Exploring Novel Inhibitory Compounds Against Phosphatase Gamma 2: A Therapeutic Target for Male Contraceptives
Abstract
1. Introduction
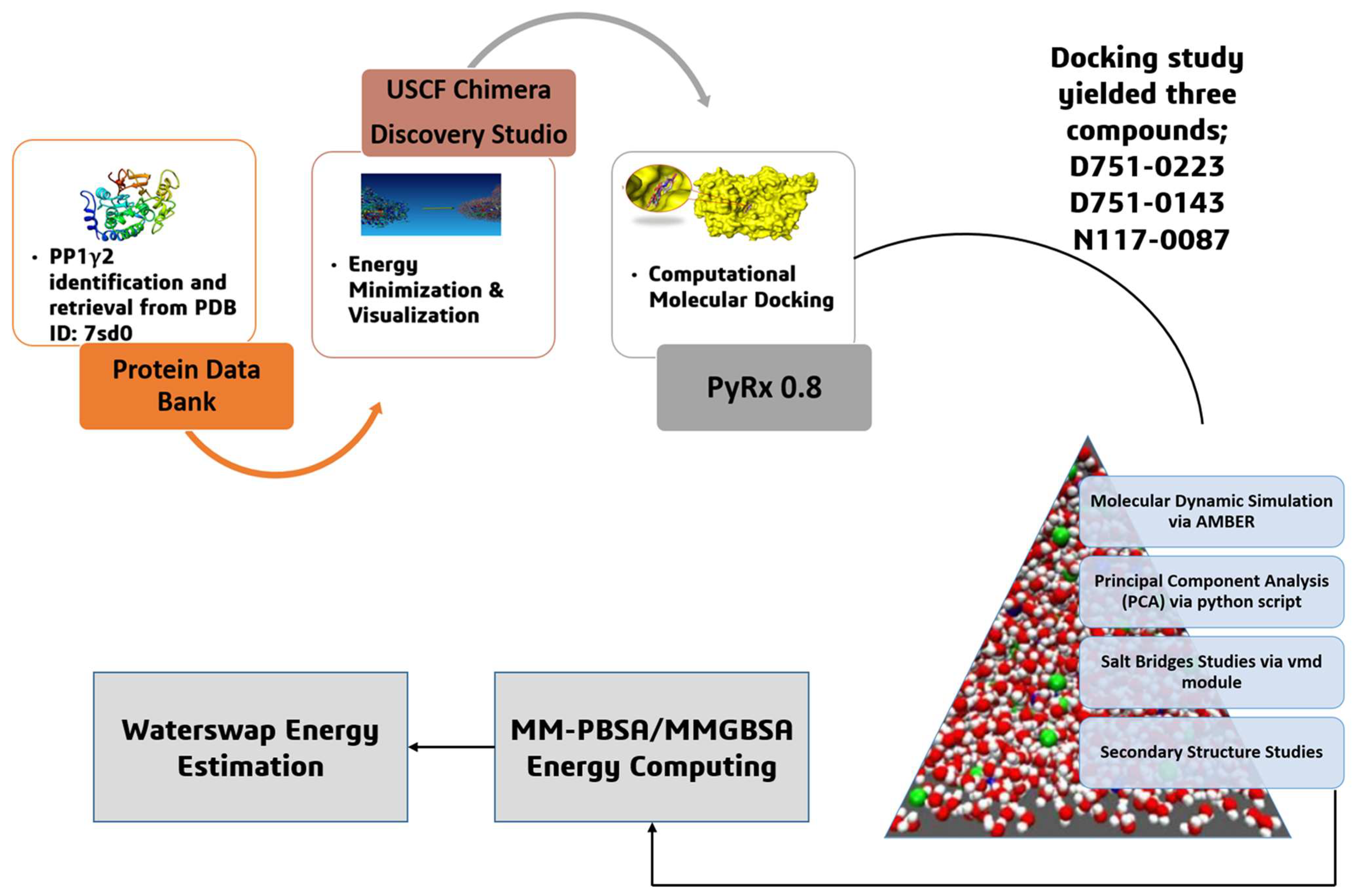
2. Materials and Methods
2.1. Identification, Retrieval, and Preparation of Target Receptor
2.2. Identification of Binding Pockets
2.3. Identification and Preparation of Compound Library
2.4. Screening of Compound Library Against the Receptor
2.5. Interpretation and Interaction Analysis of Docking Findings
2.6. Validating Lipinski Rule of 5 and Pharmacokinetics Assessment
2.7. Molecular Dynamics Simulation
2.8. Salt Bridges (SS) Analysis
2.9. Principal Component Analysis (PCA)
2.10. Secondary Structure Analysis
2.11. MMPB/GBSA Calculations
2.12. WaterSwap Energy Estimation
3. Results
3.1. Identification, Retrieval, and Preparation of Male Contraceptives with the Main PP1γ2
3.2. Molecular Docking Analysis
3.3. Docking Analysis and Interpretation of Selected Hits
3.4. Lipinski Rule of 5 and Pharmacokinetics Characteristics
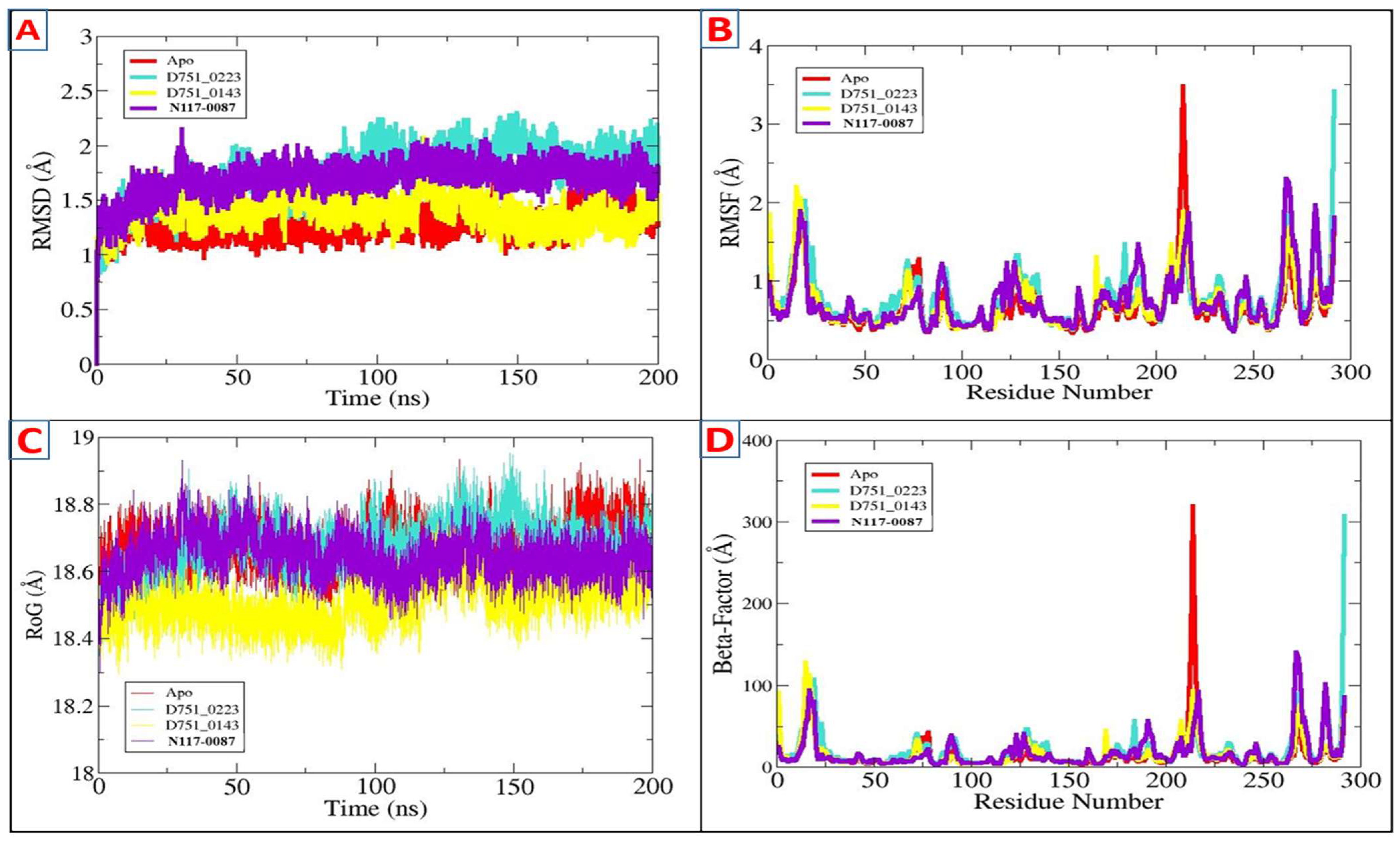
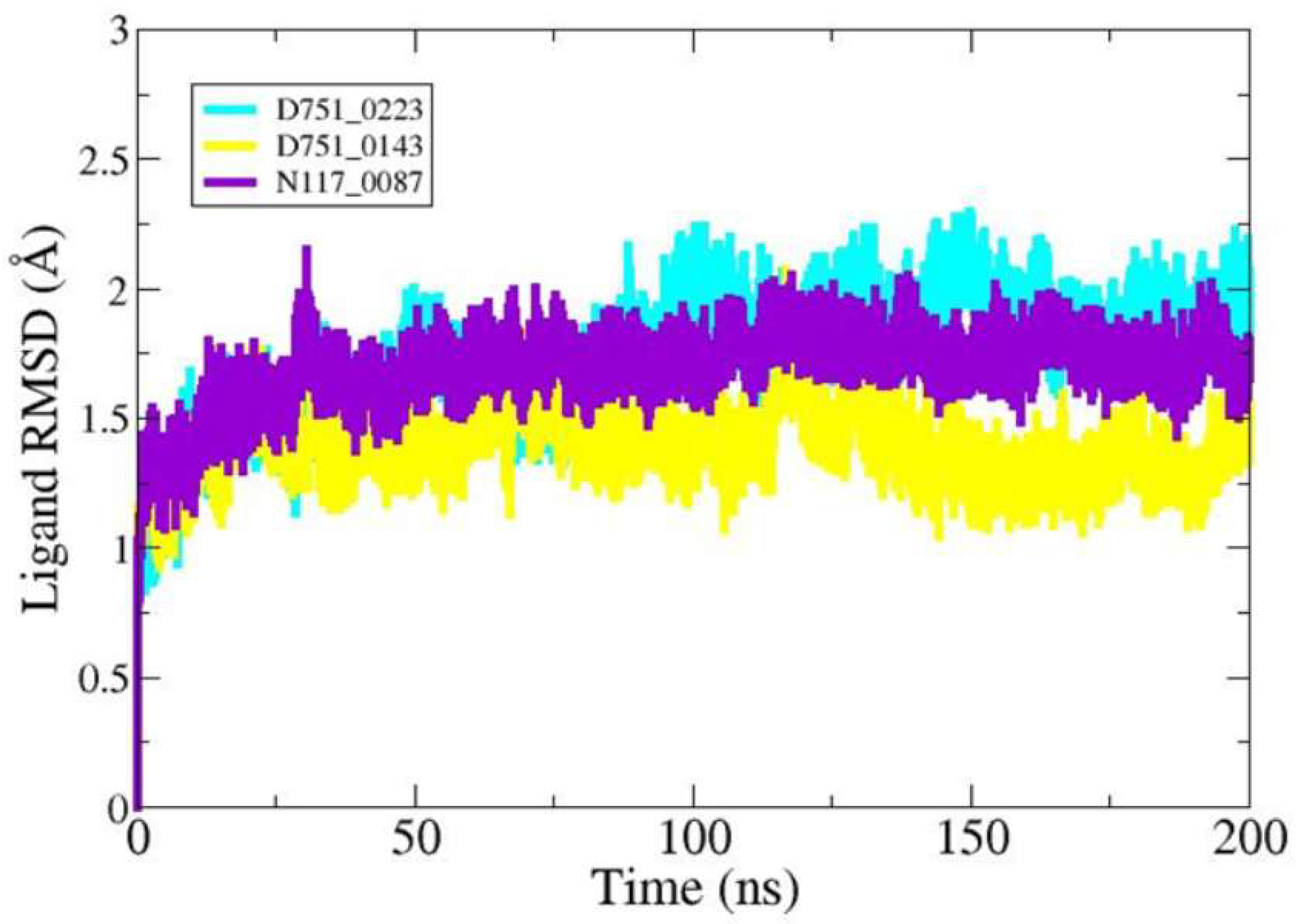
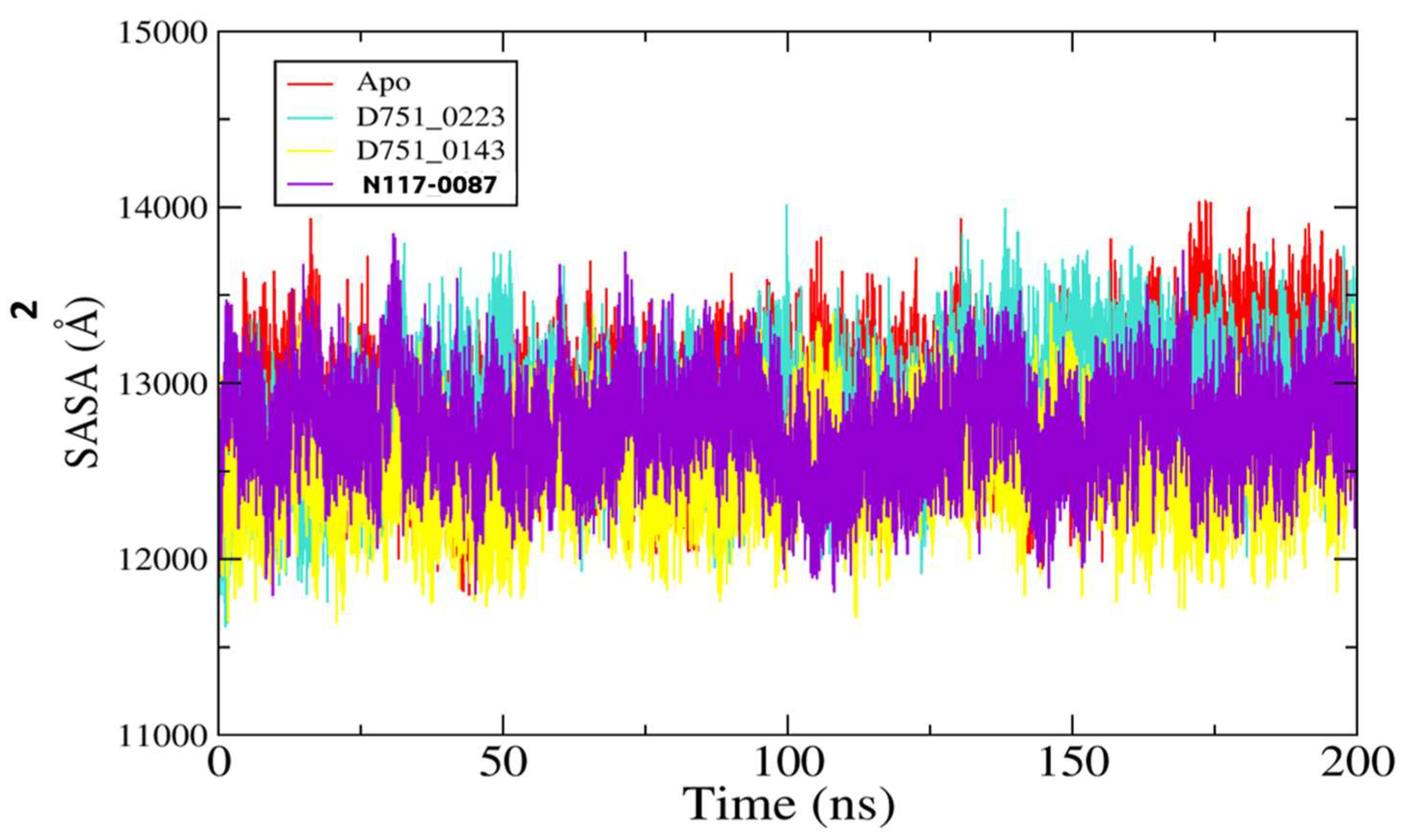
3.5. Molecular Dynamic Simulation of PP1γ2–Ligand Complexes and Apo
3.6. Salt Bridge Studies: (SB)
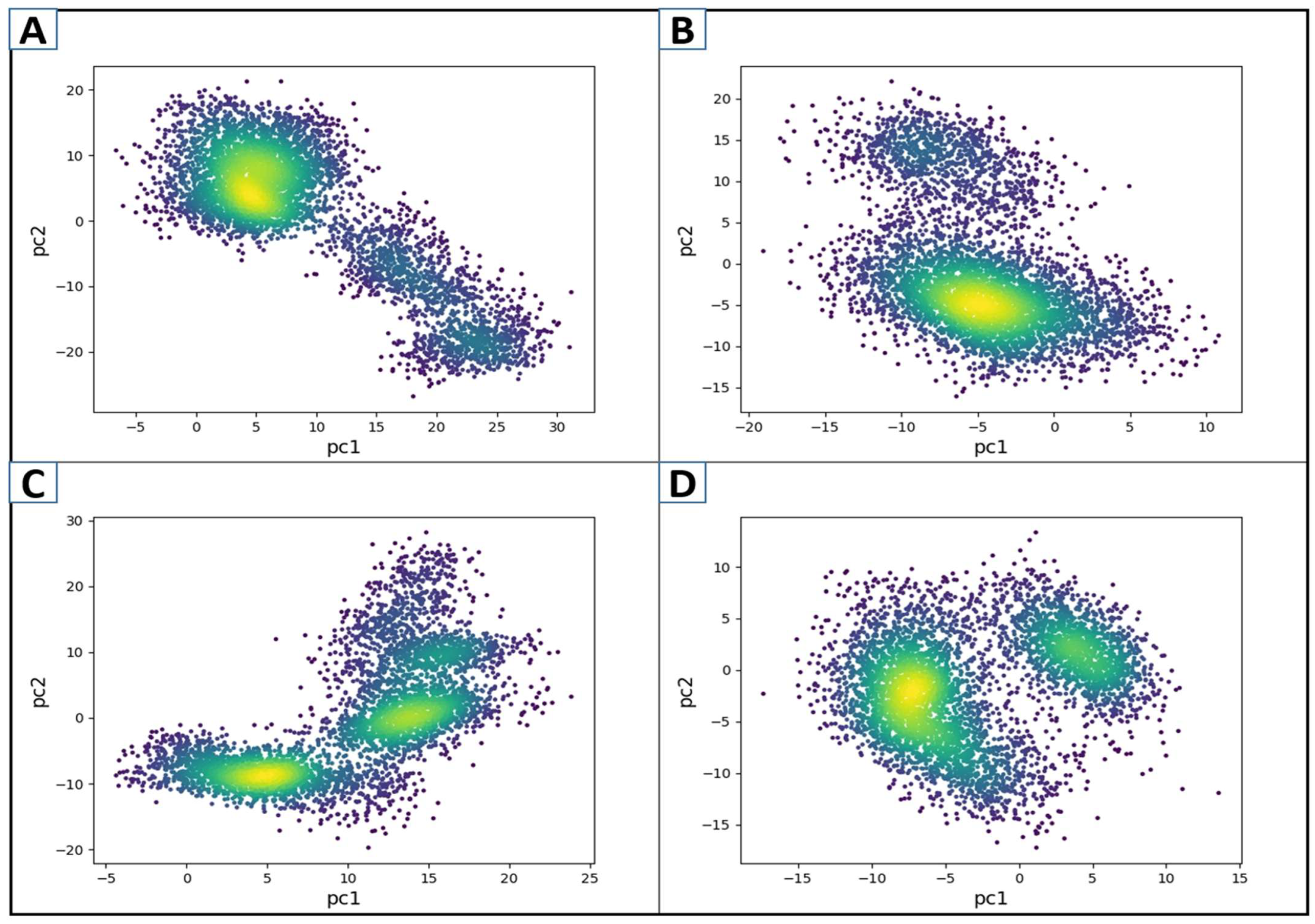
3.7. Principal Component Analysis (PCA)
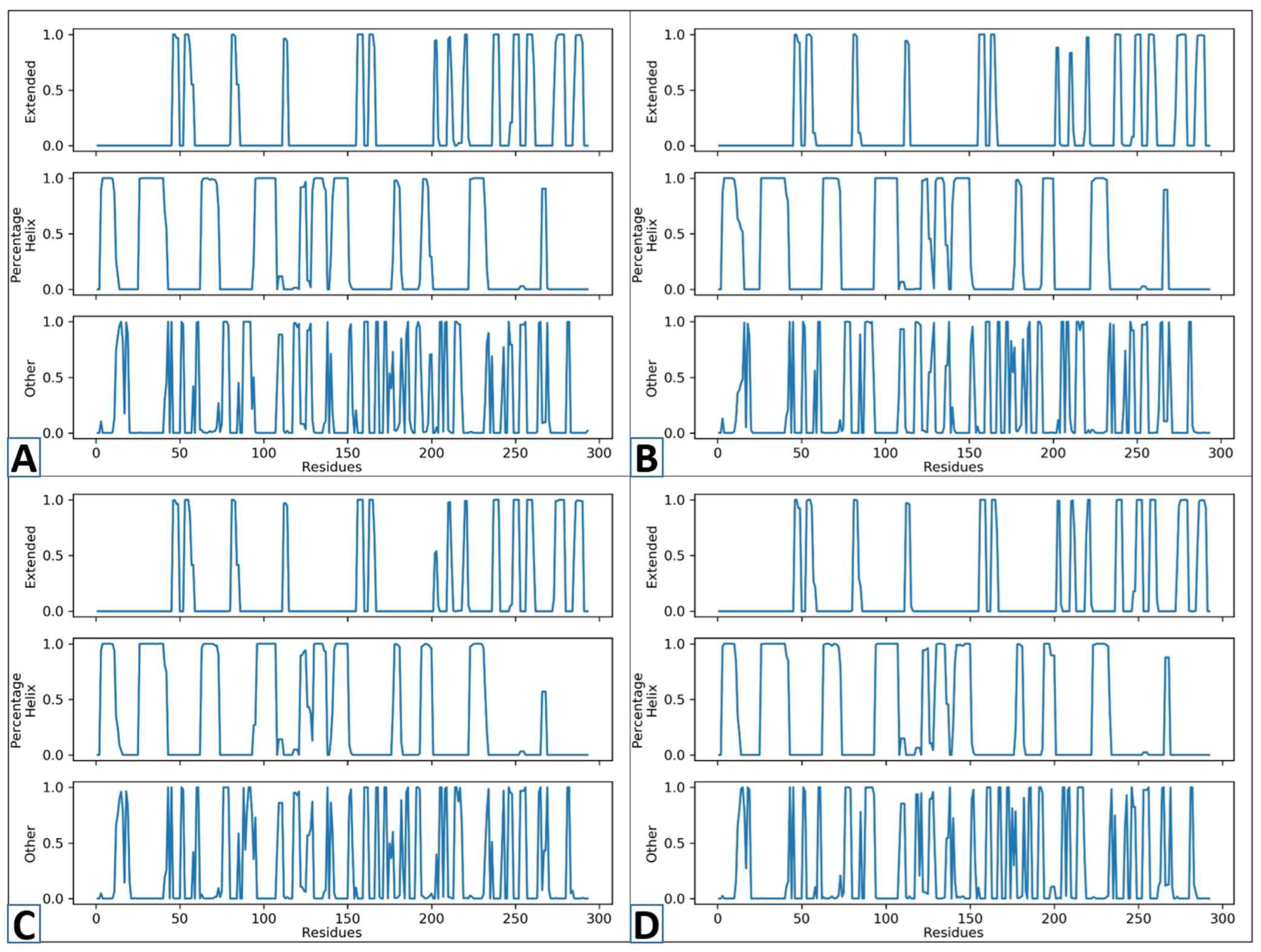
3.8. Secondary Structure Studies (SS)
3.9. MM/GBSA and MM/PBSA Calculations
3.10. WaterSwap Energy Estimation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Coulson, J.; Sharma, V.; Wen, H. Understanding the global dynamics of continuing unmet need for family planning and unintended pregnancy. China Popul. Dev. Stud. 2023, 7, 1–14. [Google Scholar] [CrossRef]
- Barton, B.E.; Erickson, J.A.; Allred, S.I.; Jeffries, J.M.; Stephens, K.K.; Hunter, M.I.; Woodall, K.A.; Winuthayanon, W. Reversible female contraceptives: Historical, current, and future perspectives. Biol. Reprod. 2024, 110, 14–32. [Google Scholar] [CrossRef]
- Le Guen, M.; Schantz, C.; Régnier-Loilier, A.; de La Rochebrochard, E. Reasons for rejecting hormonal contraception in Western countries: A systematic review. Soc. Sci. Med. 2021, 284, 114247. [Google Scholar] [CrossRef]
- Page, S.T.; Amory, J.K. The world needs better male contraceptives: What is taking so long? FASEB J. 2022, 36, e22658. [Google Scholar] [CrossRef] [PubMed]
- Darko Okyere, C. The Conundrum of Unsafe Abortion Among the Youth in Ghana: A Case of Awutu Senya East Municipal Assembly. Master’s Thesis, OsloMet-Storbyuniversitetet, Oslo, Norway, 2022. [Google Scholar]
- Cleland, J. The Contraceptive Revolution. In International Handbook of Population Policies; Springer: Berlin/Heidelberg, Germany, 2022; pp. 595–615. [Google Scholar]
- Pyo, Y.; Kwon, K.H. A review of various types of male contraception. J. Mens. Health 2024, 20, 1–8. [Google Scholar]
- Jacobstein, R.; Radloff, S.; Khan, F.; Mimno, K.; Pal, M.; Snell, J.; Stafford, R.; Touré, C.; Tripathi, V. Down but not out: Vasectomy is faring poorly almost everywhere—we can do better to make it a true method option. Glob. Heal. Sci. Pract. 2023, 11, e2200369. [Google Scholar] [CrossRef] [PubMed]
- Kerk, D.; White-Gloria, C.; Johnson, J.J.; Moorhead, G.B. Eukaryotic-like phosphoprotein phosphatase (PPP) enzyme evolution: Interactions with environmental toxins and regulatory proteins. Biosci. Rep. 2023, 43, BSR20230378. [Google Scholar] [CrossRef]
- Ferreira, A.F.; Santiago, J.; Silva, J.V.; Oliveira, P.F.; Fardilha, M. PP1, PP2A and PP2B interplay in the regulation of sperm motility: Lessons from protein phosphatase inhibitors. Int. J. Mol. Sci. 2022, 23, 15235. [Google Scholar] [CrossRef]
