Yeast Oral Delivery of DAF16 shRNAs Results in Effective Gene Silencing in C. elegans
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains, Growth Conditions
2.2. Yeast Transformation
2.3. Plasmids and Recombinant Yeast Construction
2.4. Yeast RNA Extraction and Primer Extension
2.5. C. elegans Fluorescence Microscopy and Lifespan Analysis
2.6. Real Time-qPCR
3. Results
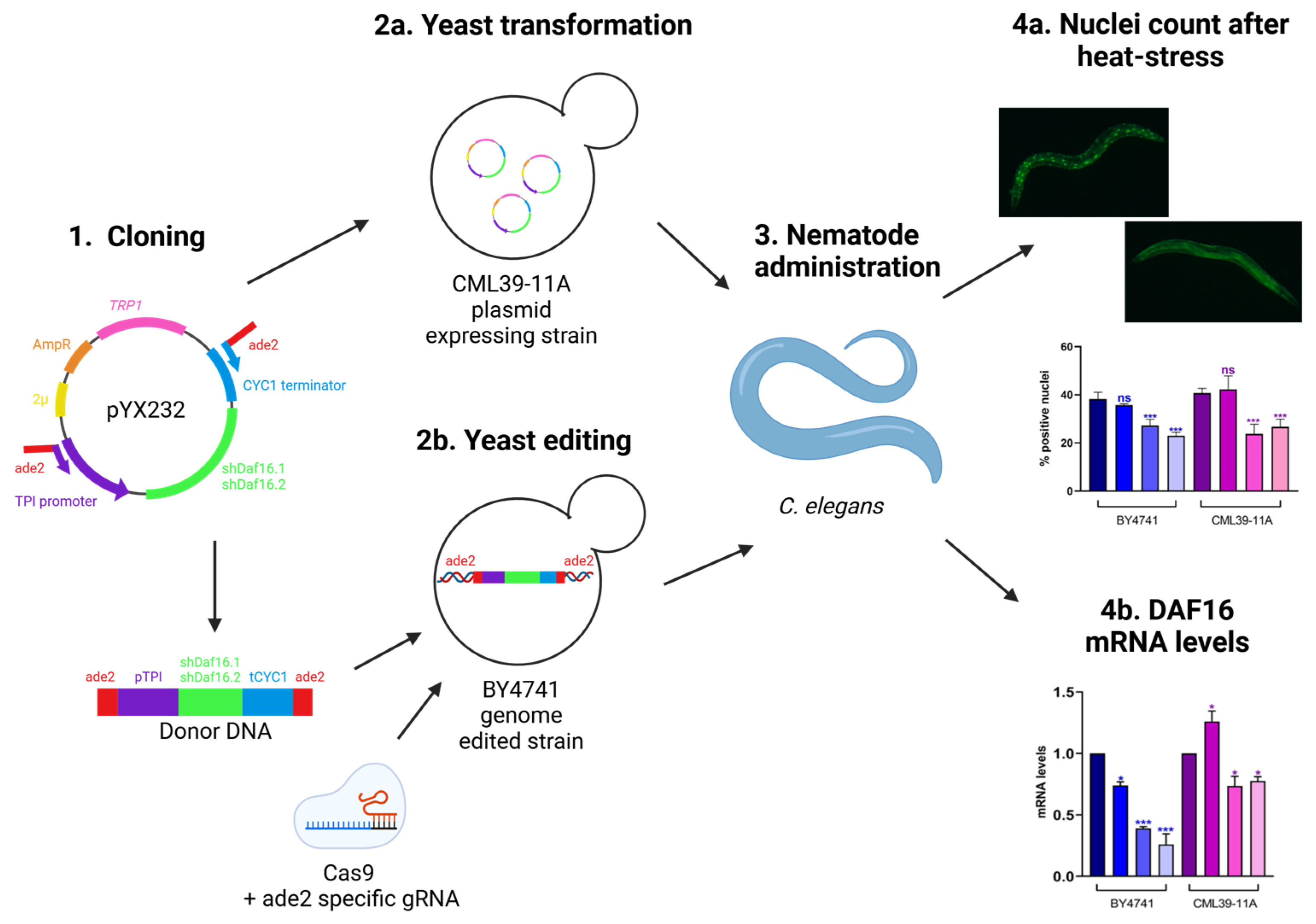
3.1. Experimental Set up
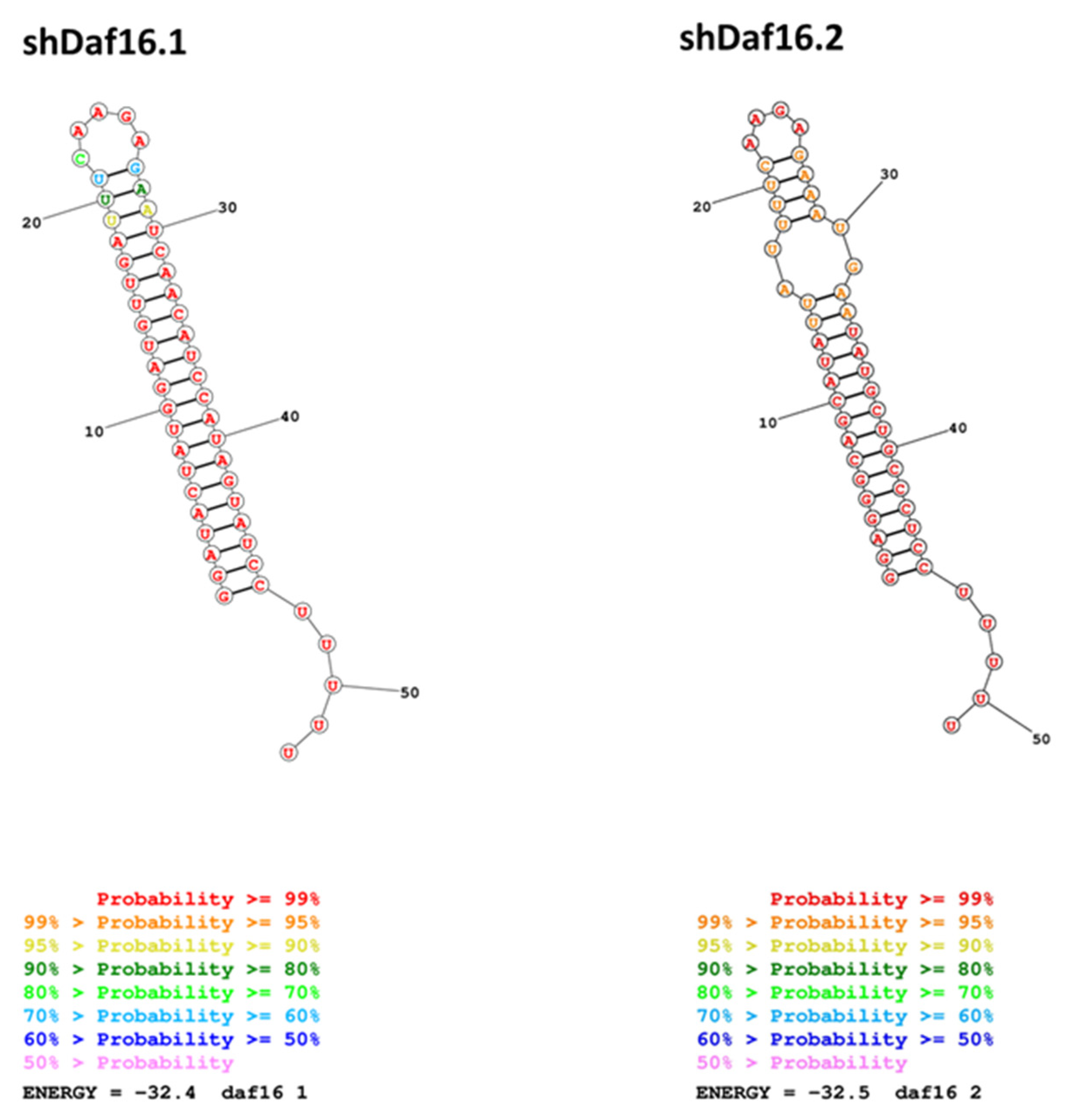
3.2. Design of shRNAs and Yeast Expression
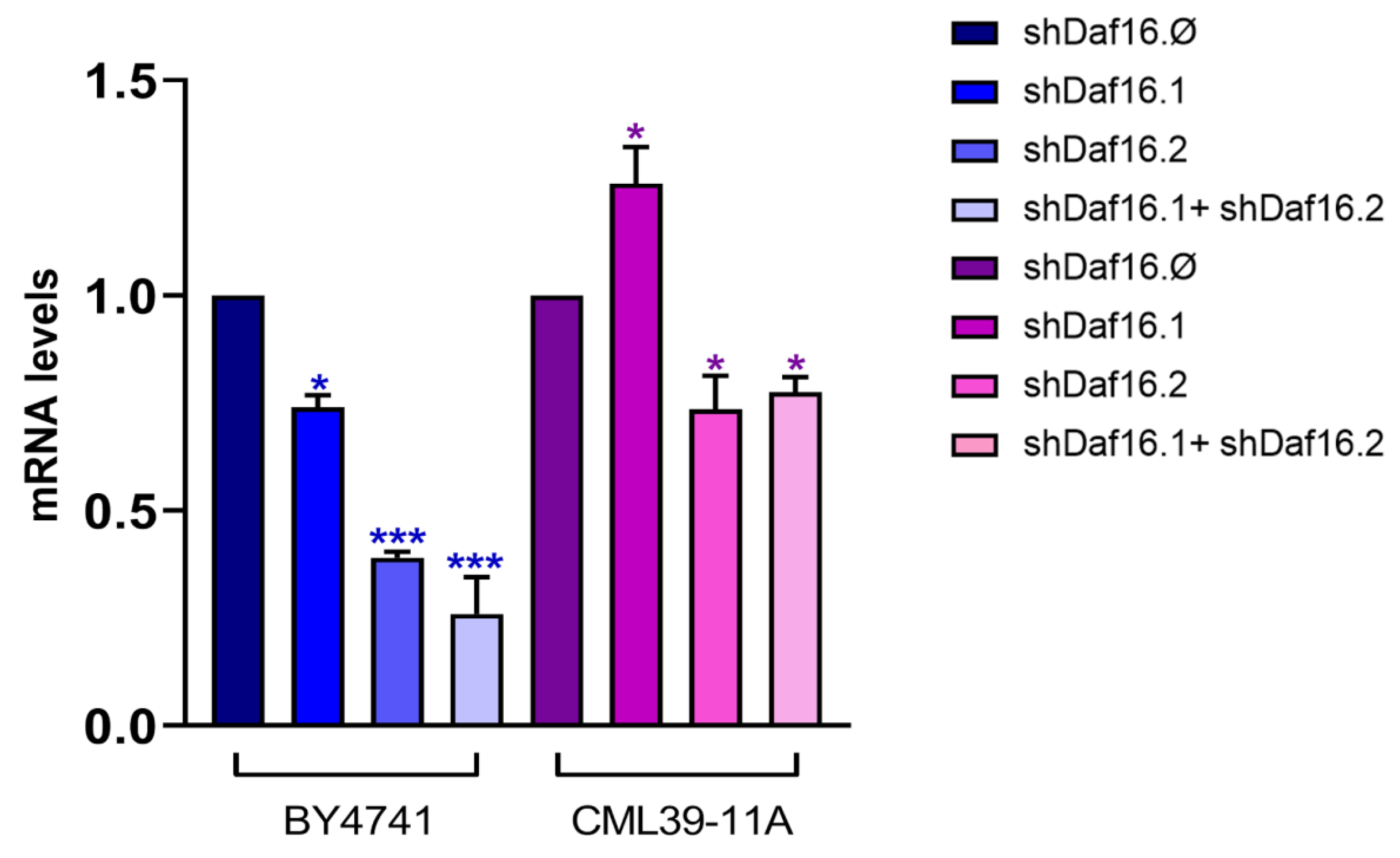
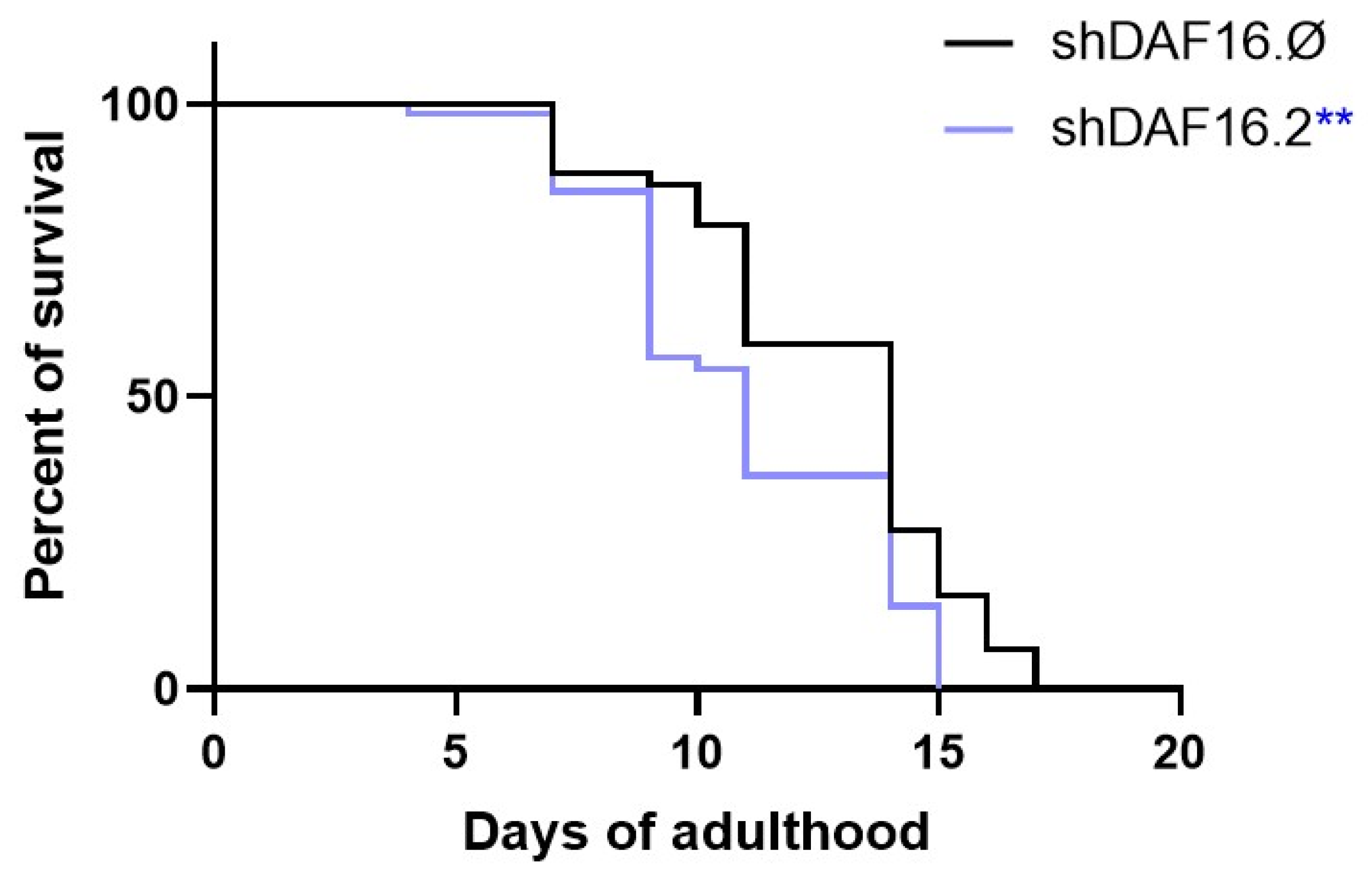
3.3. C. elegans shRNAs Administration and RNA Interference
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PPNs | Plant Parasitic Nematodes |
| BCAs | Biocontrol Agents |
| GRAS | Generally Recognized As Safe |
| shRNAs | Short Hairpin RNAs |
| RNAi | RNA Interference |
| dsRNA | Double-Stranded RNA |
| mRNA | Messenger RNA |
| GM | Genetically Modified |
| qPCR | Quantitative PCR |
| Ct | Threshold Cycle |
| TSS | Transcription Start Site |
References
- Hannon, G.J. RNA interference. Nature 2002, 418, 244–251. [Google Scholar] [CrossRef] [PubMed]
- Gordon, K.H.J.; Waterhouse, P.M. RNAi for insect-proof plants. Nat. Biotechnol. 2007, 25, 1231–1232. [Google Scholar] [CrossRef]
- Whyard, S.; Singh, A.D.; Wong, S. Ingested double-stranded RNAs can act as species-specific insecticides. Insect Biochem. Mol. Biol. 2009, 39, 824–832. [Google Scholar] [CrossRef]
- Huvenne, H.; Smagghe, G. Mechanisms of dsRNA uptake in insects and potential of RNAi for pest control: A review. J. Insect Physiol. 2010, 56, 227–235. [Google Scholar] [CrossRef]
- Bolognesi, R.; Ramaseshadri, P.; Anderson, J.; Bachman, P.; Clinton, W.; Flannagan, R.; Ilagan, O.; Lawrence, C.; Levine, S.; Moar, W.; et al. Characterizing the mechanism of action of double-stranded RNA activity against western corn rootworm (Diabrotica virgiferavirgifera LeConte). PLoS ONE 2012, 7, e47534. [Google Scholar] [CrossRef]
