The Transcription Machinery and the Driving Force of the Transcriptional Molecular Condensate: The Role of Phosphates
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. Characterization of the Pulse During the Transcription Process
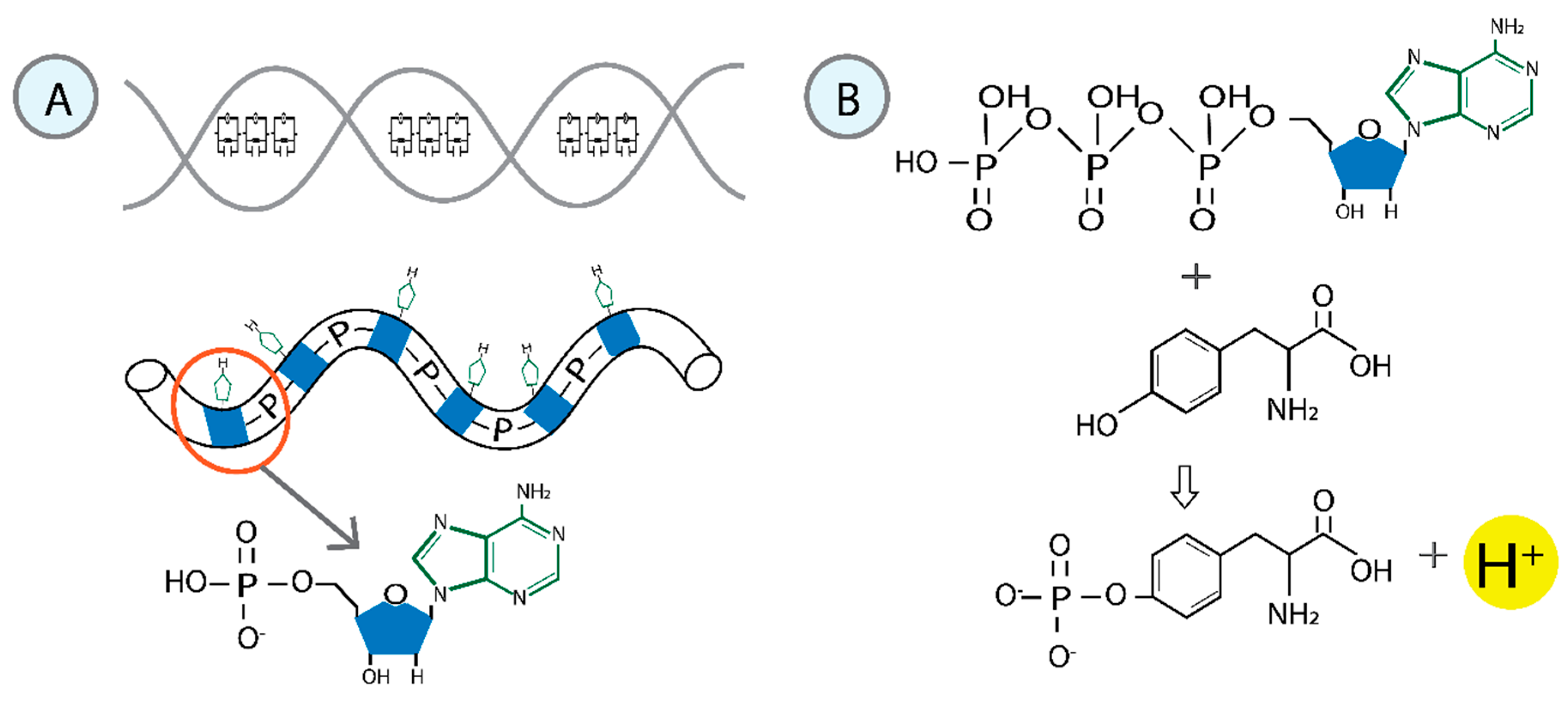
3.1.1. Phosphates as the Driving Force
3.1.2. Model of Phosphorylation-Inducing Interactions Between RNA Pol II and DNA
3.2. Water Molecules in DNA and Intrinsically Disordered Proteins
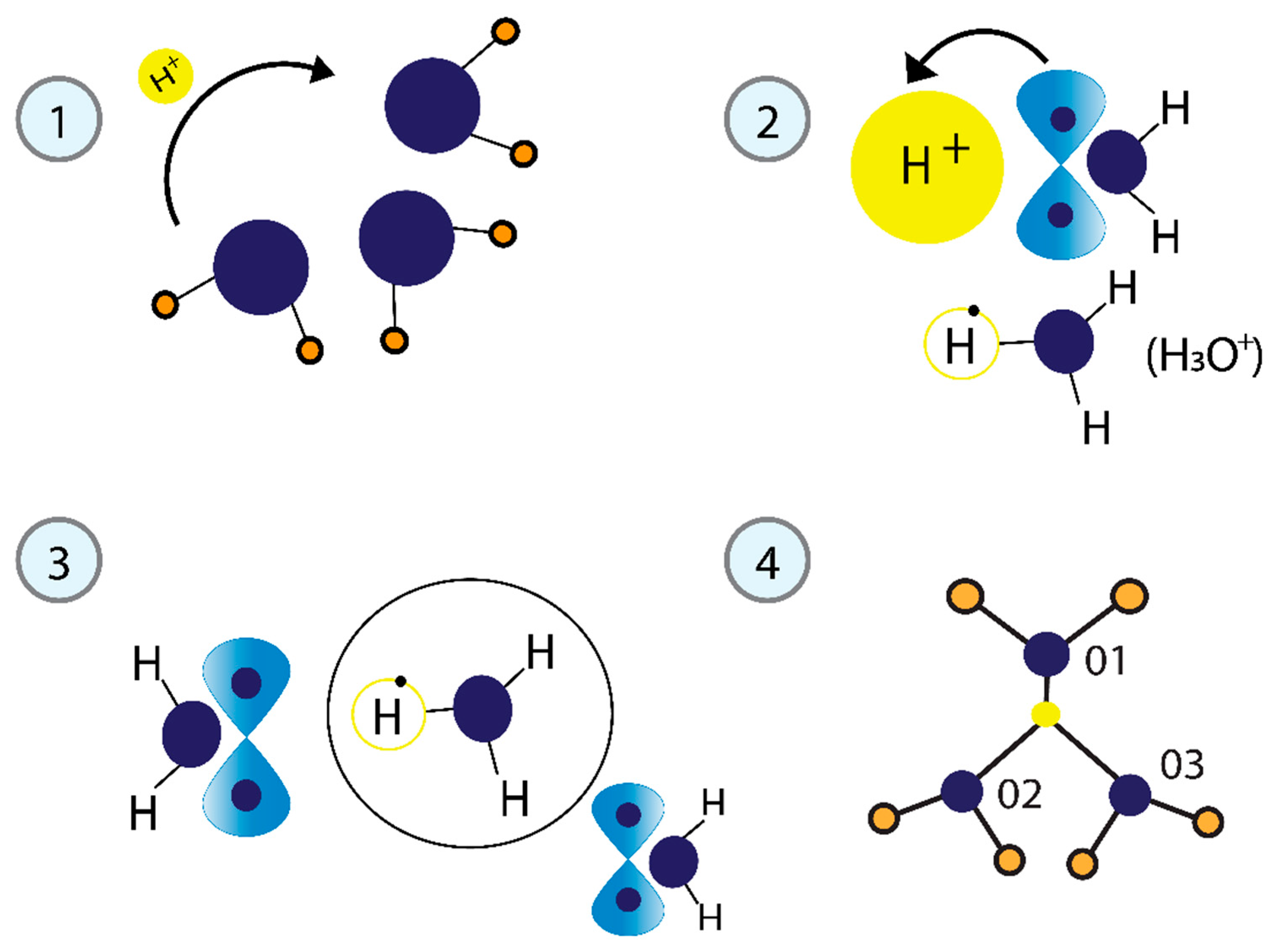
3.2.1. Physical Model of Phosphorylation Mediating H+ Proton Transfer
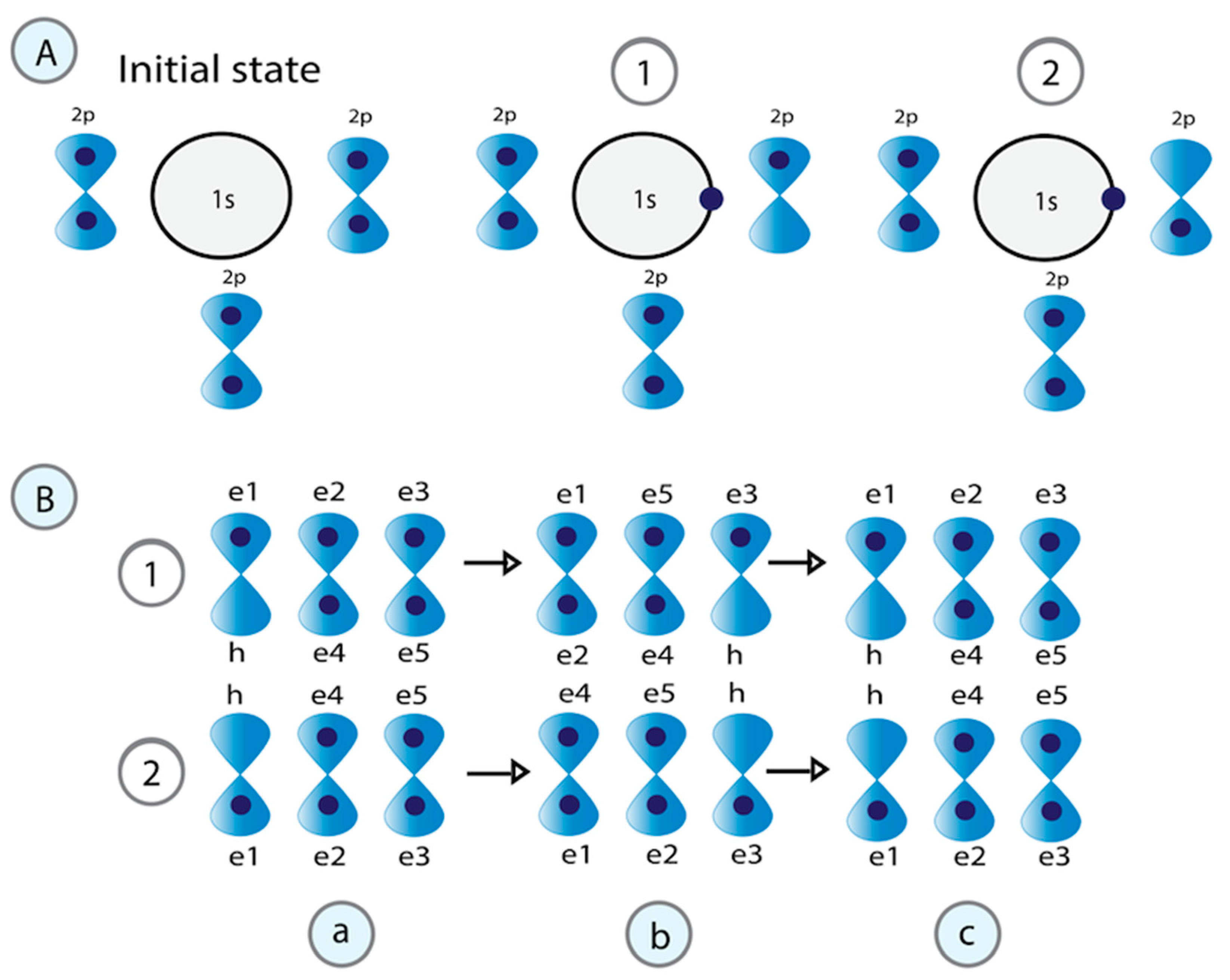
3.2.2. Superconductor Character of Water Induced by H+ Proton Transfer
3.2.3. Application of the Physical-Mathematical Model for the Correlation of Pairs of the Same Type of Fermions (Two Electrons) That Exchange a Boson to Explain Superconductivity in Water
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Correction Statement
References
- Tian, T.; Chu, X.-Y.; Yang, Y.; Zhang, X.; Liu, Y.-M.; Gao, J.; Ma, B.-G.; Zhang, H.-Y. Phosphates as Energy Sources to Expand Metabolic Networks. Life 2019, 9, 43. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.L.; Yaron, T.M.; Huntsman, E.M.; Kerelsky, A.; Song, J.; Regev, A.; Lin, T.-Y.; Liberatore, K.; Cizin, D.M.; Cohen, B.M.; et al. An Atlas of Substrate Specificities for the Human Serine/Threonine Kinome. Nature 2023, 613, 759–766. [Google Scholar] [CrossRef] [PubMed]
- Ubersax, J.A.; Ferrell Jr, J.E. Mechanisms of Specificity in Protein Phosphorylation. Nat. Rev. Mol. Cell Biol. 2007, 8, 530–541, Erratum in Nat. Rev. Mol. Cell Biol. 2007, 8, 665. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Alqahtani, S.; Hu, X. Aromatic Rings as Molecular Determinants for the Molecular Recognition of Protein Kinase Inhibitors. Molecules 2021, 26, 1776. [Google Scholar] [CrossRef] [PubMed]
- Bubon, T.L.; Perepelytsya, S.M. Low-Frequency Vibrations of Water Molecules in DNA Minor Groove. Eur. Phys. J. E 2021, 44, 84. [Google Scholar] [CrossRef] [PubMed]
- Bubon, T.; Zdorevskyi, O.; Perepelytsya, S. Molecular Dynamics Study of Collective Water Vibrations in a DNA Hydration Shell. Eur. Biophys. J. 2023, 52, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Dragan, A.I.; Read, C.M.; Crane-Robinson, C. Enthalpy–Entropy Compensation: The Role of Solvation. Eur. Biophys. J. 2017, 46, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Duboué-Dijon, E.; Fogarty, A.C.; Hynes, J.T.; Laage, D. Dynamical Disorder in the DNA Hydration Shell. J. Am. Chem. Soc. 2016, 138, 7610–7620. [Google Scholar] [CrossRef] [PubMed]
- Tereshko, V.; Minasov, G.; Egli, M. A “Hydrat-Ion” Spine in a B-DNA Minor Groove. J. Am. Chem. Soc. 1999, 121, 3590–3595. [Google Scholar] [CrossRef]
- Riera Aroche, R.; Ortiz García, Y.M.; Martínez Arellano, M.A.; Riera Leal, A. DNA as a Perfect Quantum Computer Based on the Quantum Physics Principles. Sci. Rep. 2024, 14, 11636. [Google Scholar] [CrossRef] [PubMed]
- Riera Aroche, R.; Ortiz García, Y.M.; Sánchez Moreno, E.C.; Enriquez Cervantes, J.S.; Machado Sulbaran, A.C.; Riera Leal, A. DNA Gene’s Basic Structure as a Nonperturbative Circuit Quantum Electrodynamics: Is RNA Polymerase II the Quantum Bus of Transcription? Curr. Issues Mol. Biol. 2024, 46, 12152–12173. [Google Scholar] [CrossRef] [PubMed]