- Mehta, V.; Chamousset, D.; Law, J.; Ooi, S.; Campuzano, D.; Nguyen, V.; Boisvert, F.-M.; Moorhead, G.B.; Trinkle-Mulcahy, L. Subcellular distribution of PP1 isoforms in holoenzyme complexes. bioRxiv 2022. [Google Scholar] [CrossRef]
- Silva, J.V.; Freitas, M.J.; Santiago, J.; Jones, S.; Guimarães, S.; Vijayaraghavan, S.; Publicover, S.; Colombo, G.; Howl, J.; Fardilha, M. Disruption of protein phosphatase 1 complexes with the use of bioportides as a novel approach to target sperm motility. Fertil. Steril. 2021, 115, 348–362. [Google Scholar] [CrossRef]
- Marques, L.; Costa, B.; Pereira, M.; Silva, A.; Santos, J.; Saldanha, L.; Silva, I.; Magalhães, P.; Schmidt, S.; Vale, N. Advancing precision medicine: A review of innovative In Silico approaches for drug development, clinical pharmacology and personalized healthcare. Pharmaceutics 2024, 16, 332. [Google Scholar] [CrossRef]
- Nagarajan, K.; Sundaram, D.P.; Marimuthu, S.K. Innovations In Pharmaceutical Biotechnology; Academic Guru Publishing House: Bhopal, India, 2024; ISBN 8197059136. [Google Scholar]
- Rajaei, F.; Minoccheri, C.; Wittrup, E.; Wilson, R.C.; Athey, B.D.; Omenn, G.S.; Najarian, K. AI-based Computational Methods in Early Drug Discovery and Post Market Drug Assessment: A Survey. IEEE/ACM Trans. Comput. Biol. Bioinforma. 2024, 22, 97–115. [Google Scholar] [CrossRef] [PubMed]
- Shah, A.; Jain, M. Limitations and future challenges of computer-aided drug design methods. In Computer Aided Drug Design (CADD): From Ligand-Based Methods to Structure-Based Approaches; Elsevier: Amsterdam, The Netherlands, 2022; pp. 283–297. [Google Scholar]
- Bouribab, A.; Errougui, A.; Chtita, S. CADD Methods for Developing Novel Compounds Synthesized to Inhibit Tyrosine Kinase Receptors. Curr. Top. Med. Chem. 2024, 25, 1141–1164. [Google Scholar] [CrossRef] [PubMed]
- Giménez, B.G.; Santos, M.S.; Ferrarini, M.; Dos Santos Fernandes, J.P. Evaluation of blockbuster drugs under the rule-of-five. Pharmazie 2010, 65, 148–152. [Google Scholar] [CrossRef] [PubMed]
- Niazi, S.K.; Mariam, Z. Computer-aided drug design and drug discovery: A prospective analysis. Pharmaceuticals 2023, 17, 22. [Google Scholar] [CrossRef]
- Nisius, B.; Sha, F.; Gohlke, H. Structure-based computational analysis of protein binding sites for function and druggability prediction. J. Biotechnol. 2012, 159, 123–134. [Google Scholar] [CrossRef]
- Liau, N.P.D.; Johnson, M.C.; Izadi, S.; Gerosa, L.; Hammel, M.; Bruning, J.M.; Wendorff, T.J.; Phung, W.; Hymowitz, S.G.; Sudhamsu, J. Structural basis for SHOC2 modulation of RAS signalling. Nature 2022, 609, 400–407. [Google Scholar] [CrossRef]
- Aloliqi, A.A. Towards identification of therapeutics against multi-infections and cancers causing Propionibacterium acnes: Molecular modeling and dynamics simulation investigation. J. Mol. Liq. 2024, 126373. [Google Scholar] [CrossRef]
- Abdullahi, M.; Adeniji, S.E. In-silico Molecular Docking and ADME/Pharmacokinetic Prediction Studies of Some Novel Carboxamide Derivatives as Anti-tubercular Agents. Chem. Afr. 2020, 3, 989–1000. [Google Scholar] [CrossRef]
- Kondapuram, S.K.; Sarvagalla, S.; Coumar, M.S. Docking-based virtual screening using PyRx Tool: Autophagy target Vps34 as a case study. In Molecular Docking for Computer-Aided Drug Design; Elsevier: Amsterdam, The Netherlands, 2021; pp. 463–477. [Google Scholar]
- Sabe, V.T.; Ntombela, T.; Jhamba, L.A.; Maguire, G.E.M.; Govender, T.; Naicker, T.; Kruger, H.G. Current trends in computer aided drug design and a highlight of drugs discovered via computational techniques: A review. Eur. J. Med. Chem. 2021, 224, 113705. [Google Scholar] [CrossRef]
