Abstract
The objective of this work was to analyse the genetic diversity of a population of Citrus spp. in the south of the State of Espírito Santo, Brazil, for pre-breeding studies. For that, a total of sixty genotypes were analysed, including ten citrus varieties from four species of the Citrus genus. The methodology involved DNA extraction, amplification via polymerase chain reaction, and the use of a set of 16 Simple Sequence Repeat markers. These markers identified 42 alleles, with a variation of one to four alleles per locus, an average heterozygosity value of 0.53, and an average polymorphic information content of up to 0.29 per species. After the analysis, a dissimilarity matrix was generated using Jaccard distance and a dendrogram, revealing the formation of two groups: Group I, comprising Citrus sinensis varieties, and Group II, comprising varieties of Citrus latifolia, Citrus aurantifolia, and Citrus reticulata. Our study demonstrated that the combination of these markers allowed for the differentiation of genotypes within the collection. The results obtained are valuable for the future management of the collection and the efficient use of genetic diversity estimation in Citrus spp.
1. Introduction
The Citrus genus belongs to the Rutaceae family and comprises various species, such as Citrus reticulata Blanco (mandarins), Citrus sinensis L. Osbeck (oranges), Citrus limon L., Citrus latifolia Tanaka, Citrus aurantifolia (C.) Swingle (lemons, limes), Citrus paradisi L. (grapefruits), and others [1,2]. These citrus fruits are widely consumed worldwide due to their aromas, flavours, and nutritional value [3]. They are rich in vitamins, fibre, minerals, and phytonutrients essential for human health [4]. Additionally, they play a significant role as a food source and raw material in various industrial sectors, including the production of juices, beverages, and processed foods, and for fresh consumption [5,6].
Citrus fruits are available on global markets, with China being the largest producer (26%), followed by Brazil (13%) [7]. Among the citrus species, C. sinensis is the most widely cultivated and has a high profile in Brazil, which is one of the largest producers and exporters of orange juice. In addition, other species such as C. reticulata, C. latifolia, and C. aurantifolia are also economically important, used both for fresh consumption and by industry [8,9]. To ensure the efficiency of the production chain and food safety, detailed knowledge of morphological characteristics is essential for genetic improvement [10,11]. However, the genus Citrus shows significant phenotypic variation both within and between species [12].
This phenotypic variation is not always sufficient to accurately estimate the genetic distance between accessions. Therefore, the incorporation of molecular markers serves as an auxiliary tool that provides greater discriminatory power between genotypes [13,14]. Microsatellites, or simple sequence repeats (SSRs), are markers characterised by high levels of polymorphism, codominant inheritance, and multiallelism [15]. SSRs are widely used in diversity studies between populations, including cultivars, due to their simplicity, speed, and efficiency in genotypic and germplasm characterisation in citrus [3,16,17,18].
Germplasm conservation and knowledge are fundamental for citrus cultivation, especially in places where this activity is of great economic and social importance. In the state of Espírito Santo, citrus farming is particularly important in the southern region and in the Caparaó area, where soil and climatic conditions are favourable for citrus cultivation [19,20]. However, although the state is a centre of citrus production, the low productivity of the fruit and the lack of genetic material recommended for the region have contributed to a reduction in the area planted and a decrease in producers’ income. Although producers traditionally maintain genotypes, it is essential to evaluate the genetic diversity of these citrus varieties to ensure their sustainability and continuous improvement. Therefore, the present study aimed to evaluate the genetic diversity of a citrus germplasm collection in southern Espírito Santo using SSR molecular markers.
2. Material and Methods
A total of 60 accessions of 10 citrus varieties were evaluated, comprising the following species: I-Citrus sinensis (L.) Osbeck, II-Citrus reticulata Blanco, III-Citrus latifolia and IV-Citrus aurantifolia (C.) Swingle. These accessions originated from the germplasm collection of the Federal Institute of Education, Science and Technology of Espírito Santo (IFES)-Campus Alegre, located in the southern region of the State of Espírito Santo, Brazil (latitude 20°45′20′′ S and longitude 41°27′43′′ W) (Figure 1).
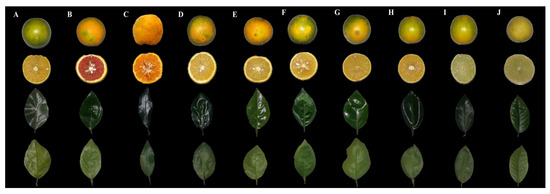
Figure 1.