- Valiev, M.; Yang, J.; Adams, J.A.; Taylor, S.S.; Weare, J.H. Phosphorylation Reaction in CAPK Protein Kinase-Free Energy Quantum Mechanical/Molecular Mechanics Simulations. J. Phys. Chem. B 2007, 111, 13455–13464. [Google Scholar] [CrossRef] [PubMed]
- Batishcheva, E.V.; Smirnov, N.N.; Bobrova, N.V.; Sokolova, M.P.; Smirnov, M.A. Ion-Conducting Membranes Based on Bacterial Cellulose Nanofibers Modified by Poly(Sodium Acrylate-Co-2-Acrylamido-2-Methylpropanesulfonic Acid). Chin. J. Polym. Sci. 2024, 42, 333–343. [Google Scholar] [CrossRef]
- Xie, C.; Yang, R.; Wan, X.; Li, H.; Ge, L.; Li, X.; Zhao, G. A High-Proton Conductivity All-Biomass Proton Exchange Membrane Enabled by Adenine and Thymine Modified Cellulose Nanofibers. Polymers 2024, 16, 1060. [Google Scholar] [CrossRef] [PubMed]
- Riera Aroche, R.; Ortiz García, Y.M.; Sánchez Moreno, E.C.; Riera Leal, L.; Machado Sulbarán, A.C.; Riera Leal, A. The π–π Architectures Reveal a Hidden Quantum Code Linking Aromaticity to Light Interaction. Sci. Rep. 2025, 15, 25110. [Google Scholar] [CrossRef] [PubMed]
- Patikoglou, G.A.; Kim, J.L.; Sun, L.; Yang, S.-H.; Kodadek, T.; Burley, S.K. TATA Element Recognition by the TATA Box-Binding Protein Has Been Conserved throughout Evolution. Genes Dev. 1999, 13, 3217–3230. [Google Scholar] [CrossRef] [PubMed]
- Savinkova, L.K.; Sharypova, E.B.; Kolchanov, N.A. On the Role of TATA Boxes and TATA-Binding Protein in Arabidopsis Thaliana. Plants 2023, 12, 1000. [Google Scholar] [CrossRef] [PubMed]
- Vanaja, A.; Yella, V.R. Delineation of the DNA Structural Features of Eukaryotic Core Promoter Classes. ACS Omega 2022, 7, 5657–5669. [Google Scholar] [CrossRef] [PubMed]
- Sabari, B.R.; Dall’Agnese, A.; Boija, A.; Klein, I.A.; Coffey, E.L.; Shrinivas, K.; Abraham, B.J.; Hannett, N.M.; Zamudio, A.V.; Manteiga, J.C.; et al. Coactivator Condensation at Super-Enhancers Links Phase Separation and Gene Control. Science 2018, 361, eaar3958. [Google Scholar] [CrossRef] [PubMed]
- Yasuhara, T.; Xing, Y.-H.; Bauer, N.C.; Lee, L.; Dong, R.; Yadav, T.; Soberman, R.J.; Rivera, M.N.; Zou, L. Condensates Induced by Transcription Inhibition Localize Active Chromatin to Nucleoli. Mol. Cell 2022, 82, 2738–2753.e6. [Google Scholar] [CrossRef] [PubMed]
- Cho, W.-K.; Spille, J.-H.; Hecht, M.; Lee, C.; Li, C.; Grube, V.; Cisse, I.I. Mediator and RNA Polymerase II Clusters Associate in Transcription-Dependent Condensates. Science 2018, 361, 412–415. [Google Scholar] [CrossRef] [PubMed]
- Pei, G.; Lyons, H.; Li, P.; Sabari, B.R. Transcription Regulation by Biomolecular Condensates. Nat. Rev. Mol. Cell Biol. 2024, 26, 213–236. [Google Scholar] [CrossRef] [PubMed]
- Gui, T.; Fleming, C.; Manzato, C.; Bourgeois, B.; Sirati, N.; Heuer, J.; Papadionysiou, I.; Montfort, D.I.v.; Gijzen, M.v.; Smits, L.M.M.; et al. Targeted Perturbation of Signaling-Driven Condensates. Mol. Cell 2023, 83, 4141–4157.e11. [Google Scholar] [CrossRef] [PubMed]
- Cramer, P. Organization and Regulation of Gene Transcription. Nature 2019, 573, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Henninger, J.E.; Oksuz, O.; Shrinivas, K.; Sagi, I.; LeRoy, G.; Zheng, M.M.; Andrews, J.O.; Zamudio, A.V.; Lazaris, C.; Hannett, N.M.; et al. RNA-Mediated Feedback Control of Transcriptional Condensates. Cell 2021, 184, 207–225.e24. [Google Scholar] [CrossRef] [PubMed]