- Crampon, K.; Giorkallos, A.; Deldossi, M.; Baud, S.; Steffenel, L.A. Machine-learning methods for ligand-protein molecular docking. Drug Discov. Today 2022, 27, 151–164. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Lin, S.; Yang, F.; Chen, Z.; Guo, L.; Yang, J.; Lin, X.; Wang, L.; Duan, Y.; Wen, A. Structural and functional basis of low-affinity SAM/SAH-binding in the conserved MTase of the multi-segmented Alongshan virus distantly related to canonical unsegmented flaviviruses. PLoS Pathog. 2023, 19, e1011694. [Google Scholar] [CrossRef]
- Wong, F.; Krishnan, A.; Zheng, E.J.; Stärk, H.; Manson, A.L.; Earl, A.M.; Jaakkola, T.; Collins, J.J. Benchmarking AlphaFold-enabled molecular docking predictions for antibiotic discovery. Mol. Syst. Biol. 2022, 18, e11081. [Google Scholar] [CrossRef]
- Acharya, A.; Agarwal, R.; Baker, M.B.; Baudry, J.; Bhowmik, D.; Boehm, S.; Byler, K.G.; Chen, S.Y.; Coates, L.; Cooper, C.J.; et al. Supercomputer-Based Ensemble Docking Drug Discovery Pipeline with Application to COVID-19. J. Chem. Inf. Model. 2020, 60, 5832–5852. [Google Scholar] [CrossRef]
- Alamri, M.A.; Mirza, M.U.; Adeel, M.M.; Ashfaq, U.A.; Tahir Ul Qamar, M.; Shahid, F.; Ahmad, S.; Alatawi, E.A.; Albalawi, G.M.; Allemailem, K.S.; et al. Structural Elucidation of Rift Valley Fever Virus L Protein towards the Discovery of Its Potential Inhibitors. Pharmaceuticals 2022, 15, 659. [Google Scholar] [CrossRef]
- Bender, B.J.; Gahbauer, S.; Luttens, A.; Lyu, J.; Webb, C.M.; Stein, R.M.; Fink, E.A.; Balius, T.E.; Carlsson, J.; Irwin, J.J. A practical guide to large-scale docking. Nat. Protoc. 2021, 16, 4799–4832, Erratum in Nat. Protoc. 2022, 17, 177. [Google Scholar] [CrossRef]
- Jejurikar, B.L.; Rohane, S.H. Drug designing in discovery studio. Asian J. Res. Chem. 2021, 14, 135–138. [Google Scholar]
- Makhlouf, J.; El Bakri, Y.; Valkonen, A.; Saravanan, K.; Ahmad, S.; Smirani, W. Growth, single crystal investigations, hirshfeld surface analysis, DFT studies, molecular dynamics simulations, molecular docking, physico-chemical characterization and biological activity of novel thiocyanic complex with zinc transition metal precursor. Polyhedron 2022, 222, 115937. [Google Scholar] [CrossRef]
- Kamble, A.N.S.; Mitkar, A.A. Swiss ADME predictions of pharmacokinetics and drug-likeness properties of secondary metabolites present in trigonella foenum-graecum. J. Pharmacogn. Phytochem. 2023, 12, 341–349. [Google Scholar] [CrossRef]
- Šegota, S.B.; Anđelić, N.; Lorencin, I.; Musulin, J.; Štifanić, D.; Car, Z. Preparation of simplified molecular input line entry system notation datasets for use in convolutional neural networks. In Proceedings of the 2021 IEEE 21st International Conference on Bioinformatics and Bioengineering (BIBE), Kragujevac, Serbia, 25–27 October 2021; IEEE: New York, NY, USA, 2021; pp. 1–6. [Google Scholar]
- Akash, S.; Bayıl, I.; Rahman, M.A.; Mukerjee, N.; Maitra, S.; Islam, M.R.; Rajkhowa, S.; Ghosh, A.; Al-Hussain, S.A.; Zaki, M.E.A.; et al. Target specific inhibition of West Nile virus envelope glycoprotein and methyltransferase using phytocompounds: An in silico strategy leveraging molecular docking and dynamics simulation. Front. Microbiol. 2023, 14, 1189786. [Google Scholar] [CrossRef]
- Bhrdwaj, A.; Abdalla, M.; Pande, A.; Madhavi, M.; Chopra, I.; Soni, L.; Vijayakumar, N.; Panwar, U.; Khan, M.A.; Prajapati, L. Structure-based virtual screening, molecular docking, molecular dynamics simulation of EGFR for the clinical treatment of glioblastoma. Appl. Biochem. Biotechnol. 2023, 195, 5094–5119. [Google Scholar] [CrossRef] [PubMed]