- Scott, J.G.; Michel, K.; Bartholomay, L.C.; Siegfried, B.D.; Hunter, W.B.; Smagghe, G.; Zhu, K.Y.; Douglas, A.E. Towards the elements of successful insect RNAi. J. Insect Physiol. 2013, 59, 1212–1221. [Google Scholar] [CrossRef]
- Kola, V.S.R.; Renuka, P.; Madhav, M.S.; Mangrauthia, S.K. Key enzymes and proteins of crop insects as candidate for RNAi based gene silencing. Front. Physiol. 2015, 6, 119. [Google Scholar] [CrossRef]
- Lilley, C.J.; Davies, L.J.; Urwin, P.E. RNA interference in plant parasitic nematodes: A summary of the current status. Parasitology 2012, 139, 630–640. [Google Scholar] [CrossRef]
- Nunes, C.C.; Dean, R.A. Host-induced gene silencing: A tool for understanding fungal host interaction and for developing novel disease control strategies. Mol. Plant Pathol. 2012, 13, 519–529. [Google Scholar] [CrossRef]
- Koch, A.; Kogel, K.H. New wind in the sails: Improving the agronomic value of crop plants through RNAi-mediated gene silencing. Plant Biotechnol. J. 2014, 12, 821–831. [Google Scholar] [CrossRef]
- Mitter, N.; Worrall, E.A.; Robinson, K.E.; Li, P.; Jain, R.G.; Taochy, C.; Fletcher, S.J.; Carroll, B.J.; Lu, G.Q.; Xu, Z.P. Clay nanosheets for topical delivery of RNAi for sustained protection against plant viruses. Nat. Plants 2017, 3, 16207. [Google Scholar] [CrossRef]
- Mao, Y.B.; Cai, W.J.; Wang, J.W.; Hong, G.J.; Tao, X.Y.; Wang, L.J.; Huang, Y.P.; Chen, X.Y. Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat. Biotechnol. 2007, 25, 1307–1313. [Google Scholar] [CrossRef] [PubMed]
- Miguel, K.S.; Scott, J.G. The next generation of insecticides: dsRNA is stable as a foliar-applied insecticide. Pest. Manag. Sci. 2016, 72, 801–809. [Google Scholar] [CrossRef] [PubMed]
- Auer, C.; Frederick, R. Crop improvement using small RNAs: Applications and predictive ecological risk assessments. Trends Biotechnol. 2009, 27, 644–651. [Google Scholar] [CrossRef] [PubMed]