- Lyons, H.; Veettil, R.T.; Pradhan, P.; Fornero, C.; De La Cruz, N.; Ito, K.; Eppert, M.; Roeder, R.G.; Sabari, B.R. Functional Partitioning of Transcriptional Regulators by Patterned Charge Blocks. Cell 2023, 186, 327–345.e28. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Molina, J.B.; West, S.; Passmore, L.A. Knowing When to Stop: Transcription Termination on Protein-Coding Genes by Eukaryotic RNAPII. Mol. Cell 2023, 83, 404–415. [Google Scholar] [CrossRef] [PubMed]
- Vervoort, S.J.; Devlin, J.R.; Kwiatkowski, N.; Teng, M.; Gray, N.S.; Johnstone, R.W. Targeting Transcription Cycles in Cancer. Nat. Rev. Cancer 2022, 22, 5–24. [Google Scholar] [CrossRef] [PubMed]
- Ehara, H.; Yokoyama, T.; Shigematsu, H.; Yokoyama, S.; Shirouzu, M.; Sekine, S. Structure of the Complete Elongation Complex of RNA Polymerase II with Basal Factors. Science 2017, 357, 921–924. [Google Scholar] [CrossRef] [PubMed]
- Lassila, J.K.; Zalatan, J.G.; Herschlag, D. Biological Phosphoryl-Transfer Reactions: Understanding Mechanism and Catalysis. Annu. Rev. Biochem. 2011, 80, 669–702. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, Y.; Wang, Y.; Lin, C.; Zhang, D.; Chen, J.; Ouyang, L.; Wu, F.; Zhang, J.; Chen, L. Recent Progress on Vascular Endothelial Growth Factor Receptor Inhibitors with Dual Targeting Capabilities for Tumor Therapy. J. Hematol. Oncol. 2022, 15, 89. [Google Scholar] [CrossRef] [PubMed]
- Kamerlin, S.C.L.; Sharma, P.K.; Prasad, R.B.; Warshel, A. Why Nature Really Chose Phosphate. Q. Rev. Biophys. 2013, 46, 1–132. [Google Scholar] [CrossRef] [PubMed]
- Robertson, A.J.; Wilson, A.L.; Burn, M.J.; Cliff, M.J.; Popelier, P.L.A.; Waltho, J.P. The Relationship between Enzyme Conformational Change, Proton Transfer, and Phosphoryl Transfer in β-Phosphoglucomutase. ACS Catal. 2021, 11, 12840–12849. [Google Scholar] [CrossRef]
- Banani, S.F.; Lee, H.O.; Hyman, A.A.; Rosen, M.K. Biomolecular Condensates: Organizers of Cellular Biochemistry. Nat. Rev. Mol. Cell Biol. 2017, 18, 285–298. [Google Scholar] [CrossRef] [PubMed]
- Visser, B.S.; Lipiński, W.P.; Spruijt, E. The Role of Biomolecular Condensates in Protein Aggregation. Nat. Rev. Chem. 2024, 8, 686–700. [Google Scholar] [CrossRef] [PubMed]
- Kaufman, N.E.M.; Dhingra, S.; Jois, S.D.; Vicente, M.d.G.H. Molecular Targeting of Epidermal Growth Factor Receptor (EGFR) and Vascular Endothelial Growth Factor Receptor (VEGFR). Molecules 2021, 26, 1076. [Google Scholar] [CrossRef] [PubMed]
- Dopkins, N.; Nixon, D.F. Activation of Human Endogenous Retroviruses and Its Physiological Consequences. Nat. Rev. Mol. Cell Biol. 2024, 25, 212–222. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, M.; Rizzo, S.; Paz, W.E.; Kresinsky, A.; Thévenin, D.; Müller, J.P. Disrupting PTPRJ Transmembrane-Mediated Oligomerization Counteracts Oncogenic Receptor Tyrosine Kinase FLT3 ITD. Front. Oncol. 2022, 12, 1017947. [Google Scholar] [CrossRef] [PubMed]
- Burevschi, E.; Alonso, E.R.; Sanz, M.E. Binding Site Switch by Dispersion Interactions: Rotational Signatures of Fenchone–Phenol and Fenchone–Benzene Complexes. Chem. A Eur. J. 2020, 26, 11327–11333. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, C.A.; Weber, D.S.; Warren, J.J. Clustering of Aromatic Amino Acid Residues around Methionine in Proteins. Biomolecules 2021, 12, 6. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, O.; Odani, A. Structure-Stability Relationship in Ternary Copper(II) Complexes Involving Aromatic Amines and Tyrosine or Related Amino Acids. Intramolecular Aromatic Ring Stacking and Its Regulation through Tyrosine Phosphorylation. J. Am. Chem. Soc. 1985, 107, 5938–5945. [Google Scholar] [CrossRef]
