Genome-Wide Identification and Characterization of TaCRY Gene Family and Its Expression in Seed Aging Process of Wheat
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of TaCRY Members
2.2. Physicochemical Properties Analysis of TaCRY Members
2.3. Phylogenetic Analysis of TaCRY Members
2.4. Gene Motif, Conserved Domain and Structure Analysis of TaCRY Members
2.5. Chromosomal Mapping and Collinearity Analysis of TaCRY Members
2.6. Cis-Acting Elements Analysis of TaCRY Members
2.7. Expression Analysis of TaCRY Members
2.8. RNA Extraction and RT-qPCR Analysis
3. Results
3.1. Identification and Characterization of TaCRY Members
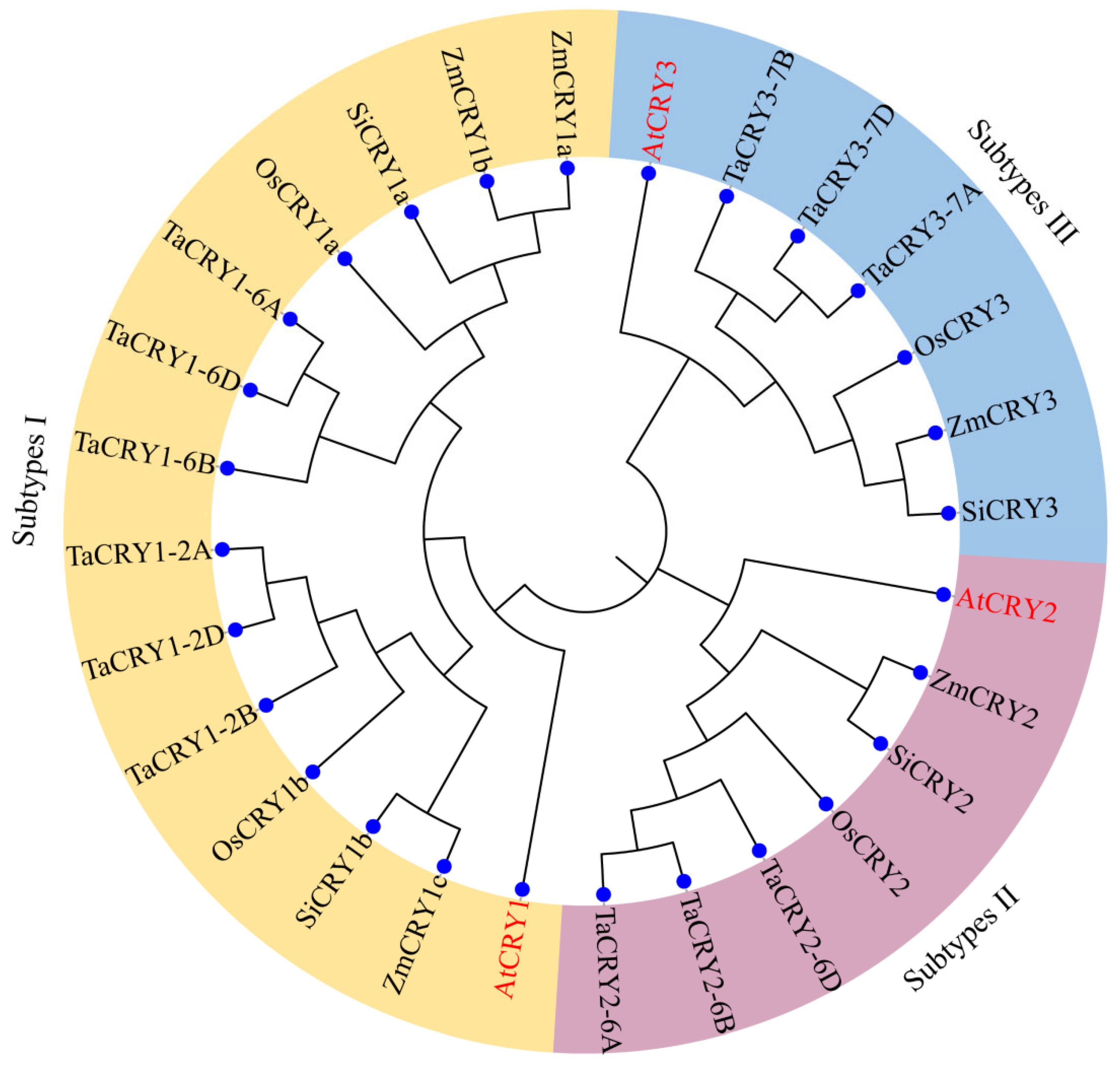