- Lundgren, J.G.; Duan, J.J. RNAi-Based Insecticidal Crops: Potential Effects on Nontarget Species. BioScience 2013, 63, 657–665. [Google Scholar] [CrossRef]
- Parker, K.M.; Sander, M. Environmental Fate of Insecticidal Plant-Incorporated Protectants from Genetically Modified Crops: Knowledge Gaps and Research Opportunities. Environ. Sci. Technol. 2017, 51, 12049–12057. [Google Scholar] [CrossRef]
- Parker, K.M.; Barraganń Borrero, V.; van Leeuwen, D.M.; Lever, M.A.; Mateescu, B.; Sander, M. Environmental fate of RNA interference pesticides: Adsorption and degradation of double- stranded RNA molecules in agricultural soils. Environ. Sci. Technol. 2019, 53, 3027–3036. [Google Scholar] [CrossRef]
- Zhang, K.; Wei, J.; Huff Hartz, K.E.; Lydy, M.J.; Moon, T.S.; Sander, M.; Parker, K.M. Analysis of RNA Interference (RNAi) Biopesticides: Double-Stranded RNA (dsRNA) Extraction from Agricultural Soils and Quantification by RT-qPCR. Environ. Sci. Technol. 2020, 54, 4893–4902. [Google Scholar] [CrossRef]
- Chen, Y.; De Schutter, K. Biosafety aspects of RNAi-based pests control. Pest. Manag. Sci. 2024, 8, 3697–3706. [Google Scholar] [CrossRef]
- Hernández-Fernández, M.; Cordero-Bueso, G.; Ruiz-Muñoz, M.; Cantoral, J.M. Culturable Yeasts as Biofertilizers and Biopesticides for a Sustainable Agriculture: A Comprehensive Review. Plants 2021, 10, 822. [Google Scholar] [CrossRef]
- Drinnenberg, I.A.; Fink, G.R.; Bartel, D.P. Compatibility with killer explains the rise of RNAi-deficient fungi. Science 2011, 333, 1592. [Google Scholar] [CrossRef] [PubMed]
- Paddison, P.J.; Silva, J.M.; Conklin, D.S.; Schlabach, M.; Li, M.; Aruleba, S.; Balija, V.; O’Shaughnessy, A.; Gnoj, L.; Scobie, K.; et al. A resource for large-scale RNA-interference-based screens in mammals. Nature 2004, 428, 427–431. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Liu, Z.; Zhang, L.; Zhang, T.; Zhang, Z. siRNA In Vivo-targeted delivery to murine dendritic cells by oral administration of recombinant yeast. Methods Mol. Biol. 2016, 1364, 165–181. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Peng, H.; Feng, M.; Zhang, W.; Li, Y. Yeast microcapsule-mediated oral delivery of IL-1β shRNA for post-traumatic osteoarthritis therapy. Mol. Ther. Nucleic Acids 2020, 23, 336–346. [Google Scholar] [CrossRef]