- Zhang, J.; Hao, M.; Zhang, D.; Zhang, X.; Guo, S.; Wang, B.; Xiao, J.; Gao, Y.; Li, X. Enhanced Polyacrylamide Degradation via OH Radical-Initiated Single-Electron Transfer. ACS Omega 2023, 8, 46589–46597. [Google Scholar] [CrossRef] [PubMed]
- Blumberger, J. Recent Advances in the Theory and Molecular Simulation of Biological Electron Transfer Reactions. Chem. Rev. 2015, 115, 11191–11238. [Google Scholar] [CrossRef] [PubMed]
- Matyushov, D.V. Reorganization Energy of Electron Transfer. Phys. Chem. Chem. Phys. 2023, 25, 7589–7610. [Google Scholar] [CrossRef] [PubMed]
- Nakane, K.; Nagasawa, H.; Fujimura, C.; Koyanagi, E.; Tomoshige, S.; Ishikawa, M.; Sato, S. Switching of Photocatalytic Tyrosine/Histidine Labeling and Application to Photocatalytic Proximity Labeling. Int. J. Mol. Sci. 2022, 23, 11622. [Google Scholar] [CrossRef] [PubMed]
- Qiao, H.; Xia, M.; Cheng, Y.; Zhou, J.; Zheng, L.; Li, W.; Wang, J.; Fang, P. Tyrosine-Targeted Covalent Inhibition of a TRNA Synthetase Aided by Zinc Ion. Commun. Biol. 2023, 6, 107. [Google Scholar] [CrossRef] [PubMed]
- Ghoshal, S.; Hazra, M.K. Impact of OH Radical-Initiated H2CO3 Degradation in the Earth’s Atmosphere via Proton-Coupled Electron Transfer Mechanism. J. Phys. Chem. A 2016, 120, 562–575. [Google Scholar] [CrossRef] [PubMed]
- Mahmudov, K.T.; Kopylovich, M.N.; Guedes da Silva, M.F.C.; Pombeiro, A.J.L. Non-Covalent Interactions in the Synthesis of Coordination Compounds: Recent Advances. Coord. Chem. Rev. 2017, 345, 54–72. [Google Scholar] [CrossRef]
- Gray, T.M.; Matthews, B.W. Intrahelical Hydrogen Bonding of Serine, Threonine and Cysteine Residues within α-Helices and Its Relevance to Membrane-Bound Proteins. J. Mol. Biol. 1984, 175, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Hubbard, E.E.; Julian, R.R. Unravelling the Favorability of Radical-Directed Xn-H2O Dissociation at Serine and Threonine. Int. J. Mass Spectrom. 2025, 507, 117363. [Google Scholar] [CrossRef]
- Zhang, L.; Reilly, J.P. Radical-Driven Dissociation of Odd-Electron Peptide Radical Ions Produced in 157 Nm Photodissociation. J. Am. Soc. Mass Spectrom. 2009, 20, 1378–1390. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. The Intrinsic Disorder Alphabet. III. Dual Personality of Serine. Intrinsically Disord. Proteins 2015, 3, e1027032. [Google Scholar] [CrossRef] [PubMed]
- Basile, W.; Salvatore, M.; Bassot, C.; Elofsson, A. Why Do Eukaryotic Proteins Contain More Intrinsically Disordered Regions? PLoS Comput. Biol. 2019, 15, e1007186. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, R.; Nagarajaram, H.A. Intrinsically Disordered Proteins: An Overview. Int. J. Mol. Sci. 2022, 23, 14050. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Lessing, J.; Meisl, G.; Ganim, Z.; Tokmakoff, A.; Knoester, J.; Jansen, T.L.C. Solvent and Conformation Dependence of Amide I Vibrations in Peptides and Proteins Containing Proline. J. Chem. Phys. 2011, 135, 234507. [Google Scholar] [CrossRef] [PubMed]