- Guterres, H.; Park, S.; Zhang, H.; Perone, T.; Kim, J.; Im, W. CHARMM-GUI high-throughput simulator for efficient evaluation of protein–ligand interactions with different force fields. Protein Sci. 2022, 31, e4413. [Google Scholar] [CrossRef]
- Raguette, L.E.; Cuomo, A.E.; Belfon, K.A.A.; Tian, C.; Hazoglou, V.; Witek, G.; Telehany, S.M.; Wu, Q.; Simmerling, C. phosaa14SB and phosaa19SB: Updated Amber Force Field Parameters for Phosphorylated Amino Acids. J. Chem. Theory Comput. 2024, 20, 7199–7209. [Google Scholar] [CrossRef]
- Liao, J.; Wang, Q.; Wu, F.; Huang, Z. In silico methods for identification of potential active sites of therapeutic targets. Molecules 2022, 27, 7103. [Google Scholar] [CrossRef]
- Szél, V.; Zsidó, B.Z.; Jeszenői, N.; Hetényi, C. Target–ligand binding affinity from single point enthalpy calculation and elemental composition. Phys. Chem. Chem. Phys. 2023, 25, 31714–31725. [Google Scholar] [CrossRef]
- Rawat, R.; Kant, K.; Kumar, A.; Bhati, K.; Verma, S.M. HeroMDAnalysis: An automagical tool for GROMACS-based molecular dynamics simulation analysis. Future Med. Chem. 2021, 13, 447–456. [Google Scholar] [CrossRef]
- Pan, X.; Van, R.; Epifanovsky, E.; Liu, J.; Pu, J.; Nam, K.; Shao, Y. Accelerating ab initio quantum mechanical and molecular mechanical (QM/MM) molecular dynamics simulations with multiple time step integration and a recalibrated semiempirical QM/MM Hamiltonian. J. Phys. Chem. B 2022, 126, 4226–4235. [Google Scholar] [CrossRef] [PubMed]
- Pederson, J.P.; McDaniel, J.G. DFT-based QM/MM with particle-mesh Ewald for direct, long-range electrostatic embedding. J. Chem. Phys. 2022, 156, 174105. [Google Scholar] [CrossRef] [PubMed]
- Maszota-Zieleniak, M.; Samsonov, S.A. Molecular Dynamics Simulation-Based Prediction of Glycosaminoglycan Interactions with Drug Molecules. In Computational Drug Discovery and Design; Springer: Berlin/Heidelberg, Germany, 2023; pp. 143–153. [Google Scholar]
- Spassov, D.S.; Atanasova, M.; Doytchinova, I. A role of salt bridges in mediating drug potency: A lesson from the N-myristoyltransferase inhibitors. Front. Mol. Biosci. 2023, 9, 1066029. [Google Scholar] [CrossRef]
- Bibi, S.; Khan, M.S.; El-Kafrawy, S.A.; Alandijany, T.A.; El-Daly, M.M.; Yousafi, Q.; Fatima, D.; Faizo, A.A.; Bajrai, L.H.; Azhar, E.I. Virtual screening and molecular dynamics simulation analysis of Forsythoside A as a plant-derived inhibitor of SARS-CoV-2 3CLpro. Saudi Pharm. J. 2022, 30, 979–1002. [Google Scholar] [CrossRef]
- Hasan, B.M.S.; Abdulazeez, A.M. A review of principal component analysis algorithm for dimensionality reduction. J. Soft Comput. Data Min. 2021, 2, 20–30. [Google Scholar] [CrossRef]
- Montgomerie, S.; Sundararaj, S.; Gallin, W.J.; Wishart, D.S. Improving the accuracy of protein secondary structure prediction using structural alignment. BMC Bioinform. 2006, 7, 301. [Google Scholar] [CrossRef]
- Micsonai, A.; Moussong, E.; Wien, F.; Boros, E.; Vadászi, H.; Murvai, N.; Lee, Y.-H.; Molnár, T.; Réfrégiers, M.; Goto, Y. BeStSel: Webserver for secondary structure and fold prediction for protein CD spectroscopy. Nucleic Acids Res. 2022, 50, W90–W98. [Google Scholar] [CrossRef]
- Miandad, K.; Ullah, A.; Bashir, K.; Khan, S.; Abideen, S.A.; Shaker, B.; Alharbi, M.; Alshammari, A.; Ali, M.; Haleem, A.; et al. Virtual Screening of Artemisia annua Phytochemicals as Potential Inhibitors of SARS-CoV-2 Main Protease Enzyme. Molecules 2022, 27, 8103. [Google Scholar] [CrossRef]
- Acikgoz, A.; Demircan, G.; Yılmaz, D.; Aktas, B.; Yalcin, S.; Yorulmaz, N. Structural, mechanical, radiation shielding properties and albedo parameters of alumina borate glasses: Role of CeO2 and Er2O3. Mater. Sci. Eng. B 2022, 276, 115519. [Google Scholar] [CrossRef]