- Murphy, K.A.; Tabuloc, C.A.; Cervantes, K.R.; Chiu, J.C. Ingestion of genetically modified yeast symbiont reduces fitness of an insect pest via RNA interference. Sci. Rep. 2016, 6, 22587. [Google Scholar] [CrossRef]
- Hapairai, L.K.; Mysore, K.; Chen, Y.; Harper, E.I.; Scheel, M.P.; Lesnik, A.M.; Sun, L.; Severson, D.W.; Wei, N.; Duman-Scheel, M. Lure-and-kill yeast interfering RNA larvicides targeting neural genes in the human disease vector mosquito Aedes aegypti. Sci. Rep. 2017, 7, 13223. [Google Scholar] [CrossRef]
- Gillet, F.X.; Bournaud, C.; Antonino de Souza Júnior, J.D.; Grossi-de-Sa, M.F. Plant-parasitic nematodes: Towards understanding molecular players in stress responses. Ann. Bot. 2017, 119, 775–789. [Google Scholar] [CrossRef]
- Cuko, L.; Cale, A.R.; Rambo, L.; Knoblock, M.L.; Karp, X. Distinct daf-16 isoforms regulate specification of vulval precursor cells in Caenorhabditis elegans. MicroPubl. Biol. 2022. [Google Scholar] [CrossRef]
- McHugh, D.R.; Koumis, E.; Jacob, P.; Goldfarb, J.; Schlaubitz-Garcia, M.; Bennani, S.; Regan, P.; Patel, P.; Youngman, M.J. DAF-16 and SMK-1 Contribute to Innate Immunity During Adulthood in Caenorhabditis elegans. G3 2020, 10, 1521–1539. [Google Scholar] [CrossRef]
- Basso, M.F.; Lourenço-Tessutti, I.T.; Mendes, R.A.G.; Pinto, C.E.M.; Bournaud, C.; Gillet, F.X.; Togawa, R.C.; de Macedo, L.L.P.; de Almeida Engler, J.; Grossi-de-Sa, M.F. MiDaf16-like and MiSkn1-like gene families are reliable targets to develop biotechnological tools for the control and management of Meloidogyne incognita. Sci. Rep. 2020, 10, 6991. [Google Scholar] [CrossRef]