- Kubyshkin, V.; Rubini, M. Proline Analogues. Chem. Rev. 2024, 124, 8130–8232. [Google Scholar] [CrossRef] [PubMed]
- Byun, B.J.; Kang, Y.K. Conformational Preferences and Prolyl Cis-Trans Isomerization of Phosphorylated Ser/Thr-Pro Motifs. Biopolymers 2010, 93, 330–339. [Google Scholar] [CrossRef] [PubMed]
- Kubyshkin, V.; Mykhailiuk, P.K. Proline Analogues in Drug Design: Current Trends and Future Prospects. J. Med. Chem. 2024, 67, 20022–20055. [Google Scholar] [CrossRef] [PubMed]
- Joshi, A.; Avni, A.; Walimbe, A.; Rai, S.K.; Sarkar, S.; Mukhopadhyay, S. Hydrogen-Bonded Network of Water in Phase-Separated Biomolecular Condensates. J. Phys. Chem. Lett. 2024, 15, 7724–7734. [Google Scholar] [CrossRef] [PubMed]
- Codorniu-Hernández, E.; Kusalik, P.G. Mobility Mechanism of Hydroxyl Radicals in Aqueous Solution via Hydrogen Transfer. J. Am. Chem. Soc. 2012, 134, 532–538. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Tang, C.; Jiao, N. Recent Advances in Copper-Catalyzed Dehydrogenative Functionalization via a Single Electron Transfer (SET) Process. Chem. Soc. Rev. 2012, 41, 3464. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Wen, C.; Cheng, S.; Vinh, N.Q. Long-Range DNA-Water Interactions. Biophys. J. 2021, 120, 4966–4979. [Google Scholar] [CrossRef] [PubMed]
- McDermott, M.L.; Vanselous, H.; Corcelli, S.A.; Petersen, P.B. DNA’s Chiral Spine of Hydration. ACS Cent. Sci. 2017, 3, 708–714. [Google Scholar] [CrossRef] [PubMed]
- Perets, E.A.; Konstantinovsky, D.; Santiago, T.; Videla, P.E.; Tremblay, M.; Velarde, L.; Batista, V.S.; Hammes-Schiffer, S.; Yan, E.C.Y. Beyond the “Spine of Hydration”: Chiral SFG Spectroscopy Detects DNA First Hydration Shell and Base Pair Structures. J. Chem. Phys. 2024, 161, 095104. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Mondal, S.; Acharya, S.; Bagchi, B. DNA Solvation Dynamics. J. Phys. Chem. B 2018, 122, 11743–11761. [Google Scholar] [CrossRef] [PubMed]
- Gavrilov, Y.; Leuchter, J.D.; Levy, Y. On the Coupling between the Dynamics of Protein and Water. Phys. Chem. Chem. Phys. 2017, 19, 8243–8257. [Google Scholar] [CrossRef] [PubMed]
- Conti Nibali, V.; D’Angelo, G.; Paciaroni, A.; Tobias, D.J.; Tarek, M. On the Coupling between the Collective Dynamics of Proteins and Their Hydration Water. J. Phys. Chem. Lett. 2014, 5, 1181–1186. [Google Scholar] [CrossRef] [PubMed]
- Wood, K.; Plazanet, M.; Gabel, F.; Kessler, B.; Oesterhelt, D.; Tobias, D.J.; Zaccai, G.; Weik, M. Coupling of Protein and Hydration-Water Dynamics in Biological Membranes. Proc. Natl. Acad. Sci. USA 2007, 104, 18049–18054. [Google Scholar] [CrossRef] [PubMed]
- Otake, K.; Kitagawa, H. Control of Proton-Conductive Behavior with Nanoenvironment within Metal–Organic Materials. Small 2021, 17, 2006189. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Wang, C. Surface Confinement of Finite-Size Water Droplets for SO3 Hydrolysis Reaction Revealed by Molecular Dynamics Simulations Based on a Machine Learning Force Field. J. Am. Chem. Soc. 2023, 145, 10631–10640. [Google Scholar] [CrossRef] [PubMed]
- Kornyshev, A.A.; Kuznetsov, A.M.; Spohr, E.; Ulstrup, J. Kinetics of Proton Transport in Water. J. Phys. Chem. B 2003, 107, 3351–3366. [Google Scholar] [CrossRef]