- Rani, P.; Chahal, S.; Kataria, R.; Kumar, P.; Kumar, S.; Sindhu, J. Unravelling the thermodynamics and binding interactions of bovine serum albumin (BSA) with thiazole based carbohydrazide: Multi-spectroscopic, DFT and molecular dynamics approach. J. Mol. Struct. 2022, 1270, 133939. [Google Scholar] [CrossRef]
- Karnik, K.S.; Sarkate, A.P.; Jambhorkar, V.S.; Wakte, P. WaterSwap Analysis, a Computation-based Method for the Discovery of Effective and Stable Binding Compounds for Mutant EGFR Inhibition. arXiv 2021. [Google Scholar] [CrossRef]
- Fratev, F.; Sirimulla, S. An improved free energy perturbation FEP+ sampling protocol for flexible ligand-binding domains. Sci. Rep. 2019, 9, 16829. [Google Scholar] [CrossRef] [PubMed]
- Mariani, N.A.P.; Silva, J.V.; Fardilha, M.; Silva, E.J.R. Advances in non-hormonal male contraception targeting sperm motility. Hum. Reprod. Update 2023, 29, 545–569. [Google Scholar] [CrossRef]
- Uddin, A.; Gupta, S.; Mohammad, T.; Shahi, D.; Hussain, A.; Alajmi, M.F.; El-Seedi, H.R.; Hassan, I.; Singh, S.; Abid, M. Target-based virtual screening of natural compounds identifies a potent antimalarial with selective falcipain-2 inhibitory activity. Front. Pharmacol. 2022, 13, 850176. [Google Scholar] [CrossRef]
- Li, L.; Mohammed, A.H.; Auda, N.A.; Alsallameh, S.M.S.; Albekairi, N.A.; Muhseen, Z.T.; Butch, C.J. Network Pharmacology, Molecular Docking, and Molecular Dynamics Simulation Analysis Reveal Insights into the Molecular Mechanism of Cordia myxa in the Treatment of Liver Cancer. Biology 2024, 13, 315. [Google Scholar] [CrossRef]
- Altharawi, A.; Ahmad, S.; Alamri, M.A.; ul Qamar, M.T. Structural insight into the binding pattern and interaction mechanism of chemotherapeutic agents with Sorcin by docking and molecular dynamic simulation. Colloids Surf. B Biointerfaces 2021, 208, 112098. [Google Scholar] [CrossRef]
- Roy, S.; Ghosh, P.; Bandyopadhyay, A.; Basu, S. Capturing a crucial ‘disorder-to-order transition’at the heart of the coronavirus molecular pathology—triggered by highly persistent, interchangeable salt-bridges. Vaccines 2022, 10, 301. [Google Scholar] [CrossRef]
- Reeda, V.S.J.; Sakthivel, S.; Divya, P.; Javed, S.; Jothy, V.B. Conformational stability, quantum computational (DFT), vibrational, electronic and non-covalent interactions (QTAIM, RDG and IGM) of antibacterial compound N-(1-naphthyl) ethylenediamine dihydrochloride. J. Mol. Struct. 2024, 1298, 137043. [Google Scholar] [CrossRef]
- Padilla-Bernal, G.; Vargas, R.; Martínez, A. Salt bridge: Key interaction between antipsychotics and receptors. Theor. Chem. Acc. 2023, 142, 65. [Google Scholar] [CrossRef]
- Furkan, M.; Khan, M.S.; Shahwan, M.; Hassan, N.; Yadav, D.K.; Anwar, S.; Khan, R.H.; Shamsi, A. Identifying repurposed drugs as potential inhibitors of Apolipoprotein E: A bioinformatics approach to target complex diseases associated with lipid metabolism and neurodegeneration. Int. J. Biol. Macromol. 2024, 259, 129167. [Google Scholar] [CrossRef] [PubMed]
- Bharadwaj, K.K.; Ahmad, I.; Pati, S.; Ghosh, A.; Rabha, B.; Sarkar, T.; Bhattacharjya, D.; Patel, H.; Baishya, D. Screening of phytocompounds for identification of prospective histone deacetylase 1 (HDAC1) inhibitor: An in silico molecular docking, molecular dynamics simulation, and MM-GBSA approach. Appl. Biochem. Biotechnol. 2024, 196, 3747–3764. [Google Scholar] [CrossRef] [PubMed]
- Miles, A.J.; Ramalli, S.G.; Wallace, B.A. DichroWeb, a website for calculating protein secondary structure from circular dichroism spectroscopic data. Protein Sci. 2022, 31, 37–46. [Google Scholar] [CrossRef]
