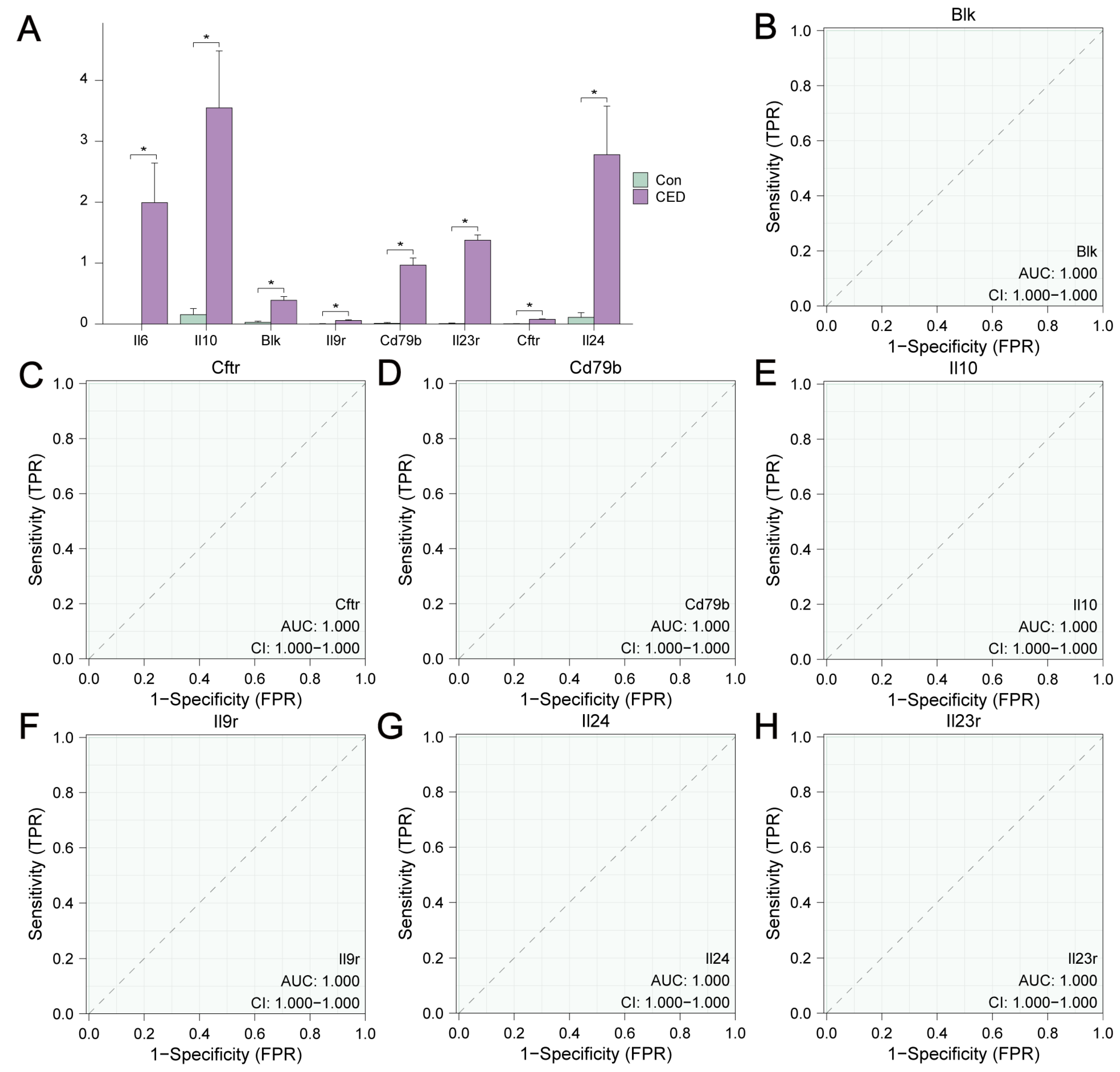
2.7. ROC Curve Analysis
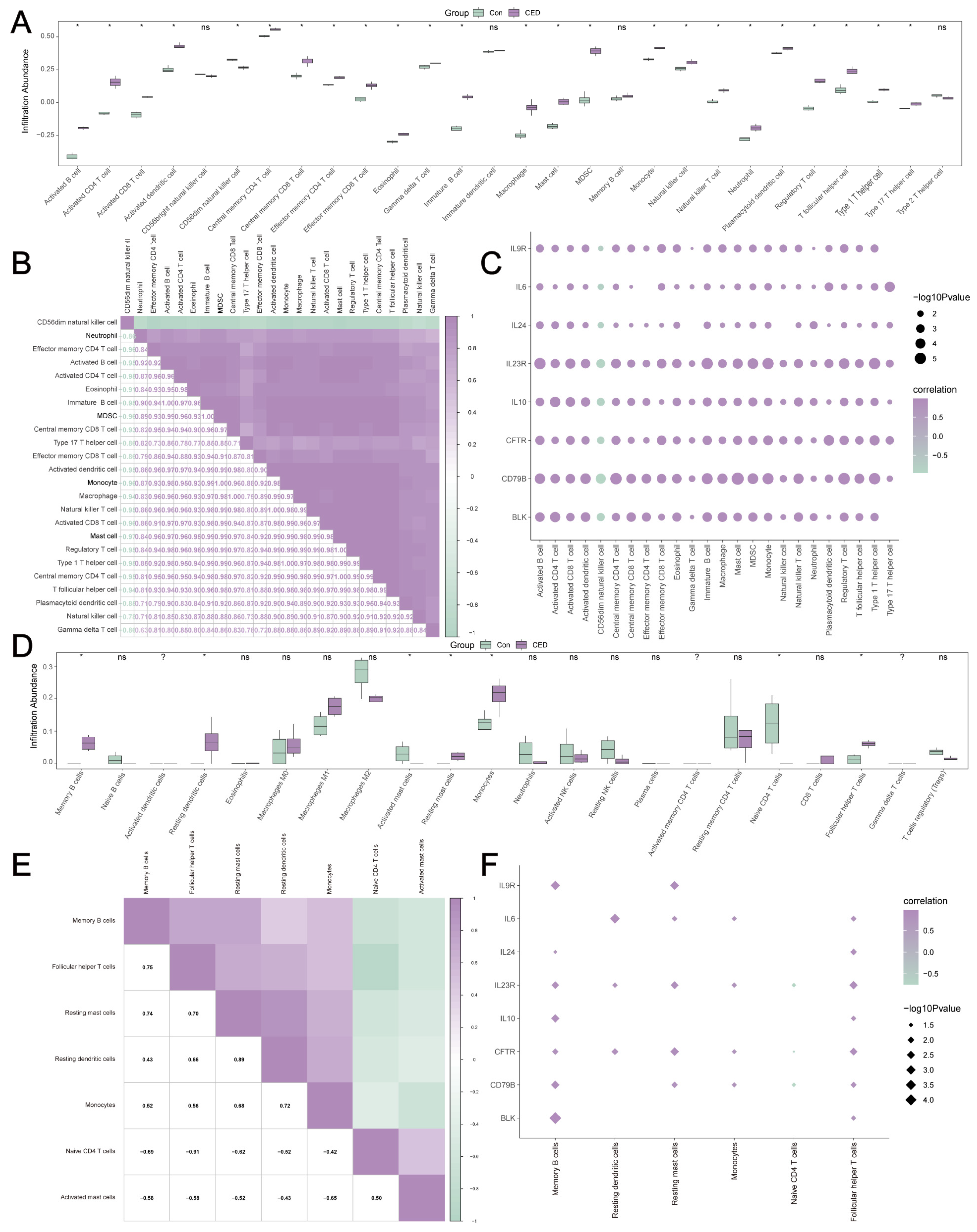
2.8. Immune Infiltration Analysis
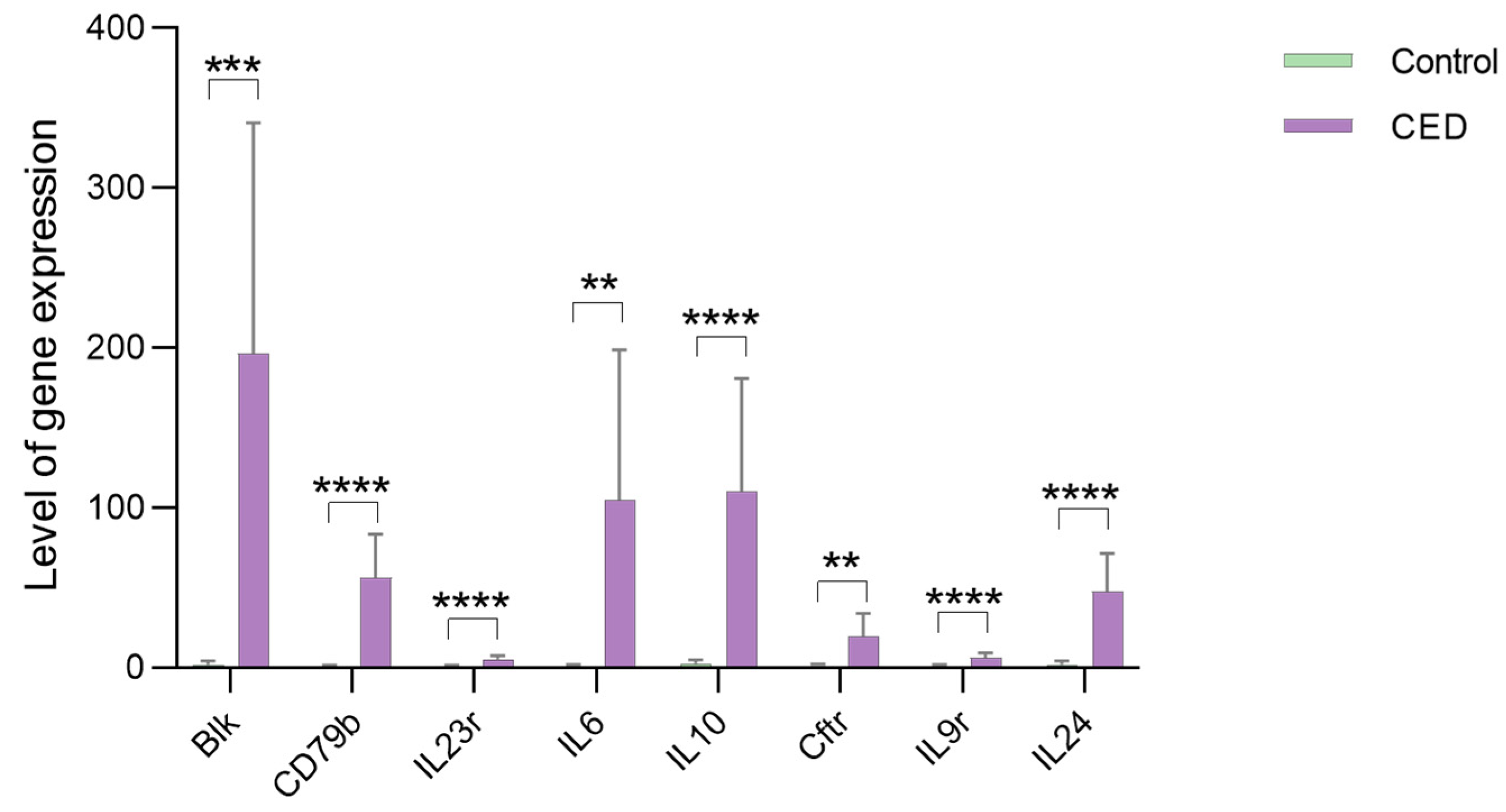
2.9. qRT-PCR Validation of the Hub Genes
2.10. Statistical Analysis
3. Results
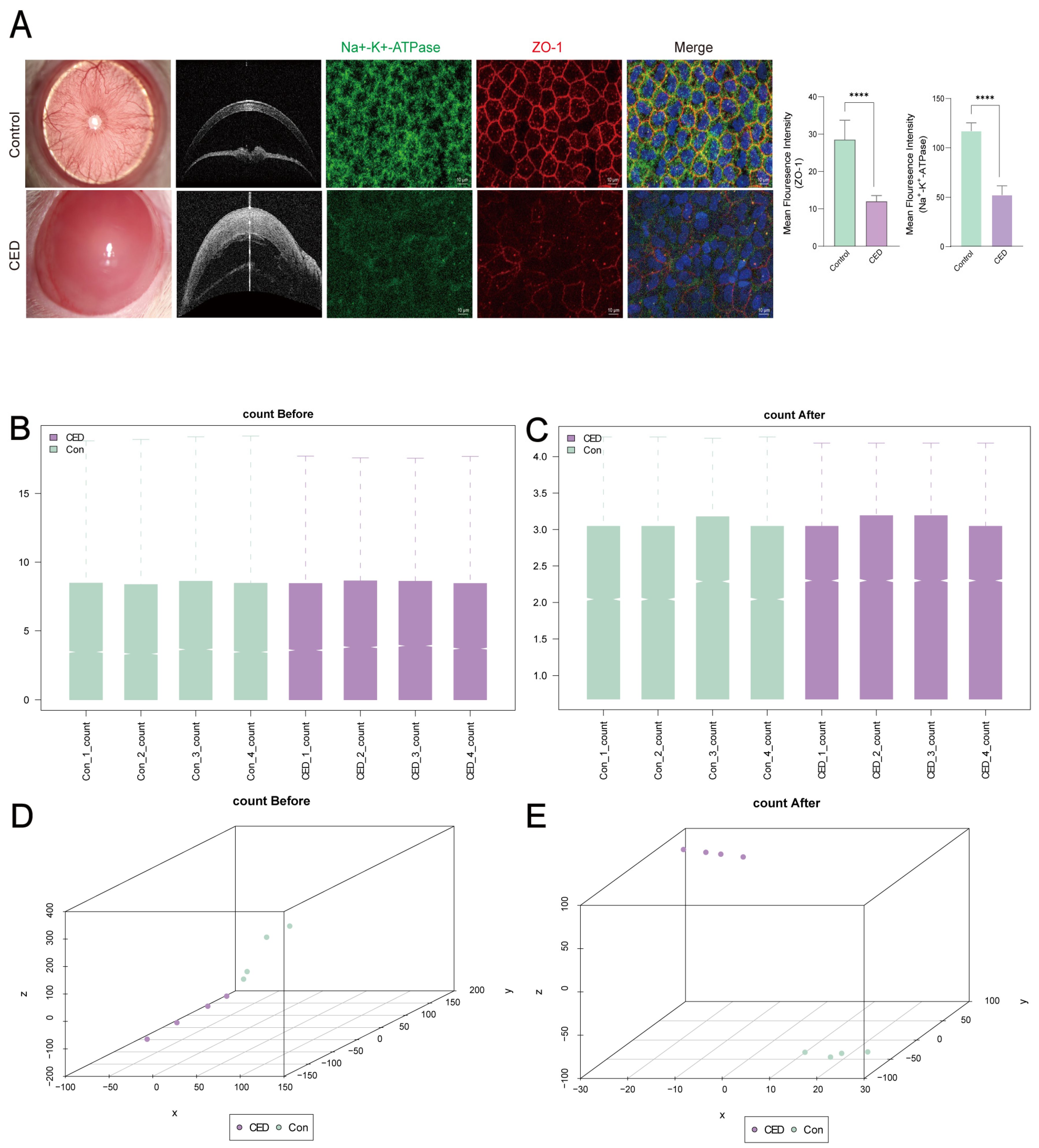
3.1. TNF-α and IFN-γ Induced CED
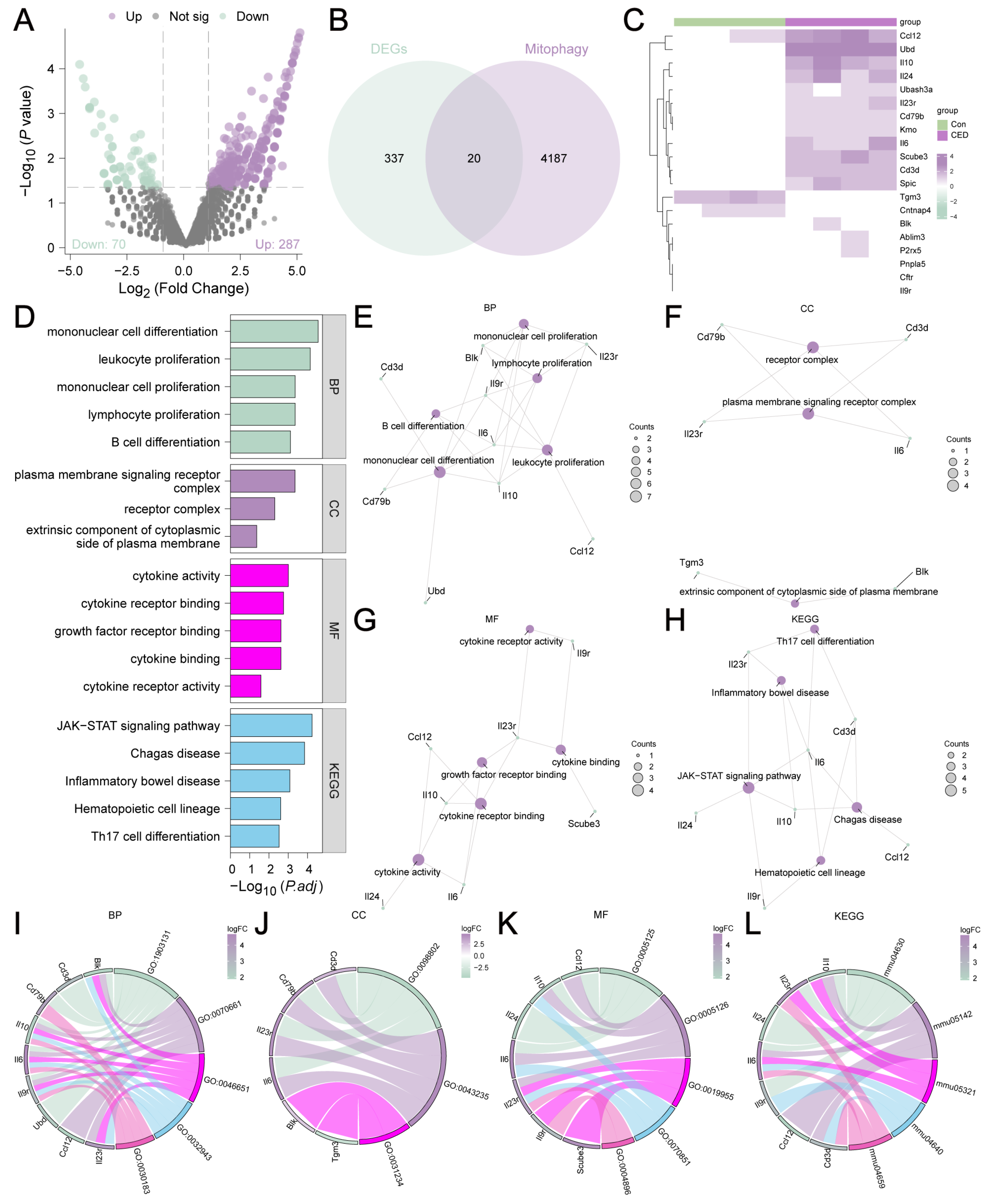
3.2. Analysis of DEGs Related to CED
3.3. GO and KEGG Analyses of MRDEGs
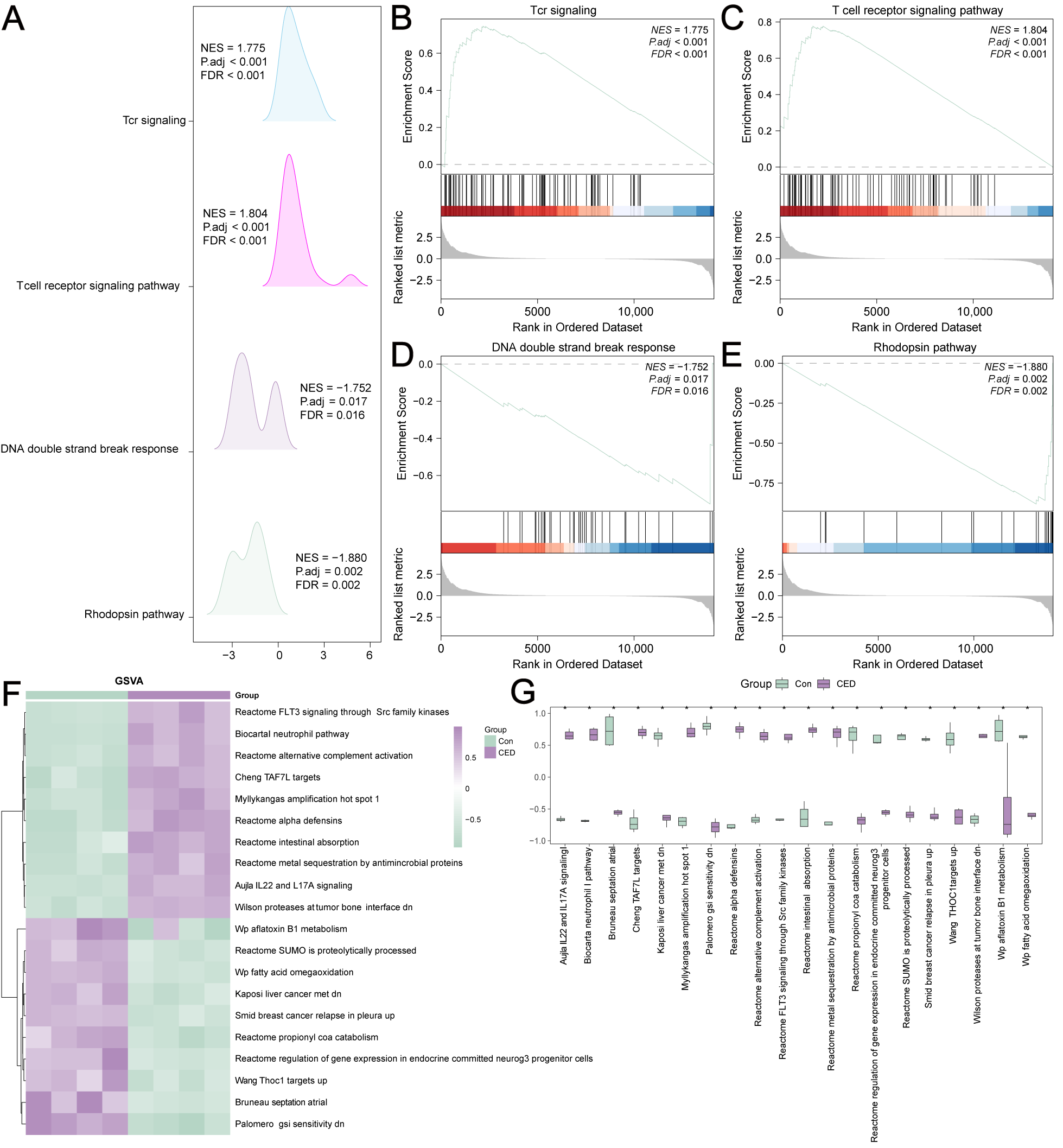
3.4. GSEA and GSVA of Data from the Self-Testing CED Dataset
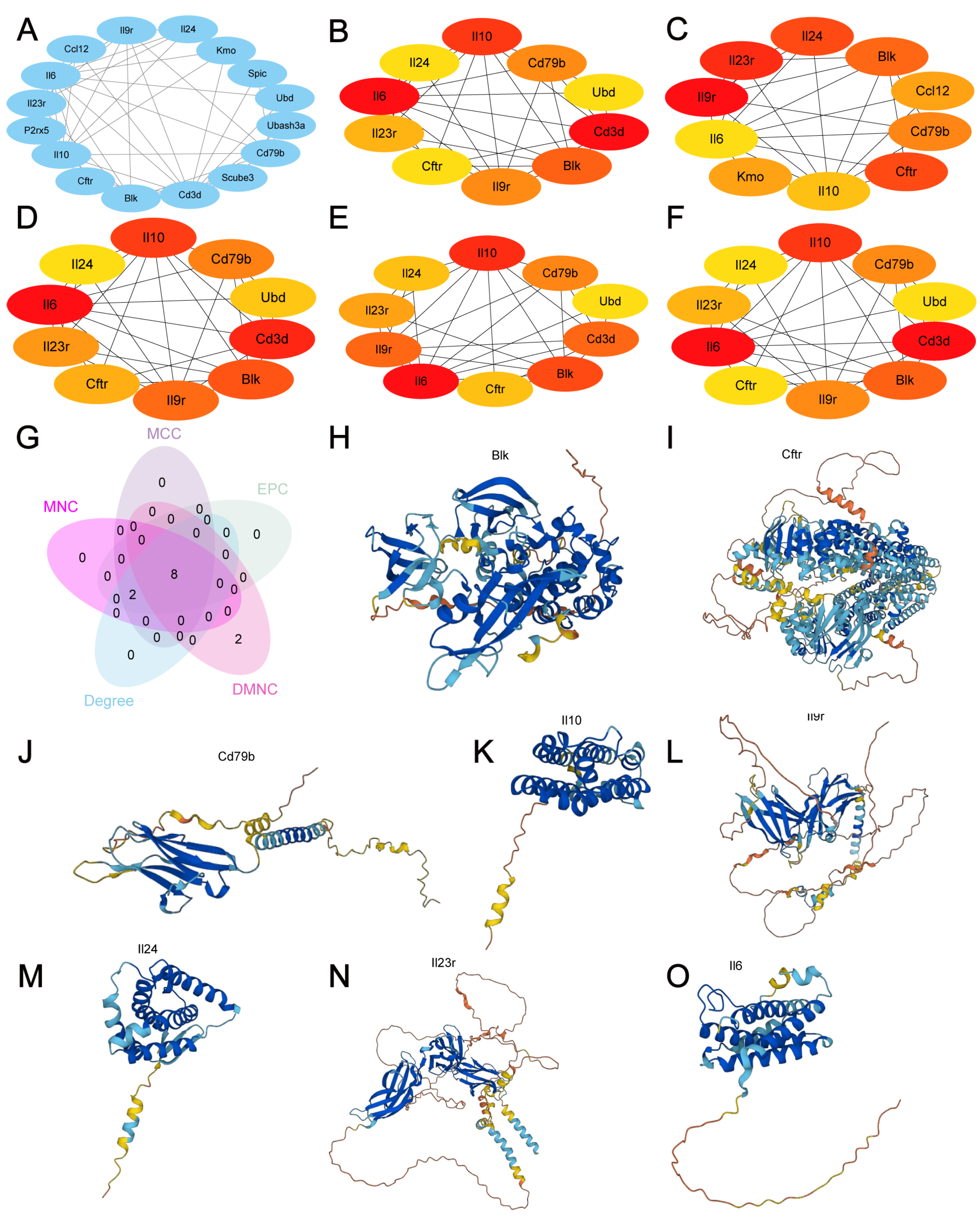
3.5. Construction of PPI Networks
3.6. mRNA-miRNA, mRNA-TF, and mRNA-RBP Interaction Networks
3.7. Differential Expression Analysis of MRDEGs
3.8. Immune Characteristic Analysis of MRDEGs in the Self-Testing CED Dataset
3.9. qPCR Validation of the Hub Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CED | Corneal endothelial dysfunction |
| MRDEGs | Mitophagy-related differentially expressed genes |
| DEGs | Differentially expressed genes |
| GSEA | Gene set enrichment analysis |
| GSVA | Gene set variation analysis |
| PPI | Protein–protein interaction |
| AS-OCT | Anterior segment optical coherence tomography |
| CCT | Central corneal thickness |
| MRGs | Mitophagy-related genes |
| MCC | Matthews correlation coefficient |
| DMNC | Differential metabolic network construction |
| MNC | Maximal neighborhood coefficient |
| EPC | Edge percolated component |
| TFs | Transcription factors |
| RBPs | RNA-binding proteins |
| ROC | Receiver operating characteristic curve |
References
- Xia, X.; Atkins, M.; Dalal, R.; Kuzmenko, O.; Chang, K.-C.; Sun, C.B.; Benatti, C.A.; Rak, D.J.; Nahmou, M.; Kunzevitzky, N.J.; et al. Magnetic Human Corneal Endothelial Cell Transplant: Delivery, Retention, and Short-Term Efficacy. Investig. Ophthalmol. Vis. Sci. 2019, 60, 2438–2448. [Google Scholar] [CrossRef] [PubMed]