- Lee, S.H.; Rasaiah, J.C. Proton Transfer and the Mobilities of the H+ and OH− Ions from Studies of a Dissociating Model for Water. J. Chem. Phys. 2011, 135, 124505. [Google Scholar] [CrossRef] [PubMed]
- Adhav, V.A.; Shelke, S.S.; Balanarayan, P.; Saikrishnan, K. Sulfur-Mediated Chalcogen versus Hydrogen Bonds in Proteins: A See-Saw Effect in the Conformational Space. QRB Discov. 2023, 4, e5. [Google Scholar] [CrossRef] [PubMed]
- Nishio, M.; Umezawa, Y.; Fantini, J.; Weiss, M.S.; Chakrabarti, P. CH–π Hydrogen Bonds in Biological Macromolecules. Phys. Chem. Chem. Phys. 2014, 16, 12648–12683. [Google Scholar] [CrossRef] [PubMed]
- Mohan, N.; Vijayalakshmi, K.P.; Koga, N.; Suresh, C.H. Comparison of Aromatic NH···π, OH···π, and CH···π Interactions of Alanine Using MP2, CCSD, and DFT Methods. J. Comput. Chem. 2010, 31, 2874–2882. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Raj, P.; Dubowski, J.J.; Singh, N. ATP Induced Modulation in π–π Stacking Interactions in Pyrene Based Zinc Complexes: Chemosensor Study and Quantitative Investigation of Apyrase Activity. Cryst. Growth Des. 2018, 18, 4320–4333. [Google Scholar] [CrossRef]
- Brandl, M.; Weiss, M.S.; Jabs, A.; Sühnel, J.; Hilgenfeld, R. C-H⋯π-Interactions in Proteins. J. Mol. Biol. 2001, 307, 357–377. [Google Scholar] [CrossRef] [PubMed]
- Krone, M.W.; Travis, C.R.; Lee, G.Y.; Eckvahl, H.J.; Houk, K.N.; Waters, M.L. More Than π–π–π Stacking: Contribution of Amide−π and CH−π Interactions to Crotonyllysine Binding by the AF9 YEATS Domain. J. Am. Chem. Soc. 2020, 142, 17048–17056. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Tian, W.; Severance, Z.C.; Chaudhary, S.K.; Anokhina, V.; Mondal, B.; Pergu, R.; Singh, P.; Dhawa, U.; Singha, S.; et al. Proximity-Inducing Modalities: The Past, Present, and Future. Chem. Soc. Rev. 2023, 52, 5485–5515. [Google Scholar] [CrossRef] [PubMed]
- Bremer, A.; Farag, M.; Borcherds, W.M.; Peran, I.; Martin, E.W.; Pappu, R.V.; Mittag, T. Deciphering How Naturally Occurring Sequence Features Impact the Phase Behaviours of Disordered Prion-like Domains. Nat. Chem. 2022, 14, 196–207. [Google Scholar] [CrossRef] [PubMed]
- Villegas, J.A.; Heidenreich, M.; Levy, E.D. Molecular and Environmental Determinants of Biomolecular Condensate Formation. Nat. Chem. Biol. 2022, 18, 1319–1329. [Google Scholar] [CrossRef] [PubMed]
- Pappu, R.V.; Cohen, S.R.; Dar, F.; Farag, M.; Kar, M. Phase Transitions of Associative Biomacromolecules. Chem. Rev. 2023, 123, 8945–8987. [Google Scholar] [CrossRef] [PubMed]
- Martin, E.W.; Holehouse, A.S.; Peran, I.; Farag, M.; Incicco, J.J.; Bremer, A.; Grace, C.R.; Soranno, A.; Pappu, R.V.; Mittag, T. Valence and Patterning of Aromatic Residues Determine the Phase Behavior of Prion-like Domains. Science 2020, 367, 694–699. [Google Scholar] [CrossRef] [PubMed]
- Vernon, R.M.; Forman-Kay, J.D. First-Generation Predictors of Biological Protein Phase Separation. Curr. Opin. Struct. Biol. 2019, 58, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Nott, T.J.; Petsalaki, E.; Farber, P.; Jervis, D.; Fussner, E.; Plochowietz, A.; Craggs, T.D.; Bazett-Jones, D.P.; Pawson, T.; Forman-Kay, J.D.; et al. Phase Transition of a Disordered Nuage Protein Generates Environmentally Responsive Membraneless Organelles. Mol. Cell 2015, 57, 936–947. [Google Scholar] [CrossRef] [PubMed]