- Brachmann, C.; Davies, A.; Cost, G.J.; Caputo, E.; Li, J.; Hieter, P.; Boeke, J.D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 1998, 14, 115–132. [Google Scholar] [CrossRef]
- Mazzoni, C.; Herker, E.; Palermo, V.; Jungwirth, H.; Eisenberg, T.; Madeo, F.; Falcone, C. Yeast caspase 1 links messenger RNA stability to apoptosis in yeast. EMBO Rep. 2005, 6, 1076–1081. [Google Scholar] [CrossRef]
- Taylor, R.G.; Walker, D.C.; McInnes, R.R. E. coli host strains significantly affect the quality of small scale plasmid DNA preparations used for sequencing. Nucleic Acids Res. 1993, 21, 1677–1678. [Google Scholar] [CrossRef]
- Chen, D.C.; Yang, B.C.; Kuo, T.T. One-step transformation of yeast in stationary phase. Curr Genet. 1992, 1, 83–84. [Google Scholar] [CrossRef] [PubMed]
- Westermann, B.; Neupert, W. Mitochondria-targeted green fluorescent proteins: Convenient tools for the study of organelle biogenesis in Saccharomyces cerevisiae. Yeast 2000, 15, 1421–1427. [Google Scholar] [CrossRef]
- Stovicek, V.; Borodina, I.; Forster, J. CRISPR-Cas system enables fast and simple genome editing of industrial Saccharomyces cerevisiae strains. Metab. Eng. Commun. 2015, 2, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Gietz, R.D.; Woods, R.A. Yeast transformation by the LiAc/SS Carrier DNA/PEG method. Methods Mol. Biol. 2006, 313, 107–120. [Google Scholar] [CrossRef]
- Schmitt, M.E.; Brown, T.A.; Trumpower, B.L. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990, 18, 3091–3092. [Google Scholar] [CrossRef]
- Schifano, E.; Cicalini, I.; Pieragostino, D.; Heipieper, H.J.; Del Boccio, P.; Uccelletti, D. In Vitro and In Vivo lipidomics as a tool for probiotics evaluation. Appl. Microbiol. Biotechnol. 2020, 104, 8937–8948. [Google Scholar] [CrossRef]
- Schifano, E.; Conta, G.; Preziosi, A.; Ferrante, C.; Batignani, G.; Mancini, P.; Tomassini, A.; Sciubba, F.; Scopigno, T.; Uccelletti, D.; et al. 2-hydroxyisobutyric acid (2-HIBA) modulates ageing and fat deposition in Caenorhabditis elegans. Front. Mol. Biosci. 2022, 9, 986022. [Google Scholar] [CrossRef]