3.2. Phylogenetic Analyses and Classification of TaCRY Members
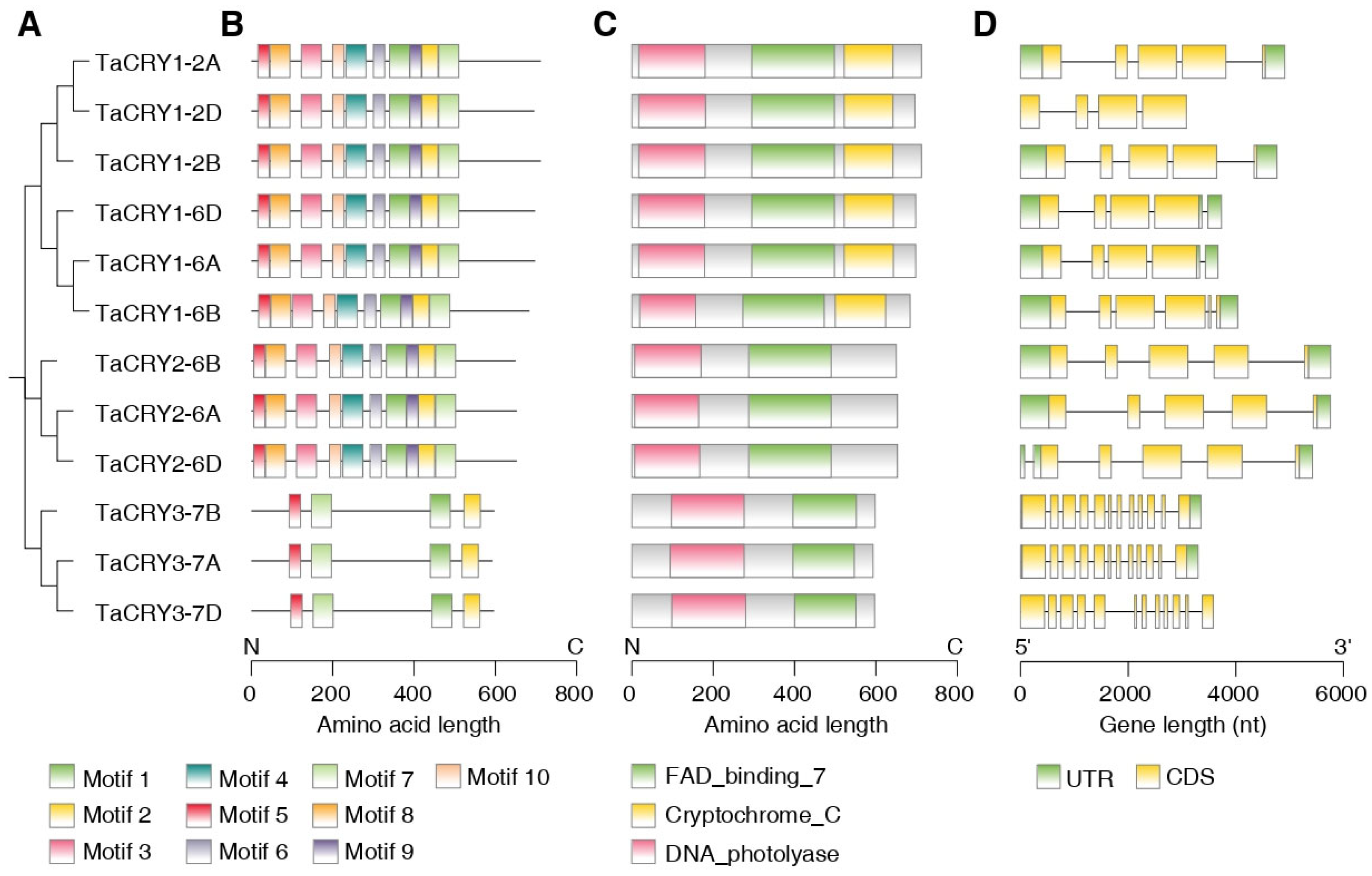
3.3. Conserved Motifs, Domains, and Gene Structure Analysis of TaCRY Members
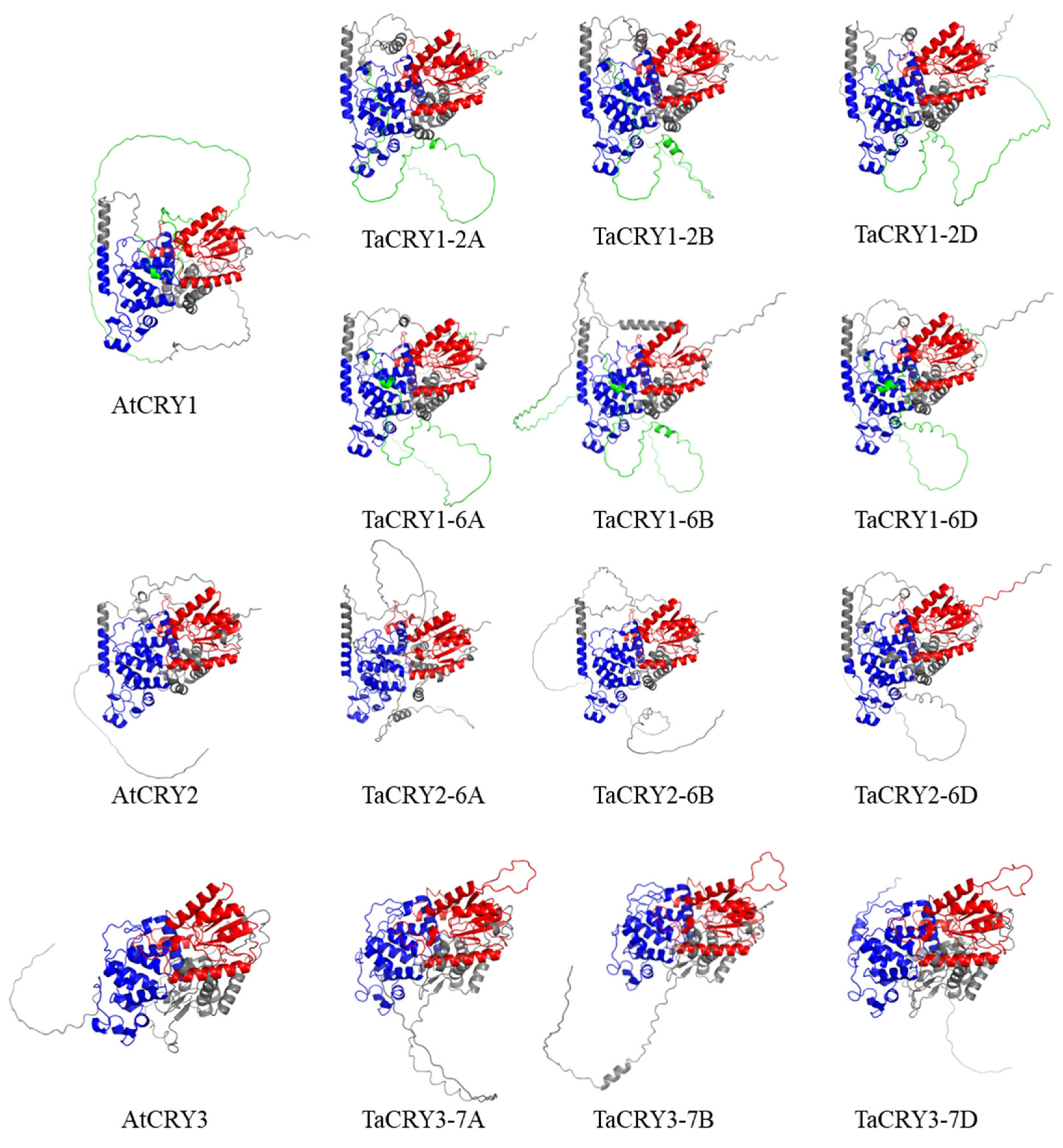