- Gutowsk, M.; Kowalczyk, S. A Study of Free Radical Chemistry: Their Role and Pathophysiological Significance. Acta Biochim. Pol. 2016, 60, 1–16. [Google Scholar] [CrossRef]
- Meng, X.; Qi, J. Manipulating Tyrosine Phosphorylation by Heterobifunctional Small Molecules. ACS Cent. Sci. 2023, 9, 1512–1514. [Google Scholar] [CrossRef] [PubMed]
- Winsor, T.S.; Bartkowiak, B.; Bennett, C.B.; Greenleaf, A.L. A DNA Damage Response System Associated with the PhosphoCTD of Elongating RNA Polymerase II. PLoS ONE 2013, 8, e60909. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.; Park, S.; Park, S.-H.; Lee, H.G.; Park, J.H. A Double-Edged Sword: The Two Faces of PARylation. Int. J. Mol. Sci. 2022, 23, 9826. [Google Scholar] [CrossRef] [PubMed]
- Groslambert, J.; Prokhorova, E.; Ahel, I. ADP-Ribosylation of DNA and RNA. DNA Repair 2021, 105, 103144. [Google Scholar] [CrossRef] [PubMed]
- Páhi, Z.G.; Borsos, B.N.; Pantazi, V.; Ujfaludi, Z.; Pankotai, T. PARylation During Transcription: Insights into the Fine-Tuning Mechanism and Regulation. Cancers 2020, 12, 183. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Tian, S.; Guo, Q.; Bao, K.; Yu, G.; Wang, X.; Shen, X.; Zhang, J.; Chen, J.; Yang, Y.; et al. A PARylation-Phosphorylation Cascade Promotes TOPBP1 Loading and RPA-RAD51 Exchange in Homologous Recombination. Mol. Cell. 2022, 82, 2571–2587.e9. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Joachimiak, G.; Bigelow, L.; Babnigg, G.; Joachimiak, A. How Aromatic Compounds Block DNA Binding of HcaR Catabolite Regulator. J. Biol. Chem. 2016, 291, 13243–13256. [Google Scholar] [CrossRef] [PubMed]
- Czernek, J.; Brus, J.; Czerneková, V.; Kobera, L. Quantifying the Intrinsic Strength of C–H⋯O Intermolecular Interactions. Molecules 2023, 28, 4478. [Google Scholar] [CrossRef] [PubMed]
- Biswas, R.; Tse, Y.-L.S.; Tokmakoff, A.; Voth, G.A. Role of Presolvation and Anharmonicity in Aqueous Phase Hydrated Proton Solvation and Transport. J. Phys. Chem. B 2016, 120, 1793–1804. [Google Scholar] [CrossRef] [PubMed]
- Dutta, A.; Lazaridis, T. Classical Models of Hydroxide for Proton Hopping Simulations. J. Phys. Chem. B 2024, 128, 12161–12170. [Google Scholar] [CrossRef] [PubMed]
- Kier, L.B. Proton Hopping in Living Systems. Curr. Comput. Aided Drug Des. 2021, 17, 333–336. [Google Scholar] [CrossRef] [PubMed]
- Farrell, K.M.; Zanni, M.T. Observing Aqueous Proton Transfer Dynamics. ACS Cent. Sci. 2019, 5, 1114–1116. [Google Scholar] [CrossRef] [PubMed]
- Popov, I.; Zhu, Z.; Young-Gonzales, A.R.; Sacci, R.L.; Mamontov, E.; Gainaru, C.; Paddison, S.J.; Sokolov, A.P. Search for a Grotthuss Mechanism through the Observation of Proton Transfer. Commun. Chem. 2023, 6, 77. [Google Scholar] [CrossRef] [PubMed]
- Bogot, A.; Poline, M.; Ji, M.; Dochain, A.; Simonsson, A.; Rosén, S.; Zettergren, H.; Schmidt, H.T.; Thomas, R.D.; Strasser, D. The Mutual Neutralization of Hydronium and Hydroxide. Science 2024, 383, 285–289. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Matta, C.F.; Wallace, S.; Massa, L.; Bernal, I. A Unique Trapping by Crystal Forces of a Hydronium Cation Displaying a Transition State Structure. Comptes Rendus. Chim. 2015, 18, 511–515. [Google Scholar] [CrossRef]
- Yu, Q.; Bowman, J.M. Tracking Hydronium/Water Stretches in Magic H3O+(H2O)20 Clusters through High-Level Quantum VSCF/VCI Calculations. J. Phys. Chem. A 2020, 124, 1167–1175. [Google Scholar] [CrossRef] [PubMed]