- Pompa, L.; Montanari, A.; Tomassini, A.; Bianchi, M.M.; Aureli, W.; Miccheli, A.; Uccelletti, D.; Schifano, E. In Vitro Probiotic Properties and In Vivo Anti-Ageing Effects of Lactoplantibacillus plantarum PFA2018AU Strain Isolated from Carrots on Caenorhabditis elegans. Microorganisms 2023, 11, 1087. [Google Scholar] [CrossRef]
- Boutla, A.; Delidakis, C.; Livadaras, I.; Tabler, M. Variations of the 3′ protruding ends in synthetic short interfering RNA (siRNA) tested by microinjection in Drosophila embryos. Oligonucleotides 2003, 13, 295–301. [Google Scholar] [CrossRef]
- Bofill-De Ros, X.; Gu, S. Guidelines for the optimal design of miRNA-based shRNAs. Methods 2016, 103, 157–166. [Google Scholar] [CrossRef]
- Terasawa, K.; Shimizu, K.; Tsujimoto, G. Synthetic Pre-miRNA-Based shRNA as Potent RNAi Triggers. J. Nucleic Acids 2011, 2011, 131579. [Google Scholar] [CrossRef]
- Lu, Z.; Lin, Z. Pervasive and dynamic transcription initiation in Saccharomyces cerevisiae. Genome Res. 2019, 29, 1198–1210. [Google Scholar] [CrossRef] [PubMed]
- Duman-Scheel, M. Saccharomyces cerevisiae (Baker’s Yeast) as an Interfering RNA Expression and Delivery System. Curr. Drug Targets 2019, 20, 942–952. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, W.; Peng, H.; Li, Y.; Leng, T.; Xie, C.; Zhang, L. Oral Gene Therapy of HFD-Obesity via Nonpathogenic Yeast Microcapsules Mediated shRNA Delivery. Pharmaceutics 2021, 13, 1536. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Li, Q.X.; Song, B. Chemical Nematicides: Recent Research Progress and Outlook. J. Agric. Food Chem. 2020, 68, 12175–12188. [Google Scholar] [CrossRef]
- Abd-Elgawad, M.M.M. Integrated Nematode Management Strategies: Optimization of Combined Nematicidal and Multi-Functional Inputs. Plants 2025, 14, 1004. [Google Scholar] [CrossRef]



| S. cerevisiae Strains | Genotype | Reference |
|---|---|---|
| BY4741 | Mat a, his3-Δ1, leu2-Δ0, met15-Δ0, ura3-Δ0 | [31] |