- Dirisamer, M.; Yeh, R.-Y.; van Dijk, K.; Ham, L.; Dapena, I.; Melles, G.R.J. Recipient endothelium may relate to corneal clearance in descemet membrane endothelial transfer. Am. J. Ophthalmol. 2012, 154, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Studený, P.; Hamouz, J.; Kuchynka, P. Corneal transplantations in the Czech Republic in 2012. Cesk Slov. Oftalmol. 2014, 70, 224–227. [Google Scholar]
- Wu, Q.; Ouyang, C.; Xie, L.; Ling, Y.; Huang, T. The ROCK inhibitor, thiazovivin, inhibits human corneal endothelial-to-mesenchymal transition/epithelial-to-mesenchymal transition and increases ionic transporter expression. Int. J. Mol. Med. 2017, 40, 1009–1018. [Google Scholar] [CrossRef]
- Vallabh, N.A.; Romano, V.; Willoughby, C.E. Mitochondrial dysfunction and oxidative stress in corneal disease. Mitochondrion 2017, 36, 103–113. [Google Scholar] [CrossRef]
- Kumar, V.; Jurkunas, U.V. Mitochondrial Dysfunction and Mitophagy in Fuchs Endothelial Corneal Dystrophy. Cells 2021, 10, 1888. [Google Scholar] [CrossRef]
- Liu, X.; Zheng, T.; Zhao, C.; Zhang, Y.; Liu, H.; Wang, L.; Liu, P. Genetic mutations and molecular mechanisms of Fuchs endothelial corneal dystrophy. Eye Vis. 2021, 8, 24. [Google Scholar] [CrossRef]
- Ward, K.W. Targeting the NRF2 pathway: A promising approach for corneal endothelial dysfunction. Curr. Opin. Pharmacol. 2024, 74, 102429. [Google Scholar] [CrossRef] [PubMed]
- Zeitz, C.; Roger, J.E.; Audo, I.; Michiels, C.; Sánchez-Farías, N.; Varin, J.; Frederiksen, H.; Wilmet, B.; Callebert, J.; Gimenez, M.-L.; et al. Shedding light on myopia by studying complete congenital stationary night blindness. Prog. Retin. Eye Res. 2023, 93, 101155. [Google Scholar] [CrossRef]
- Koizumi, N.; Okumura, N.; Ueno, M.; Kinoshita, S. New therapeutic modality for corneal endothelial disease using Rho-associated kinase inhibitor eye drops. Cornea 2014, 33 (Suppl. S11), S25–S31. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Yu, Y.; Xu, C.; Ma, M.; Hou, C.; Dong, X.; Wu, J.; Ouyang, C.; Ling, J.; Huang, T. Protective effects of curcumin on corneal endothelial cell PANoptosis and monocyte adhesion induced by tumor necrosis factor-alpha and interferon-gamma in rats. Exp. Eye Res. 2024, 245, 109952. [Google Scholar] [CrossRef]