- Hasan, M.R.; Alsaiari, A.A.; Fakhurji, B.Z.; Molla, M.H.R.; Asseri, A.H.; Sumon, M.A.A.; Park, M.N.; Ahammad, F.; Kim, B. Application of mathematical modeling and computational tools in the modern drug design and development process. Molecules 2022, 27, 4169. [Google Scholar] [CrossRef]
- Cournia, Z.; Chipot, C.; Roux, B.; York, D.M.; Sherman, W. Free energy methods in drug discovery—introduction. In Free Energy Methods in Drug Discovery: Current State and Future Directions; ACS Publications: Washington, DC, USA, 2021; pp. 1–38. ISBN 1947-5918. [Google Scholar]
- Sheng, Y.; Yin, Y.; Ma, Y.; Ding, H. Improving the performance of MM/PBSA in protein–protein interactions via the screening electrostatic energy. J. Chem. Inf. Model. 2021, 61, 2454–2462. [Google Scholar] [CrossRef] [PubMed]
- Sobhia, M.E.; Ghosh, K.; Kumar, G.S.; Sivangula, S.; Laddha, K.; Kumari, S.; Kumar, H. The Role of Water Network Chemistry in Proteins: A Structural Bioinformatics Perspective in Drug Discovery and Development. Curr. Top. Med. Chem. 2022, 22, 1636–1653. [Google Scholar] [CrossRef]
- Reynolds-Wright, J.J.; Cameron, N.J.; Anderson, R.A. Will men use novel male contraceptive methods and will women trust them? A systematic review. J. Sex Res. 2021, 58, 838–849. [Google Scholar] [CrossRef] [PubMed]
- Dominiak, Z.; Huras, H.; Kręcisz, P.; Krzeszowski, W.; Szymański, P.; Czarnecka, K. Promising results in development of male contraception. Bioorg. Med. Chem. Lett. 2021, 41, 128005. [Google Scholar] [CrossRef] [PubMed]
- Gunasekaran, P.; Velmurugan, Y.; Arputharaj, D.S.; Savaridasson, J.K.; Hemamalini, M.; Venkatachalam, R. In vitro contraceptive activities, molecular docking, molecular dynamics, MM-PBSA, non-covalent interaction and DFT studies of bioactive compounds from Aegle marmelos. Linn., leaves. Front. Chem. 2023, 11, 1096177. [Google Scholar] [CrossRef] [PubMed]
- Prieto-Martínez, F.D.; López-López, E.; Juárez-Mercado, K.E.; Medina-Franco, J.L. Computational drug design methods—current and future perspectives. In In Silico Drug Design; Academic Press: Cambridge, MA, USA, 2019; pp. 19–44. [Google Scholar]
- Schenone, M.; Dančík, V.; Wagner, B.K.; Clemons, P.A. Target identification and mechanism of action in chemical biology and drug discovery. Nat. Chem. Biol. 2013, 9, 232–240. [Google Scholar] [CrossRef]
- Kiriiri, G.K.; Njogu, P.M.; Mwangi, A.N. Exploring different approaches to improve the success of drug discovery and development projects: A review. Futur. J. Pharm. Sci. 2020, 6, 27. [Google Scholar] [CrossRef]
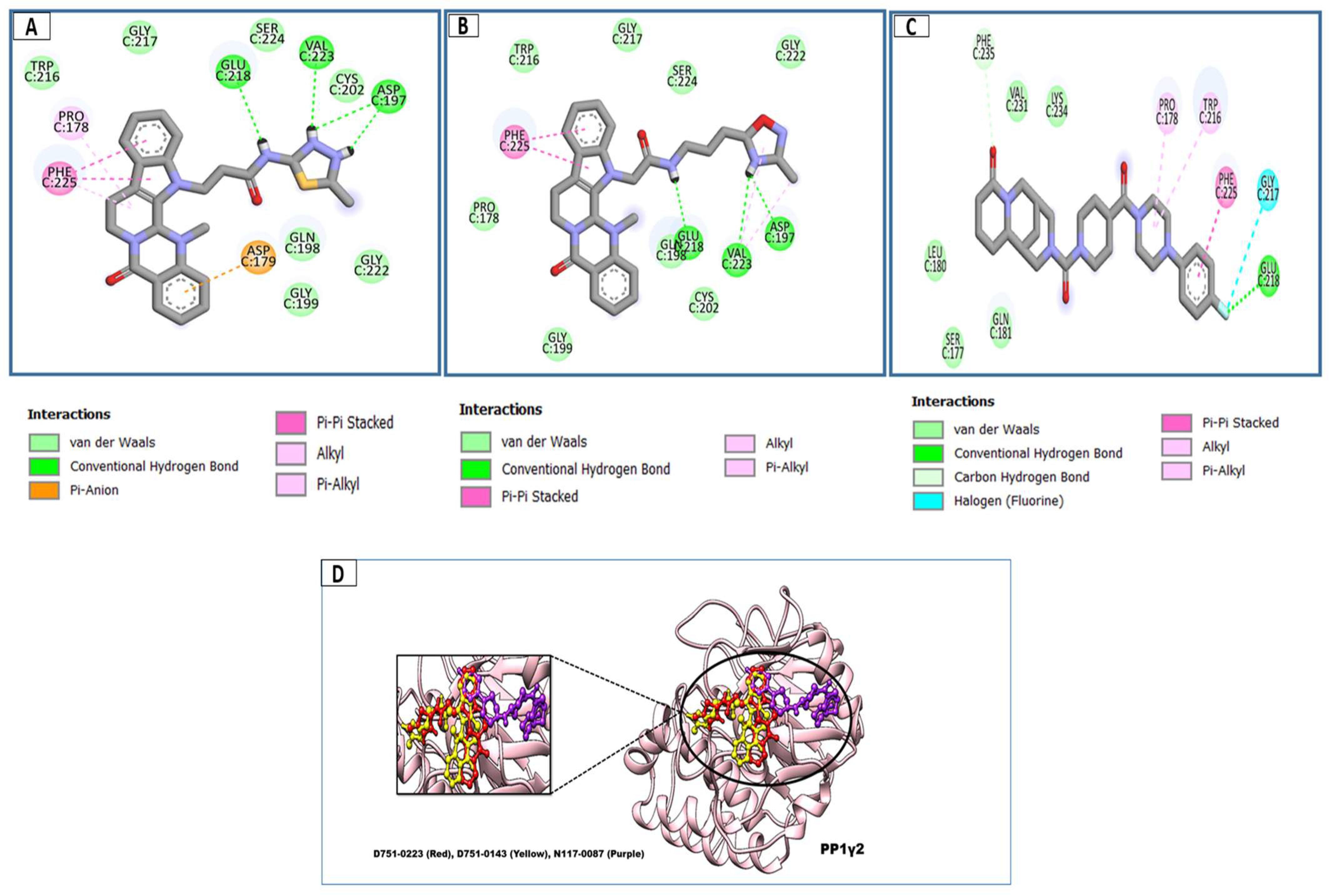
| S.No | Compound ID | Chemical Name and Structure | Binding Affinity | H-Bonds Interaction | Hydrophobic Interactions |
|---|---|---|---|---|---|
| 1. | D751-0223 | 2-methyl-5-(3-(14-methyl-5-oxo-7,8,13b,14-tetrahydroindolo[2′,3′:3,4]pyrido[2,1-b]quinazolin-13(5H)-yl)propanamido)-1,3,4-thiadiazole-3,4-diium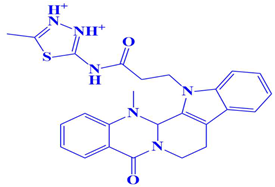 | −8.7 kcal/mol | Glu218, Val223, Asp197 | Gly217, Trp216, Ser224, Cys202, Gln198, Gly199, Gly222 |
| 2. | D751-0143 | N-(3-(3-methyl-1,2,4-oxadiazolidin-5-yl)propyl)-2-(14-methyl-5-oxo-7,8,13b,14-tetrahydroindolo[2′,3′:3,4]pyrido[2,1-b]quinazolin-13(5H)-yl)acetamide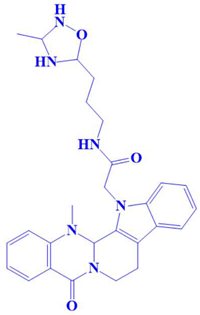 | −8.1 kcal/mol | Glu218, Asp197, Val223 | Trp216, Gly217, Ser224, Gly222, Pro178, Gln198, Cys202, Gly199 |
| 3. | N117-0087 | 3-(4-(4-(4-fluorophenyl)piperazine-1-carbonyl)piperidine-1-carbonyl)octahydro-1H-1,5-methanopyrido[1,2-a][1,5]diazocin-8(2H)-one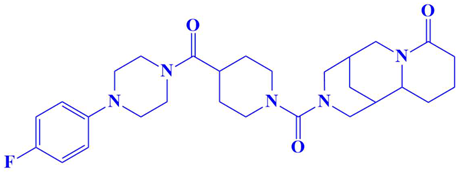 | −8 kcal/mol | Glu218 | Phe235, Val231, Lys234, Leu180, Ser177, Gln181, |