Citrus varieties from the IFES germplasm collection. (A) C. sinensis (Laranja Pêra Mel); (B) C. sinensis (Laranja Sanguínea); (C) C. reticulata (Tangerina Ponkan); (D) C. sinensis (Laranja Natal Folha Murcha); (E) C. sinensis (Laranja Bahia); (F) C. sinensis (Laranja Lima); (G) C. sinensis (Laranja Seleta Comum); (H) C. sinensis (Laranja Pêra Rio); (I) C. latifolia (Limão Taiti) and; (J) C. aurantifolia (Limão Branco).
Five vigorous leaves of each genotype were collected and preserved in liquid nitrogen. The leaves were then macerated and approximately 300 mg of the resulting plant tissue was transferred to a tube. DNA was then extracted using the CTAB protocol [21]. After removal, the samples were quantified using a spectrophotometer (NanoDrop® ND-1000, ThermoFisher, Wilmington, DE, USA) with a 260/280 nm ratio ≥ 1.8, indicating a high quality of the DNA obtained.
The PCR amplification reactions were performed in a final volume of 15 μL, containing 1.5 μL of 10× PCR buffer, 0.6 μL of MgCl2 (250 mM), 4.8 μL of dNTP (2.5 μM), 0.2 μL of Taq DNA polymerase (5 units/μL), 5 μL of genomic DNA (10 ng/μL), 0.75 μL of each oligonucleotide, 1.4 μL of ultrapure water, and 0.2 μL of Taq DNA polymerase (5 units/μL). The thermocycling programme was initiated with a denaturation step at 94 °C for 5 min, followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 60 °C for 30 s, and extension and 72 °C for 30 s, with a final extension at 72 °C for 5 min [22].
The PCR products were separated by denaturing polyacrylamide gel electrophoresis, at 10% in TBE buffer (1×) (Tris-Boric 45 mM, EDTA 1 mM pH 8.0) for 1 h and 30 min at 100 V, followed by staining with ethidium bromide (25 μL/L) for 30 min, and photographed with BioRad equipment (model Gel Doc XR+ system, Hercules, CA, USA).
Genotyping was performed using 16 SSRs provided by the Laboratory of Biochemistry and Molecular Biology of the Federal University of Espírito Santo Campus Alegre (Table 1) [23,24].

Table 1.
The name, sequence, fragment size (BP), annealing temperature (°C), and number of alleles of the 16 SSRs for Citrus spp.
The polymorphic information content (PIC) was calculated based on the 16 SSRs, employing the formula PIC = Σ1 − P2ij, allele frequency, and the expected heterozygosity within the four species. To visualise the genetic diversity relationships, the dissimilarity matrix was calculated based on the Jaccard coefficient, considering the presence and absence of alleles. Clustering was performed using the Unweighted Pair Group Method with Arithmetic Mean (UPGMA), both available in the ‘Genetic Distance Analysis’ routine of the GENES software [25]. The algorithm uses a binary matrix to generate the dissimilarity matrix and subsequently constructs the dendrogram based on the average linkage method. After obtaining the data matrix from the SSR markers, a heatmap analysis with bidimensional hierarchical clustering was performed using the pheatmap package in R version 4.4.2. [26].
3. Results and Discussion
A genetic diversity analysis was conducted on 60 citrus varieties, which were submitted to a set of 16 SSRs. The analysis identified 42 alleles, with a variation of one to four alleles between the SSRs evaluated (Table 2). The mean observed heterozygosity was 0.53, suggesting the presence of genetic variation. Seven of the SSRs exhibited higher observed heterozygosity than expected, suggesting a high frequency of heterozygotes in the population under study.

Table 2.
Allele frequency and heterozygosity of 60 citrus accessions for 16 SSRs.
The highest level of heterozygosity was observed in the SSRs CCSMEc2, CCSMEc4, CCSMEc5, CCSMEc7, CCSMEc10, CCSMEc13, and CMS-16; the latter set of markers is the most suitable for the future analysis of species belonging to the genus (Table 2). The availability of these SSR sets, together with others implemented in the citrus microsatellite database, which have polymorphic potential, can be used to infer genetic diversity and population distinction in species of the genus [15].