- Xu, C.; Guo, R.; Hou, C.; Ma, M.; Dong, X.; Ouyang, C.; Wu, J.; Huang, T. Resveratrol regulates macrophage recruitment and M1 macrophage polarization and prevents corneal allograft rejection in rats. Front. Med. 2023, 10, 1250914. [Google Scholar] [CrossRef] [PubMed]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinform. 2016, 54, 1–30. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Yu, G. Gene Ontology Semantic Similarity Analysis Using GOSemSim. Methods Mol. Biol. 2020, 2117, 207–215. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Hanzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef]
- Henrik Heiland, D.; Ravi, V.M.; Behringer, S.P.; Frenking, J.H.; Wurm, J.; Joseph, K.; Garrelfs, N.W.C.; Strähle, J.; Heynckes, S.; Grauvogel, J.; et al. Tumor-associated reactive astrocytes aid the evolution of immunosuppressive environment in glioblastoma. Nat. Commun. 2019, 10, 2541. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, C.A.; Dougherty, P.G.; Cooper, J.K.; Qian, Z.; Lindert, S.; Wang, Q.-E.; Pei, D. Cell-Permeable Bicyclic Peptidyl Inhibitors against NEMO-IκB Kinase Interaction Directly from a Combinatorial Library. J. Am. Chem. Soc. 2018, 140, 12102–12110. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Boughorbel, S.; Jarray, F.; El-Anbari, M. Optimal classifier for imbalanced data using Matthews Correlation Coefficient metric. PLoS ONE 2017, 12, e0177678. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612, Erratum in Nucleic Acids Res. 2021, 49, 10800. [Google Scholar] [CrossRef]
- Chen, C.; Lan, B.; Xie, G.; Liu, Z. Analysis and identification of ferroptosis-related genes in ulcerative colitis. Scand. J. Gastroenterol. 2023, 58, 1422–1433. [Google Scholar] [CrossRef] [PubMed]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]
- Akhtar, M.M.; Micolucci, L.; Islam, M.S.; Olivieri, F.; Procopio, A.D. Bioinformatic tools for microRNA dissection. Nucleic Acids Res. 2016, 44, 24–44. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, X. miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef] [PubMed]