| 4. | N117-0096 | 3-(4-([1,4′-bipiperidine]-1′-carbonyl)piperidine-1-carbonyl)octahydro-1H-1,5-methanopyrido[1,2-a][1,5]diazocin-8(2H)-one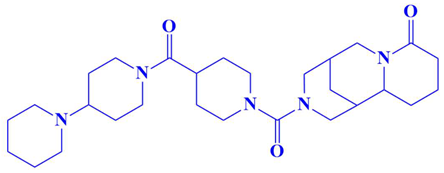 | −8 kcal/mol | Asp194, Leu210 | Pro196, Th193, Leu200, Glu199, Arg188, Gln185, Val195, Met190, Ile189, Asp179 |
| 5. | D751-0222 | 2-(3-(14-methyl-5-oxo-7,8,13b,14-tetrahydroindolo[2′,3′:3,4]pyrido[2,1-b]quinazolin-13(5H)-yl)propanamido)pyridin-1-ium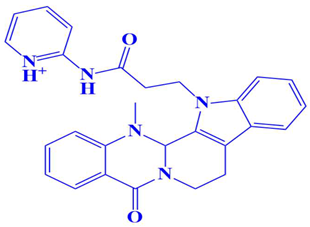 | −7.9 kcal/mol | Asp197 | Trp216, Gly217, Gly199, Glu218, Gln198, Ser224, Val223, Gly222 |
| 6. | D751-0254 | N-(2-(3-methyl-1,2,4-oxadiazolidin-5-yl)ethyl)-3-(14-methyl-5-oxo-7,8,13b,14-tetrahydroindolo[2′,3′:3,4]pyrido[2,1-b]quinazolin-13(5H)-yl)propanamide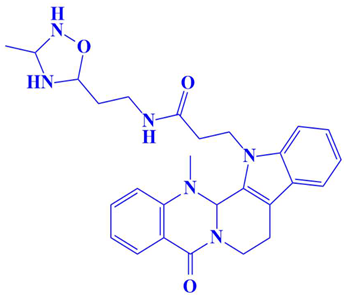 | −7.9 kcal/mol | Glu218, Asp197 | Gly217, Ser224, Glu222, Gln198, Val223, Cyc202 |
| 7. | D751-0214 | 3-(14-methyl-5-oxo-7,8,13b,14-tetrahydroindolo[2′,3′:3,4]pyrido[2,1-b]quinazolin-13(5H)-yl)-N-(2-morpholinoethyl)propanamide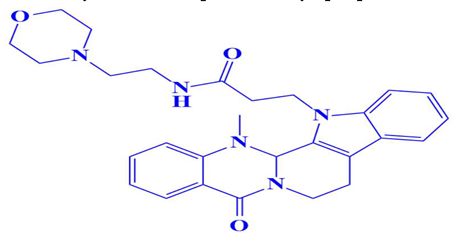 | −7.7 kcal/mol | Glu218 | Trp216, Gly217, Asp179, Glu199, Gln198, Asp197, Ser224, Val223, Gly222 |
| 8. | CM3007-1542 | (4-fluoro-1-(mesitylsulfonyl)pyrrolidin-2-yl)(1H-indol-1-yl)methanone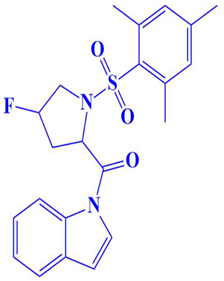 | −7.5 kcal/mol | - | Trp216, Glu218, Gly217, Ser224, Val223, Asp197, Gln198, Gly199, Leu200, Ser177, Pro178, |
| 9. | CM4579-5752 | 3-((4,4-difluoro-1-(5-(4-fluorophenyl)-2,5-dihydro-1H-pyrazole-3-carbonyl)pyrrolidin-2-yl)methoxy)pyridin-1-ium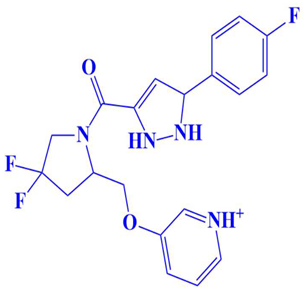 | −7 kcal/mol | Trp216, Gln181 | Lys234, Phe235, Ser177, Leu176 |
| 10 | D751-0108 | 13-(2-((3-(2,3-dihydro-1H-imidazol-1-yl)propyl)amino)-2-oxoethyl)-14-methyl-8,13-dihydro-7H-indolo[2′,3′:3,4]pyrido[2,1-b]quinazolin-6,14-diium-5-olate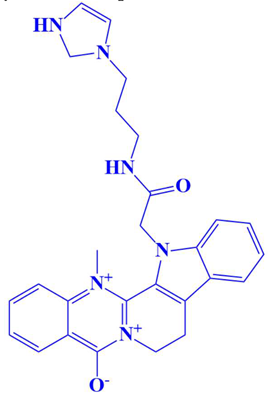 | −7 kcal/mol | Arg188 | Thr193, Met190, Ile189, Leu201, Gln198, Asp179, Gln185 |
| RMSD | |||
|---|---|---|---|
| Complexes | Minimum | Maximum | Mean |
| D751-0223 | - | 1.69 Å | 1.27 Å |
| D751-0143 | - | 2.30 Å | 1.73 Å |
| N117-0087 | - | 2.07 Å | 1.39 Å |
| Apo | - | 2.14 Å | 1.69 Å |
| RMSF | |||
| Complexes | Minimum | Maximum | Mean |
| D751-0223 | 0.34 Å | 3.48 Å | 0.66 Å |
| D751-0143 | 0.37 Å | 3.41 Å | 0.80 Å |
| N117-0087 | 0.36 Å | 2.20 Å | 0.70 Å |
| Apo | 0.35 Å | 2.30 Å | 0.73 Å |
| Beta Factor | |||
| Complexes | Minimum | Maximum | Mean |
| D751-0223 | 3.08 Å | 319.27 Å | 14.75 Å |
| D751-0143 | 3.65 Å | 307.12 Å | 20.29 Å |
| N117-0087 | 3.44 Å | 127.79 Å | 15.56 Å |
| Apo | 3.29 Å | 139.82 Å | 17.29 Å |
| RoG | |||
| Complexes | Minimum | Maximum | Mean |
| D751-0223 | 18.43 Å | 18.93 Å | 18.67 Å |
| D751-0143 | 18.34 Å | 18.95 Å | 18.67 Å |
| N117-0087 | 18.29 Å | 18.74 Å | 18.50 Å |
| Apo | 18.29 Å | 18.93 Å | 18.64 Å |
| SASA | |||
| Complexes | Minimum | Maximum | Mean |
| D751-0223 | 117.93 Å2 | 140.39 Å2 | 12,890.7 Å2 |
| D751-0143 | 11,612.1 Å2 | 14,014.4 Å2 | 12,854.5 Å2 |
| N117-0087 | 11,639.3 Å2 | 136,462.7 Å2 | 12,541.8 Å2 |
| Apo | 11,453.5 Å2 | 13,850.9 Å2 | 12,743.7 Å2 |
| Complexes | Salt Bridges Interaction | Unique SB Interactions |
|---|---|---|
| D751-0223 | Glu133-Arg137, Asp202-Arg240, Asp202-Arg215, Glu178-Arg182, Asp202-Arg215, Glu178-Arg182, Glu38-Arg37, Glu224-Lys228, Glu178-Arg182, Glu96-Arg137, Glu178-Arg182, Glu28-Arg9, Glu28-Arg9, Asp65-Arg68, Glu71-Arg14, Glu26-Lys141, Asp86-Arg90, Glu38-Lys35, Glu26-Arg30, Glu48-Arg181, Glu48-Arg181, Asp148-Arg37, Asp4-Lys107, Asp132-Lys135, Glu178-Arg181, Glu38-Lys35, Asp132-Lys135. | Glu133-Arg137 Asp4-Lys107 |