The SSRs analysed proved to be effective in discriminating diversity between species. The PIC indicated a moderate level of polymorphism, ranging from 0.21 to 0.29 (Figure 2). The C. sinensis species had the highest PIC value (0.29), suggesting greater genetic variability within the germplasm (Figure 2). A higher PIC reflects a greater variety of alleles present in the genetic material studied [27]. And the varieties of oranges (C. sinensis) exhibited greater dispersion within the group of citrus varieties compared to C. latifolia, C. aurantifolia e C. reticulata, which registered lower values (0.23, 0.21, and 0.21).
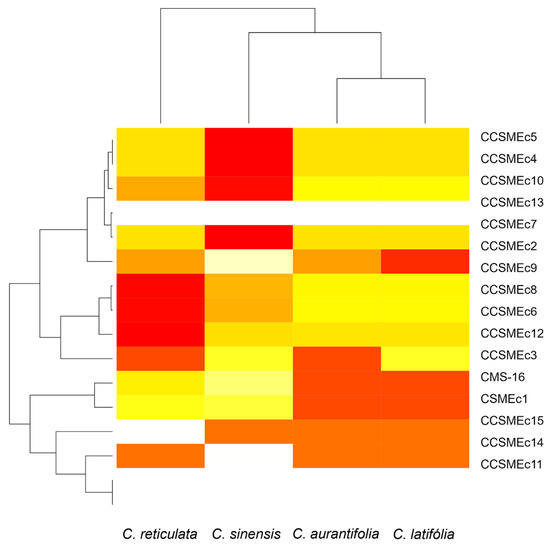
Figure 2.
Heatmap with bidimensional hierarchical clustering of the Polymorphic Information Content (PIC) analysis of 16 SSR primers in four Citrus species to generate SSR profiles in 60 genotypes. X-axis: Citrus species. Y-axis: SSRs markers.
Among the markers evaluated, the loci CCSMEc2 and CCSMEc3 were the most informative, while CCSMEc11 and CCSMEc14 showed low efficiency in polymorphism detection. The use of these SSRs was relevant for the study under analysis, as they presented high PICs, given their ability to capture variation within the genetic pool. This, in turn, facilitates a more precise discrimination between different genotypes within a species, and the inclusion of SNPs or InDels can complement low-polymorphism SSRs, providing higher resolution in genetic analyses [17].
Organising this data by species is essential for selecting and recommending the most appropriate markers [15]. Among the SSRs analysed (Figure 2), 13 were informative for C. sinensis, with PIC values ranging from (0.21 to 0.50). These values were found to be similar to those previously observed for a sweet orange population, which ranged from 0.37 to 0.43; this finding suggests that there is consistency in the patterns of genetic variability that have been observed [23]. Ten SRRs were informative for C. latifolia, and nine were informative for C. aurantifolia and C. reticulata, showing PIC values (0.30 to 0.38; 0.35 to 0.38) comparable to previous studies in a Citrus ssp. population with a PIC between 0.18 and 0.37, supporting marker recommendations based on the data obtained [28].
The UPGMA cluster analysis of the similarity matrix obtained using the 16 SSRs resulted in a dendrogram (Figure 3) with a cophenetic correlation coefficient of 0.96, indicating a well-represented genetic similarity in the structuring of the clusters between the citrus, forming two large groups of similarity. Group I is made up of seven types of orange citrus. C. sinensis demonstrates significant genetic diversity, as evidenced by the analysis of agronomic, bromatological, morphological and chemotype characteristics [19,29,30]. However, the investigation revealed minimal genetic variation when employing SSR molecular markers, such as simple sequence repeats (ISSR) [31] and random amplified polymorphic fragments (RAPD) [32].
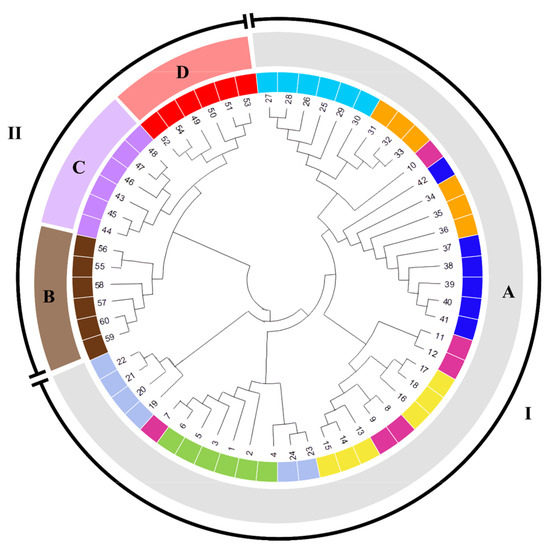
Figure 3.