- G Hendrickson, D.; Kelley, D.R.; Tenen, D.; Bernstein, B.; Rinn, J.L. Widespread RNA binding by chromatin-associated proteins. Genome Biol. 2016, 17, 28. [Google Scholar] [CrossRef] [PubMed]
- Zhou, K.R.; Liu, S.; Sun, W.J.; Zheng, L.L.; Zhou, H.; Yang, J.H.; Qu, L.H. ChIPBase v2.0: Decoding transcriptional regulatory networks of non-coding RNAs and protein-coding genes from ChIP-seq data. Nucleic Acids Res. 2017, 45, D43–D50. [Google Scholar] [CrossRef]
- Wu, D.; Huo, C.; Jiang, S.; Huang, Y.; Fang, X.; Liu, J.; Yang, M.; Ren, J.; Xu, B.; Liu, Y. Exostosin1 as a novel prognostic and predictive biomarker for squamous cell lung carcinoma: A study based on bioinformatics analysis. Cancer Med. 2021, 10, 2787–2801. [Google Scholar] [CrossRef]
- Mandrekar, J.N. Receiver operating characteristic curve in diagnostic test assessment. J. Thorac. Oncol. 2010, 5, 1315–1316. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Tosolini, M.; Kirilovsky, A.; Waldner, M.; Obenauf, A.C.; Angell, H.; Fredriksen, T.; Lafontaine, L.; Berger, A.; et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 2013, 39, 782–795. [Google Scholar] [CrossRef]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef]
- Lovatt, M.; Adnan, K.; Peh, G.S.L.; Mehta, J.S. Regulation of Oxidative Stress in Corneal Endothelial Cells by Prdx6. Antioxidants 2018, 7, 180. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Liu, Y.; Feng, Y.; Li, X.; Lu, Z.; Gu, H.; Li, W.; Hill, L.J.; Ou, S. Evolution of therapeutic strategy based on oxidant-antioxidant balance for fuchs endothelial corneal dystrophy. Ocul. Surf. 2024, 34, 247–261. [Google Scholar] [CrossRef] [PubMed]
- Gomez, A.; Serrano, A.; Salero, E.; Tovar, A.; Amescua, G.; Galor, A.; Keane, R.W.; de Rivero Vaccari, J.P.; Sabater, A.L. Tumor necrosis factor-alpha and interferon-gamma induce inflammasome-mediated corneal endothelial cell death. Exp. Eye Res. 2021, 207, 108574. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, T.; Higa, K.; Suzuki, T.; Nakayama, N.; Yagi-Yaguchi, Y.; Dogru, M.; Satake, Y.; Shimazaki, J. Elevated Cytokine Levels in the Aqueous Humor of Eyes with Bullous Keratopathy and Low Endothelial Cell Density. Investig. Ophthalmol. Vis. Sci. 2016, 57, 5954–5962. [Google Scholar] [CrossRef]