| D751-0143 | Asp214-Arg215, Asp148-ly144, Asp202-Arg215, Asp132-Arg136, Asp160-Lys162, Glu178-Arg182, Glu96-Arg137, Asp206-Lys205, Asp160-Lys162, Glu28-Arg9, Glu26-Lys141, Asp86-Arg90, Glu26-Lys141, Glu26-Arg30, Asp188-Arg116, Glu48-Arg181, Glu26-Arg30, Glu133-Arg136, Asp236-Lys162, Asp204-Lys205, Glu133-Arg136, Asp148-Arg37, Glu133-Lys92, Glu71-Arg14, Glu178-Arg181, Asp236-Lys162, Glu178-Arg181, Glu161-Lys162, Glu178-Arg181. | Asp188-Arg116 |
| N117-0087 | Glu12-Lys20, Glu28-Arg9, Glu96-Arg137, Asp148-Lys144, Glu178-Arg182, Asp202-Arg240, Asp132-Arg136, Glu178-Arg182, Glu224-Lys228, Glu178-Arg182, Glu96-Arg137, Asp132-Arg136, Glu28-Arg9, Glu26, Lys141, Glu71-Arg14, Glu71-Arg14, Asp86-Arg90, Glu71-Arg14, Asp86-Arg90, Glu38-Lys35, Glu120-Arg90, Glu133-Arg136, Glu48-Arg181, Asp234-Lys162, Asp236-Lys162, Asp132-Lys135, Glu38-Lys35, Glu26-Arg30, Glu178, Arg181 | Glu120-Arg90 |
| Apo | Glu12-Lys20, Asp148-Lys144, Glu96-Arg137, Glu28-Arg9, Asp202-Arg240, Glu38-Arg37, Glu96-Arg137, Asp132-Arg136, Glu96-Arg137, Asp132-Arg136, Glu96-Arg137, Glu28-Arg9, Asp206-Lys205, Asp65-Arg68, Glu71-Arg14, Asp86-Arg90, Glu26-Arg30, Glu48-Arg181, Glu26-Arg30, Glu48-Arg181, Glu26-Arg30, Glu120-Arg90, Glu48-Arg181, Asp148-Arg37, Glu178-Arg181, Asp4-Lys107, Asp236-Lys162, Asp148-Arg37, Glu38-Lys35, Asp234-Lys162, Glu178-Arg181. | Asp4-Lys107 Glu120-Arg90 |
| Method | Energy Section | D751-0223 | D751-0143 | N117-0087 |
|---|---|---|---|---|
| MM/GBSA | Van der Waals Energy | −75.04 | −72.36 | −65.87 |
| Electrostatic Energy | −18.01 | −15.64 | −14.88 | |
| Solvation Energy (SE) | 11.05 | 14.93 | 13.49 | |
| Gas-Phase Energy | −93.05 | −88 | −80.75 | |
| Total Binding Energy | −82 | −73.07 | −67.26 | |
| MM/PBSA | Van der Waals Energy | −75.04 | −72.36 | −65.87 |
| Electrostatic Energy | −18.01 | −15.64 | −14.88 | |
| Salvation Energy (SE) | 13.04 | 15.82 | 16.49 | |
| Gas-Phase Energy | −93.05 | −88 | −80.75 | |
| Total Binding Energy | −80.01 | −72.18 | −64.26 |
| Algorithms | PP1γ2-D751-0223 | PP1γ2-D751-0143 | PP1γ2-N117-0087 |
|---|---|---|---|
| Bennet’s | −51.42 | −47.50 | −37.45 |
| Free-Energy Perturbation (FEP) | −50.24 | −46.99 | −37.00 |
| Thermodynamic Integration (TI) | −51.49 | −47.16 | −37.15 |
| Total Mean | −51.05 | −47.21 | −37.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aljohani, H.M.; Bokhari, B.T.; Saleh, A.M.; Alyahyawi, A.Y.; Alhamawi, R.M.; Jaddah, M.M.; Alobaidy, M.A.; Eisa, A.A. Exploring Novel Inhibitory Compounds Against Phosphatase Gamma 2: A Therapeutic Target for Male Contraceptives. Curr. Issues Mol. Biol. 2025, 47, 658. https://doi.org/10.3390/cimb47080658
Aljohani HM, Bokhari BT, Saleh AM, Alyahyawi AY, Alhamawi RM, Jaddah MM, Alobaidy MA, Eisa AA. Exploring Novel Inhibitory Compounds Against Phosphatase Gamma 2: A Therapeutic Target for Male Contraceptives. Current Issues in Molecular Biology. 2025; 47(8):658. https://doi.org/10.3390/cimb47080658
Chicago/Turabian StyleAljohani, Hashim M., Bayan T. Bokhari, Alaa M. Saleh, Areej Yahya Alyahyawi, Renad M. Alhamawi, Mariam M. Jaddah, Mohammad A. Alobaidy, and Alaa Abdulaziz Eisa. 2025. "Exploring Novel Inhibitory Compounds Against Phosphatase Gamma 2: A Therapeutic Target for Male Contraceptives" Current Issues in Molecular Biology 47, no. 8: 658. https://doi.org/10.3390/cimb47080658
APA StyleAljohani, H. M., Bokhari, B. T., Saleh, A. M., Alyahyawi, A. Y., Alhamawi, R. M., Jaddah, M. M., Alobaidy, M. A., & Eisa, A. A. (2025). Exploring Novel Inhibitory Compounds Against Phosphatase Gamma 2: A Therapeutic Target for Male Contraceptives. Current Issues in Molecular Biology, 47(8), 658. https://doi.org/10.3390/cimb47080658