Circular dendrogram showing the genetic similarity between Citrus ssp. with Jaccard’s distance using the UPGMA method with a cophenetic correlation of 0.96. The numbers I and II indicate the formation of the clusters. The species are represented by A—C. sinensis, B—C. reticulata, C—C. latifolia, and D—C. aurantifolia. The citrus varieties were categorised by colour: sky blue (Laranja Lima); orange (Laranja Seleta Comum); pink (Laranja Sanguínea); dark blue (Laranja Pêra Rio); yellow (Laranja Natal Folha Murcha); light blue (Laranja Bahia); green (Laranja Pêra Mel); brown (Tangerina Ponkan); lilac (Limão Taiti), and red (Limão Branco).
Oranges have a narrow genetic base among citrus, which can be attributed to their origin from a common lineage, a genetic mutation, or domestication of the C. sinensis species [33]. The findings of this study demonstrated that all orange citruses were integrated into a single group within the dendrogram, thereby substantiating their genetic similarity (Figure 3). The formation of clusters for orange citrus can be observed in previous studies involving SSR genetic markers [34,35], confirming the hypothesis that sweet orange cultivars are monophyletic and derived from a single ancestor through mutation and the selection of desirable clones [36,37].
Group II comprises the species C. latifolia, C. aurantifolia, and C. reticulata (Figure 3). There is a high degree of genetic similarity between the genotypes of each species, suggesting a narrow genetic base or the presence of replications in the germplasm of these genotypes. The genetic similarity between C. aurantifolia and C. latifolia lemon genotypes demonstrates an intrinsic genetic relationship with other species of the genus. This finding aligns with previous studies that have indicated the classification of both species within the same cluster, owing to the presence of genetic characteristics analogous to those observed in citrus. Consequently, they are regarded as natural hybrids of this species [17,38]. The C. reticulata genotypes were grouped in this cluster with similarity to lemons. However, previous studies have demonstrated a closer genetic relationship with the orange group, as evidenced by similarity indices using SSR [17].
As was previously reported by [33,39], significant genetic variations between C. reticulata genotypes have been documented using an SRR array. However, no significant genetic dissimilarity was observed between the six genotypes analysed in the present study (Figure 3). This suggests that other SSRs are required to identify genetic variations within this species.
Considering the intraspecific diversity in the orange group, the lowest variation between the group’s accessions was found between the ‘Laranja Bahia’ and ‘Laranja Pera Mel’ citrus; this low genetic diversity may be due to clonal selection, the presence of cleistogamy, or duplicates in the germplasm [40,41]. In order to observe greater genetic variations in the gene pool among these citrus types, it is necessary to use more markers [15].
The results obtained are an important basis for the adoption of conservation measures, since understanding the genetic diversity of citrus in a germplasm collection can contribute to the control of genetic erosion and the development of citrus breeding strategies, in which the identification and selection of genetically divergent parent plants is essential to broaden the genetic base and enhance breeding efficiency [12].
4. Conclusions
This study, using a set of SSR markers as an essential tool for the analysis of genetic diversity, confirmed the existence of significant genetic variability among a citrus population from the germplasm collection of southern Espírito Santo. Furthermore, the markers CCSMEc2 and CCSMEc13 were identified as the most effective for tracking genetic variations in the gene pool at the intra- and interspecific level of Citrus spp., considering the species C. sinensis, C. latifolia, C. aurantifolia, and C. reticulata.
Author Contributions
Conceptualization, I.F.G.S. and M.M.M.; methodology, I.F.G.S.; software, I.F.G.S., C.d.M.B.O. and J.D.d.S.N.; validation, M.M.M.; formal analysis, F.C.P., C.d.M.B.O., J.D.d.S.N., R.N.d.A. and A.P.C.G.B.; investigation, T.d.O.S. and M.M.M.; resources, T.C.B.S. and M.M.M.; data curation, I.F.G.S.; writing—original draft preparation, I.F.G.S., F.C.P., C.d.M.B.O., J.D.d.S.N., T.d.O.S., R.N.d.A., A.P.C.G.B., S.d.S.B., T.C.B.S., J.O.S., A.C.S.J. and M.M.M.; writing—review and editing, I.F.G.S., F.C.P., C.d.M.B.O., J.D.d.S.N., T.d.O.S., R.N.d.A., A.P.C.G.B., S.d.S.B., T.C.B.S., J.O.S., A.C.S.J. and M.M.M.; visualization, F.C.P. and T.d.O.S.; supervision, M.M.M.; project administration, M.M.M.; funding acquisition, J.D.d.S.N. and M.M.M. All authors have read and agreed to the published version of the manuscript.