- Birgisdottir, Å.B.; Lamark, T.; Johansen, T. The LIR motif—Crucial for selective autophagy. J. Cell Sci. 2013, 126, 3237–3247. [Google Scholar] [CrossRef] [PubMed]
- Mancias, J.D.; Kimmelman, A.C. Mechanisms of Selective Autophagy in Normal Physiology and Cancer. J. Mol. Biol. 2016, 428, 1659–1680. [Google Scholar] [CrossRef]
- Chen, C.; Zhou, Q.; Li, Z.; Duan, H.; Liu, Y.; Wan, L.; Wang, H.; Xie, L. Hyperglycemia induces corneal endothelial dysfunction through attenuating mitophagy. Exp. Eye Res. 2022, 215, 108903. [Google Scholar] [CrossRef]
- Jiménez-Loygorri, J.I.; Benítez-Fernández, R.; Viedma-Poyatos, Á.; Zapata-Muñoz, J.; Villarejo-Zori, B.; Gómez-Sintes, R.; Boya, P. Mitophagy in the retina: Viewing mitochondrial homeostasis through a new lens. Prog. Retin. Eye Res. 2023, 96, 101205. [Google Scholar] [CrossRef]
- Kaarniranta, K.; Blasiak, J.; Liton, P.; Boulton, M.; Klionsky, D.J.; Sinha, D. Autophagy in age-related macular degeneration. Autophagy 2023, 19, 388–400. [Google Scholar] [CrossRef]
- Rodrigues, M.M.; Stulting, R.D.; Waring, G.O. Clinical, electron microscopic, and immunohistochemical study of the corneal endothelium and Descemet’s membrane in the iridocorneal endothelial syndrome. Am. J. Ophthalmol. 1986, 101, 16–27. [Google Scholar] [CrossRef]
- Aggarwal, S.; Cavalcanti, B.M.; Regali, L.; Cruzat, A.; Trinidad, M.; Williams, C.; Jurkunas, U.V.; Hamrah, P. In Vivo Confocal Microscopy Shows Alterations in Nerve Density and Dendritiform Cell Density in Fuchs’ Endothelial Corneal Dystrophy. Am. J. Ophthalmol. 2018, 196, 136–144. [Google Scholar] [CrossRef]
- Nesburn, A.B.; Bettahi, I.; Dasgupta, G.; Chentoufi, A.A.; Zhang, X.; You, S.; Morishige, N.; Wahlert, A.J.; Brown, D.J.; Jester, J.V.; et al. Functional Foxp3 + CD4 + CD25(Bright+) “natural” regulatory T cells are abundant in rabbit conjunctiva and suppress virus-specific CD4+ and CD8+ effector T cells during ocular herpes infection. J. Virol. 2007, 81, 7647–7661. [Google Scholar] [CrossRef]