| CML39-11A | MATa, ade1-101, his3-Δ1, leu2, ura3, trp1-289 | [32] |
| BY4741 shDaf16.Ø | Mat a, his3-Δ1, leu2-Δ0, met15-Δ0, ura3-Δ0, ade2::shDaf16.Ø | This work |
| BY4741 shDaf16.1 | Mat a, his3-Δ1, leu2-Δ0, met15-Δ0, ura3-Δ0, ade2::shDaf16.1 | This work |
| BY4741 shDaf16.2 | Mat a, his3-Δ1, leu2-Δ0, met15-Δ0, ura3-Δ0, ade2::shDaf16.2 | This work |
| CML3911A pYX232-shDaf16.Ø | MATa, ade1-101, his3-Δ1, leu2, ura3, trp1-289, pYX232- shDaf16.Ø | This work |
| CML3911A pYX232-shDaf16.1 | MATa, ade1-101, his3-Δ1, leu2, ura3, trp1-289, pYX232- shDaf16.1 | This work |
| CML3911A pYX232-shDaf16.2 | MATa, ade1-101, his3-Δ1, leu2, ura3, trp1-289, pYX232- shDaf16.2 | This work |
| E. coli Strain | Genotype | Reference |
| DH5-α | fhuA2 lac(del)U169 phoA glnV44 Φ80′ lacZ(del)M15 gyrA96 recA1 relA1 endA1 thi-1 hsdR17 | [33] |
| C. elegans Strain | Genotype | Reference |
| N2 | Caenorhabditis elegans wild isolate | https://wormbase.org |
| TJ356 | zIs356 [daf-16p::daf-16a/b::GFP + rol-6 (su1006)] | https://wormbase.org |
| Plasmid | Main Features | Reference |
|---|---|---|
| pYX232-mtGFP | 2μ expression vector–Promoter: TPI–Selection marker: TRP1 | [35] |
| pCfB2311 | 2μ expression vector–SNR52-gRNA-ADE2–Selection marker: Nat | [36] |
| pCfB2312 | Cen expression vector–Tef1-Cas9–Selection: kanMX4 | [36] |
| pYX232-shDaf16.Ø | 2μ TPI-shDaf16.Ø (empty vector) | This work |
| pYX232-shDaf16.1 | 2μ TPI-ShDaf16.1 | This work |
| pYX232-shDaf16.2 | 2μ TPI-ShDaf16.2 | This work |
| Primers | Sequence | Purpose |
|---|---|---|
| shDaf16For1 | 5′AATTCCCGGATACTATGGATGTTGA TTTCAAGAGAATCAACATCCATAGTATCCTTTTTA3′ | Cloning shDAF16.1 in pYX232 vector |
| shDaf16Rev1 | 5′AGCTTAAAAAGGATACTATGGATGT TGATTCTCTTGAAATCAACATCCATAGTATCCGGG3′ | |
| shDaf16For2 | 5′AATTCCCGGAGGGCAGCATATTAT TTTCAAGAGAAATGAATATGCTGCCCTCCTTTTTA3′ | Cloning shDAF16.2 in pYX232 vector |
| shDaf16Rev2 | 5′AGCTTAAAAAGGGAGGGCAGCATATTCATTTCTCTTGAAAATAATATGCTGCCCTCCGGG3′ | |