Funding
We would like to acknowledge the Research Support Fundação de Amparo à Pesquisa e Inovação do Espírito Santo (FAPES), the Universidade Federal do Espírito Santo Campus Alegre (UFES), and the Pró-Reitoria de Pesquisa e Pós-Graduação (PRPPG) of the Instituto Federal de Ciência e Tecnologia do Espírito Santo Campus Alegre (IFES).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Dataset available on request from the authors.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Lin, L.; Chuang, C.; Chen, H.; Yang, K. Lime (Citrus aurantifolia (Christm.) Swingle) essential oils: Volatile compounds, antioxidant capacity, and hypolipidemic effect. Foods 2019, 8, 398. [Google Scholar] [CrossRef]
- Viuda-Martos, M.; Ruiz-Navajas, Y.; Fernandes-Lopes, J.; Perez-Alvarez, J.A. Atividade antifúngica dos óleos essenciais de limão (Citrus Lemon L.), tangerina (Citrus reticulata L.), toranja (Citrus paradisi L.) e laranja (Citrus sinensis L.). Controle Aliment. 2008, 19, 1130–1138. [Google Scholar] [CrossRef]
- Luro, F.; Paoli, M.; Costantino, G.; Marchi, E. Investigation of Diversity by Analyzing the Polymorphism of SSR Markers and the Composition of Leaf and Fruit Essential Oils of 72 Mandarins (Citrus reticulata Blanco). Horticulturae 2023, 9, 577. [Google Scholar] [CrossRef]
- Vilas Boas, V.P.P.; Belico, L.A.; Lima, J.P.; Vilas Boas, E.V.B. Potencial Sensorial, Nutricional e Funcional de Diferentes Tipos de Citros. In Proceedings of the I Congresso Luso-Brasileiro de Horticultura, Lisbon, Portugal, 1–4 November 2017; pp. 231–237. [Google Scholar]
- Liu, S.; Lou, Y.; LI, Y.; Zhang, J. Review of phytochemical and nutritional characteristics and food applications of Citrus L. fruits. Front. Nutr. 2022, 9, 968604. [Google Scholar] [CrossRef] [PubMed]
- Lopes, J.M.S.; Déo, T.F.G.; Andrade, B.M.; Giroto, M.; Felipe, A.L.S.; Junior, C.E.I.; Lima, F.C.C. Importância econômica do citros no Brasil. Rev. Científica Eletrônica De Agron. 2011, 20, 1–3. [Google Scholar]
- Faostat Database. Available online: http://www.fao.org/faostat/en/#data/QC (accessed on 14 January 2024).