- Onaka, S. Periodic Arrangements of Tetrahedra Having Appearances Similar to That of the Boerdijk–Coxeter Helix. Sci. Rep. 2024, 14, 18260. [Google Scholar] [CrossRef] [PubMed]
- Limbach, H.-H. NMR Studies of Elementary Steps of Multiple Proton and Deuteron Transfers in Liquids, Crystals, and Organic Glasses. In Intermolecular Forces; Springer: Berlin/Heidelberg, Germany, 1991; pp. 281–295. [Google Scholar] [CrossRef]
- Hassanali, A.; Giberti, F.; Cuny, J.; Kühne, T.D.; Parrinello, M. Proton Transfer through the Water Gossamer. Proc. Natl. Acad. Sci. USA 2013, 110, 13723–13728. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Fang, H.; Ni, K.; Feng, Y. Water Clusters and Density Fluctuations in Liquid Water Based on Extended Hierarchical Clustering Methods. Sci. Rep. 2022, 12, 8036. [Google Scholar] [CrossRef] [PubMed]
- Frutos-Puerto, S.; Colín, M.J.; Corchado, J.C.; Sánchez, M.L.; Martín, M.E.; Aguilar, M.A. Photophysical and Photochemical Properties of 3-Hydroxyflavone in Ethanol Solution: Implicit vs Explicit Solvent Models. J. Mol. Liq. 2023, 381, 121783. [Google Scholar] [CrossRef]
- Tyburski, R.; Liu, T.; Glover, S.D.; Hammarström, L. Proton-Coupled Electron Transfer Guidelines, Fair and Square. J. Am. Chem. Soc. 2021, 143, 560–576. [Google Scholar] [CrossRef] [PubMed]
- Dubey, V.; Kumar, N.; Daschakraborty, S. Importance of Solvents’ Translational–Rotational Coupling for Translational Jump of a Small Hydrophobic Solute in Supercooled Water. J. Phys. Chem. B 2018, 122, 7569–7583. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.-C.; Chiang, K.-Y.; Okuno, M.; Seki, T.; Ohto, T.; Yu, X.; Korepanov, V.; Hamaguchi, H.; Bonn, M.; Hunger, J.; et al. Vibrational Couplings and Energy Transfer Pathways of Water’s Bending Mode. Nat. Commun. 2020, 11, 5977. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Riera Aroche, R.; Sánchez Moreno, E.C.; Ortiz García, Y.M.; Machado Sulbarán, A.C.; Riera Leal, L.; Olivas Román, L.R.; Riera Leal, A. The Transcription Machinery and the Driving Force of the Transcriptional Molecular Condensate: The Role of Phosphates. Curr. Issues Mol. Biol. 2025, 47, 571. https://doi.org/10.3390/cimb47070571
Riera Aroche R, Sánchez Moreno EC, Ortiz García YM, Machado Sulbarán AC, Riera Leal L, Olivas Román LR, Riera Leal A. The Transcription Machinery and the Driving Force of the Transcriptional Molecular Condensate: The Role of Phosphates. Current Issues in Molecular Biology. 2025; 47(7):571. https://doi.org/10.3390/cimb47070571
Chicago/Turabian StyleRiera Aroche, Raúl, Esli C. Sánchez Moreno, Yveth M. Ortiz García, Andrea C. Machado Sulbarán, Lizbeth Riera Leal, Luis R. Olivas Román, and Annie Riera Leal. 2025. "The Transcription Machinery and the Driving Force of the Transcriptional Molecular Condensate: The Role of Phosphates" Current Issues in Molecular Biology 47, no. 7: 571. https://doi.org/10.3390/cimb47070571
APA StyleRiera Aroche, R., Sánchez Moreno, E. C., Ortiz García, Y. M., Machado Sulbarán, A. C., Riera Leal, L., Olivas Román, L. R., & Riera Leal, A. (2025). The Transcription Machinery and the Driving Force of the Transcriptional Molecular Condensate: The Role of Phosphates. Current Issues in Molecular Biology, 47(7), 571. https://doi.org/10.3390/cimb47070571