| sh232insFW | 5′GTTAACGGTTTAGTGTTTTCTTACCCAATTGTAGAGACTAGGCAAGAGAGAAGACCCAGAGATG3′ | Donor amplification |
| ACR3GFPinsREV | 5′GAGTCCGGAACTCTAGCAGGCGCATAACATAAGTCACAAAGACGTTGTAAAACGACGGCC3′ | |
| SeqshRNATPIFor | 5′GGCTGCTGTAACAGGGAATATAAAGG3′ | Sequencing to verify the integration of shRNA cassette |
| Daf16.1_PriEx | 5′GGATACTATGGATGTTGATTCTCTTGAA3′ | Labeled primers for primer extension |
| Daf16.2_PriEx | 5′GGAGGGCAGCATATTCATTTCTCTTGAA3′ | |
| PRIMER daf-16 FOR | 5′TCAAGACCTCAAAGCCAATCAACTC3′ | Selective primers for daf-16 gene used in the RT-qPCR analysis. |
| PRIMER daf-16 REV | 5′ACGAGAAAGAAGGAGTAAGAGGAG-3′ | |
| PRIMER cdc-42 FOR | 5′CTGCTGGACAGGAAGATTACG3′ | Selective primers for cdc-42 gene used in the RT-qPCR analysis. |
| PRIMER cdc-42 REV | 5′CTCGGACATTCTCGAATGAAG3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caraba, B.; Montanari, A.; Schifano, E.; Stocchi, F.; Costanzo, G.; Uccelletti, D.; Mazzoni, C. Yeast Oral Delivery of DAF16 shRNAs Results in Effective Gene Silencing in C. elegans. Curr. Issues Mol. Biol. 2025, 47, 570. https://doi.org/10.3390/cimb47070570
Caraba B, Montanari A, Schifano E, Stocchi F, Costanzo G, Uccelletti D, Mazzoni C. Yeast Oral Delivery of DAF16 shRNAs Results in Effective Gene Silencing in C. elegans. Current Issues in Molecular Biology. 2025; 47(7):570. https://doi.org/10.3390/cimb47070570
Chicago/Turabian StyleCaraba, Benedetta, Arianna Montanari, Emily Schifano, Fabiana Stocchi, Giovanna Costanzo, Daniela Uccelletti, and Cristina Mazzoni. 2025. "Yeast Oral Delivery of DAF16 shRNAs Results in Effective Gene Silencing in C. elegans" Current Issues in Molecular Biology 47, no. 7: 570. https://doi.org/10.3390/cimb47070570
APA StyleCaraba, B., Montanari, A., Schifano, E., Stocchi, F., Costanzo, G., Uccelletti, D., & Mazzoni, C. (2025). Yeast Oral Delivery of DAF16 shRNAs Results in Effective Gene Silencing in C. elegans. Current Issues in Molecular Biology, 47(7), 570. https://doi.org/10.3390/cimb47070570