- Kato-Noguchi, H.; Kato, M. Pesticidal Activity of Citrus Fruits for the Development of Sustainable Fruit-Processing Waste Management and Agricultural Production. Plants 2025, 14, 754. [Google Scholar] [CrossRef] [PubMed]
- Sombra, K.E.S.; Costa, A.C.; Loureiro, F.L.C.; Uchôa, N.C. A Citricultura como Instrumento de Preservação da Agricultura Familiar no Semiárido Cearense, Brasil. Rev. Do Programa De Pós-Grad. Em Extensão Rural. (UFV) 2018, 7, 353–372. [Google Scholar]
- Langgut, D. The History of Citron: Botanical Remains and Ancient Art and Texts. In The Citron Compendium: The Citron (Etrog) Citrus medica L.: Science and Tradition; Springer International Publishing: Cham, Switzerland, 2023; pp. 473–481. [Google Scholar] [CrossRef]
- Moulin, M.M.; Santos, T.D.O.; Ramos, S.R.R.; Maxted, N.; Brehm, J.M. A crop wild relative inventory for Brazil. Crop Sci. 2025, 65, e70001. [Google Scholar] [CrossRef]
- Samarina, L.S.; Kulyan, R.V.; Koninskaya, N.; Gorshkov, V.M. Genetic diversity and phylogenetic relationships among citrus germplasm in the Western Caucasus assessed with SSR and organelle DNA markers. Sci. Hortic. 2021, 288, 110355. [Google Scholar] [CrossRef]
- de Paula, E.; Almeida, R.N.D.; Santos, T.D.O.; Souza Neto, J.D.D.; Riva-Souza, E.M.; Posse, S.C.P.; Souza, M.N.; Madella de Oliveira, A.D.F.; Santos Júnior, A.C.; Santos, J.O.; et al. Genetic Diversity of Common Bean (Phaseolus vulgaris L.) Landraces Based on Morphological Traits and Molecular Markers. Plants 2024, 13, 2584. [Google Scholar] [CrossRef]
- Mendes, D.F.J.; Rodrigues, L.M.S.; Lisboa, S.H.B. Marcadores moleculares, importância para o melhoramento genético de plantas. Sci. Gen. 2022, 2, 128. Available online: https://scientiageneralis.com.br/index.php/SG/article/view/331 (accessed on 27 January 2025).
- Duhan, N.; Meshram, M.; Orduz, C.D.L.; Kundal, R. citSATdb: Genome-wide simple sequence repeat (SSR) marker database of Citrus species for germplasm characterization and crop improvement. Genes 2020, 11, 1486. [Google Scholar] [CrossRef] [PubMed]
- Martínez, M.F.; Rodríguez-Mora, D.M.; Murcia-Riaño, N. Genetic differentiation of cultivated Citrus fruits (Citrus spp.) in Colombia using SSR molecular markers. Agrociencia 2024, 59, 1–16. [Google Scholar] [CrossRef]
- Munankarmi, N.N.; Rana, N.; Joshi, B.K.; Bhattarai, T. Characterization of the genetic diversity of Citrus species of Nepal using simple sequence repeat (SSR) markers. S. Afr. J. Bot. 2023, 156, 192–201. [Google Scholar] [CrossRef]
- Schinor, E.H.; Siviero, A.; Cristofani-Yaly, M.; Marengo, S.; Jorgino, P.; Machado, M.A. Caracterização agronômica e molecular de acessos de Citrus sunki do Banco de Germoplasma de Citros do Centro APTA Citros Sylvio Moreira. Citrus Res. Technol. 2011, 32, 27–37. [Google Scholar] [CrossRef]
- Santos, R.M.; Valadares, F.V.; Pirovani, A.A.V.; Venancio, D.F.V. Caracterização morfoagronômica e físico-química de germoplasma de Citrus. Enciclopédia Biosf. 2016, 13, 1398–1410. [Google Scholar] [CrossRef]
- da Silva, F.S.; Ferrari, J.L. Mapeamento da citricultura do município de jerônimo monteiro, ES, Brasil. Enciclopédia Biosf. 2012, 8, 90. [Google Scholar]
- Doyle, J.J.; Doyle, J.L. Isolation of plant DNA from fresh tissue. Focus v 1990, 12, 13–15. [Google Scholar]
- Sharafi, A.A.; Sharafi, A.; Abkenar, A.A. Molecular genetic diversity assessment of Citrus species grown in Iran revealed by SSR, ISSR and CAPS molecular markers. J. Sci. Res. Rev. Cienc. E Investig. 2017, 2, 22–27. [Google Scholar] [CrossRef]
- Ahmad, R.; Struss, D.; Southwick, S.M. Development and characterization of microsatellite markers in Citrus. J. Am. Soc. Hortic. Sci. 2003, 128, 584–590. [Google Scholar] [CrossRef]
- Palmieri, D.A.; Novelli, V.M.; Bastianel, M.; Cristofani-Yaly, M.; Astúa-Monge, G.; Carlos, E.F.; Oliveira, C.O.; Machado, M.A. Frequência e distribuição de microssatélites de ESTs de citros. Genet. Mol. Biol. 2007, 30, 1009–1018. [Google Scholar] [CrossRef]
- Cruz, C.D. Genes Software—Extended and integrated with the R, Matlab and Selegen. Acta Sci. 2016, 38, 547–552. [Google Scholar] [CrossRef]
- Kolde, R. Pheatmap: Pretty Heatmaps—R Package, version 1.0.13; R package. 2015. Available online: https://CRAN.R-project.org/package=pheatmap (accessed on 30 July 2025).