- Zhang, X.; Schaumburg, C.S.; Coursey, T.G.; Siemasko, K.F.; Volpe, E.A.; Gandhi, N.B.; Li, D.Q.; Niederkorn, J.Y.; Stern, M.E.; Pflugfelder, S.C.; et al. CD8⁺ cells regulate the T helper-17 response in an experimental murine model of Sjögren syndrome. Mucosal Immunol. 2014, 7, 417–427. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Sugita, S.; Horie, S.; Yamagami, S.; Mochizuki, M. Mechanisms of immune suppression for CD8+ T cells by human corneal endothelial cells via membrane-bound TGFbeta. Investig. Ophthalmol. Vis. Sci. 2010, 51, 2548–2557. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Yang, Z.; Huang, C.; McGowan, J.; Casper, T.; Sun, D.; Born, W.K.; O’Brien, R.L. γδ T Cell-Dependent Regulatory T Cells Prevent the Development of Autoimmune Keratitis. J. Immunol. 2015, 195, 5572–5581. [Google Scholar] [CrossRef] [PubMed]
- Sakowska, J.; Glasner, P.; Dukat-Mazurek, A.; Rydz, A.; Zieliński, M.; Pellowska, I.; Biernat, W.; Glasner, L.; Michalska-Małecka, K.; Trzonkowski, P. Local T cell infiltrates are predominantly associated with corneal allograft rejection. Transpl. Immunol. 2023, 79, 101852. [Google Scholar] [CrossRef]
- Wasserman, R.; Li, Y.S.; Hardy, R.R. Differential expression of the blk and ret tyrosine kinases during B lineage development is dependent on Ig rearrangement. J. Immunol. 1995, 155, 644–651. [Google Scholar] [CrossRef]
- Engel, I.; Seumois, G.; Chavez, L.; Samaniego-Castruita, D.; White, B.; Chawla, A.; Mock, D.; Vijayanand, P.; Kronenberg, M. Innate-like functions of natural killer T cell subsets result from highly divergent gene programs. Nat. Immunol. 2016, 17, 728–739, Erratum in Nat. Immunol. 2019, 20, 1700. [Google Scholar] [CrossRef]
- Yu, L.M.; Chang, T.W. Human mb-1 gene: Complete cDNA sequence and its expression in B cells bearing membrane Ig of various isotypes. J. Immunol. 1992, 148, 633–637. [Google Scholar] [CrossRef]
- Schneider-Futschik, E.K.; Zhu, Y.; Li, D.; Habgood, M.D.; Nguyen, B.N.; Pankonien, I.; Amaral, M.D.; Downie, L.E.; Chinnery, H.R. The role of CFTR in the eye, and the effect of early highly effective modulator treatment for cystic fibrosis on eye health. Prog. Retin. Eye Res. 2024, 103, 101299. [Google Scholar] [CrossRef]
- Levin, M.H.; Verkman, A.S. Aquaporins and CFTR in ocular epithelial fluid transport. J. Membr. Biol. 2006, 210, 105–115. [Google Scholar] [CrossRef]
- Pestka, S.; Krause, C.D.; Walter, M.R. Interferons, interferon-like cytokines, and their receptors. Immunol. Rev. 2004, 202, 8–32. [Google Scholar] [CrossRef]
- Heinrich, P.C.; Behrmann, I.; Müller-Newen, G.; Schaper, F.; Graeve, L. Interleukin-6-type cytokine signalling through the gp130/Jak/STAT pathway. Biochem. J. 1998, 334 Pt 2, 297–314. [Google Scholar] [CrossRef]
- Alkon, N.; Assen, F.P.; Arnoldner, T.; Bauer, W.M.; Medjimorec, M.A.; Shaw, L.E.; Rindler, K.; Holzer, G.; Weber, P.; Weninger, W.; et al. Single-cell RNA sequencing defines disease-specific differences between chronic nodular prurigo and atopic dermatitis. J. Allergy Clin. Immunol. 2023, 152, 420–435. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; David, M.; Bouchareychas, L.; Rouquier, S.; Sajuthi, S.; Ayrault, M.; Navarin, C.; Lara, G.; Lafon, A.; Saviane, G.; et al. IL23R-specific CAR Tregs for the treatment of Crohn’s disease. J. Crohns Colitis 2024, 19, jjae135. [Google Scholar] [CrossRef] [PubMed]
| CED Dataset | |
| Species | Rattus norvegicus |
| Tissue | Corneal endothelium |
| Samples in CED group | 4 |
| Samples in control group | 4 |
| Description | logFC | p Adjust |
|---|---|---|
| Palomero gsi sensitivity dn | 1.59117 | 0.0000691 |
| Reactome propionyl coa catabolism | 1.34145 | 0.000139 |
| Bruneau septation atrial | 1.28909 | 0.000257 |
| Kaposi liver cancer met dn | 1.27419 | 0.000116 |
| Wp fatty acid omegaoxidation | 1.22521 | 0.0000691 |
| Wang THOC1 targets up | 1.22475 | 0.000187 |
| Wp aflatoxin B1 metabolism | 1.21478 | 0.02236 |