- Kaur, H.; Sidhu, G.; Sarao, N.; Singh, R. Assessment of genetic diversity of mandarin cultivars grown in major citrus regions of world using morphological and microsatellite markers. Hortic. Environ. Biotechnol. 2022, 63, 425–437. [Google Scholar] [CrossRef]
- Nematollahi, A.K.; Golein, B.; Vahdati, K. Analysis of the genetic diversity in citrus (Citrus spp.) species using SSR markers. J. Plant Physiol. Breed. 2013, 3, 39–47. [Google Scholar]
- Adlini, M.N.; Umaroh, H.K. Karakterisasi Tanaman Jeruk (Citrus sp.) Di Kecamatan Nibung Hangus Kabupaten Batu Bara Sumatera Utara. KLOROFIL J. Ilmu Biol. Dan Terap. 2021, 1, 48–54. [Google Scholar] [CrossRef]
- Martinidou, E.; Michailidis, M.; Ziogas, V.; Masuero, D. Comparative Evaluation of Secondary Metabolite Chemodiversity of Citrus Genebank Collection in Greece: Can the Peel be More than Waste? J. Agric. Food Chem. 2024, 72, 9019–9032. [Google Scholar] [CrossRef]
- Rajabi, A.; Lahiji, H.S.; Golfazani, M.M. Avaliação da diversidade genética em Citrus sinensis por marcador ISSR e retrotransposon. J. Plant Prod. Res. 2022, 29, 1362022. [Google Scholar]
- Tuwo, M.; Kuswinanti, T.; Nasruddin, A. Estimating the Genetic Diversity of Oranges Citrus spp. in South Sulawesi, Indonesia, Using RAPD Markers. Scientifica 2023, 2023, 6676038. [Google Scholar] [CrossRef]
- Novelli, V.M.; Cristofani, M.; Souza, A.A. Development and characterization of polymorphic microsatellite markers for the sweet orange (Citrus sinensis L. Osbeck). Genet. Mol. Biol. 2006, 29, 90–96. [Google Scholar] [CrossRef]
- Golein, B.; Nazeryan, M.; Babakhani, B. Assessing genetic variability in male sterile and low fertile Citrus cultivars utilizing simple sequence repeat markers (SSRs). Afr. J. Biotechnol. 2012, 11, 1632–1638. [Google Scholar] [CrossRef]
- Hazarika, T.K.; Hazarika, B.N.; Shukla, A.C. Genetic variability and phylogenetic relationships studies of genus Citrus L. with the application of molecular markers. Genet. Resour. Crop Evol. 2014, 61, 1441–1454. [Google Scholar] [CrossRef]
- Fang, D.Q.; Roose, M.L. Identification of closely related Citrus cultivars with inter-simple sequence repeat markers. Theor. Appl. Genet. 1997, 95, 408–417. [Google Scholar] [CrossRef]
- Luro, F.; Laigret, F.; Bové, J.; Ollitrault, P. DNA amplified fingerprinting, a useful tool for determination of genetic origin and diversity analysis in Citrus. HortScience 1995, 30, 1063–1067. [Google Scholar] [CrossRef]
- Gulsen, O.; Roose, M.L. Lemons: Diversity and relationships with selected Citrus genotypes as measured with nuclear genome markers. J. Am. Soc. Hortic. Sci. 2001, 126, 309–317. [Google Scholar] [CrossRef]
- Golein, B.; Talaie, A.R.; Zamani, Z.; Ebadi, A. Assessment of genetic variability in some Iranian sweet oranges (Citrus sinensis [L.] Osbeck) and mandarins (Citrus reticulata Blanco) using SSR markers. Int. J. Agric. Biol. 2005, 7, 167–170. [Google Scholar]
- Caruso, M.; Smith, M.W.; Froelicher, Y.; Russo, G. Traditional breeding. In The Genus Citrus; Woodhead Publishing: Cambridge, UK, 2020; pp. 129–148. [Google Scholar] [CrossRef]
- Curk, F.; Ollitrault, F.; Garcia-Lor, A.; Luro, F.; Navarro, L.; Ollitrault, P. Phylogenetic origin of limes and lemons revealed by cytoplasmic and nuclear markers. Ann. Bot. 2016, 117, 565–583. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).