| Reactome regulation of gene expression in endocrine commited neurog3 progenitor cells | 1.21111 | 0.000192 |
| Smid breast cancer relapse in pleua up | 1.20045 | 0.000073 |
| Reactome SUMO is proteolytically processed | 1.19211 | 0.000116 |
| Reactome FLT3signaling through Src family kinases | 1.306768 | 0.000073 |
| Reactome alternative complement activation | 1.31033 | 0.000069 |
| Wilson Proteases at tumor bone interface dn | 1.312847 | 0.0000691 |
| Aujla IL22 and IL17Asignaling | 1.322637 | 0.0000691 |
| Reactome intestinal absorption | 1.354629 | 0.000131 |
| Biocarta neutrophil pathway | 1.355318 | 0.0000691 |
| Reactome metal sequestration by antimicrobial proteins | 1.376517 | 0.0000855 |
| Cheng TAF7L targets | 1.413491 | 0.0000782 |
| Myllkangas amplification hot spot 1 | 1.41585 | 0.0000691 |
| Reactome alpha defensins | 1.495057 | 0.0000691 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, R.; Xu, C.; Yu, Y.; Ma, M.; Dong, X.; Wu, J.; Ouyang, C.; Ling, J.; Huang, T. Bioinformatic Analysis of the Value of Mitophagy and Immune Responses in Corneal Endothelial Dysfunction. Curr. Issues Mol. Biol. 2025, 47, 670. https://doi.org/10.3390/cimb47080670
Guo R, Xu C, Yu Y, Ma M, Dong X, Wu J, Ouyang C, Ling J, Huang T. Bioinformatic Analysis of the Value of Mitophagy and Immune Responses in Corneal Endothelial Dysfunction. Current Issues in Molecular Biology. 2025; 47(8):670. https://doi.org/10.3390/cimb47080670
Chicago/Turabian StyleGuo, Ruilin, Chenjia Xu, Yi Yu, Minglu Ma, Xiaojuan Dong, Jing Wu, Chen Ouyang, Jie Ling, and Ting Huang. 2025. "Bioinformatic Analysis of the Value of Mitophagy and Immune Responses in Corneal Endothelial Dysfunction" Current Issues in Molecular Biology 47, no. 8: 670. https://doi.org/10.3390/cimb47080670
APA StyleGuo, R., Xu, C., Yu, Y., Ma, M., Dong, X., Wu, J., Ouyang, C., Ling, J., & Huang, T. (2025). Bioinformatic Analysis of the Value of Mitophagy and Immune Responses in Corneal Endothelial Dysfunction. Current Issues in Molecular Biology, 47(8), 670. https://doi.org/10.3390/cimb47080670