3.4. Tertiary Structure Models of TaCRY Members
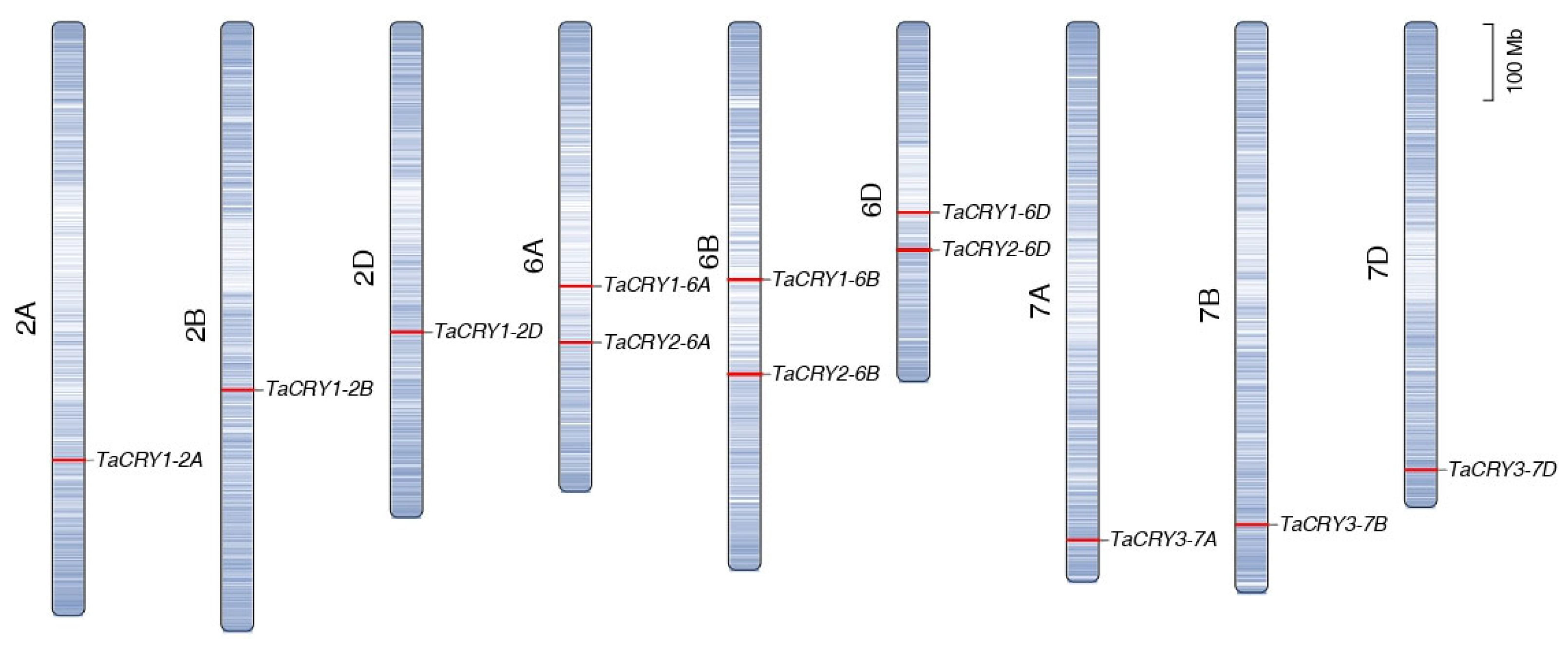
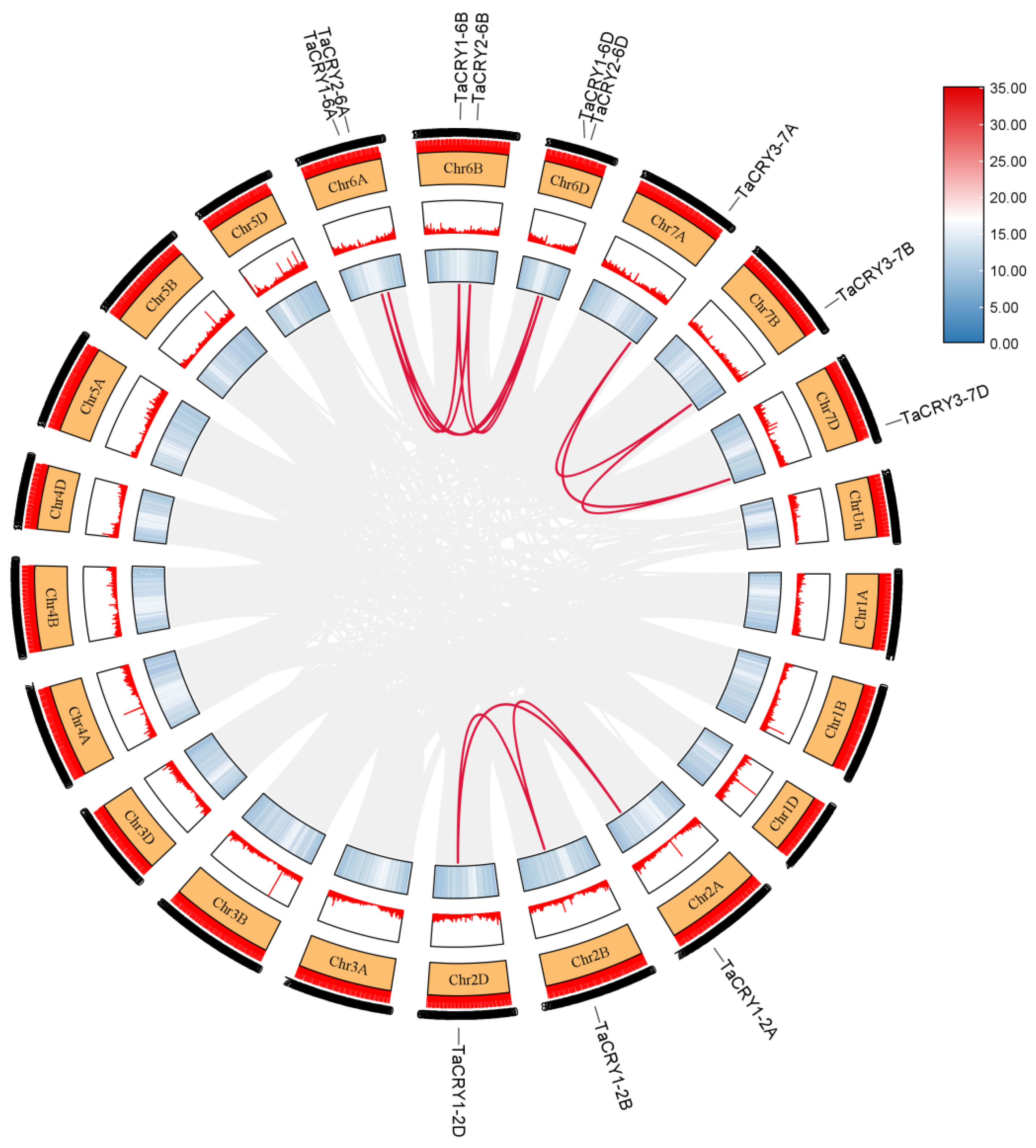
3.5. Chromosomal Distribution and Collinearity Analysis of TaCRY Members
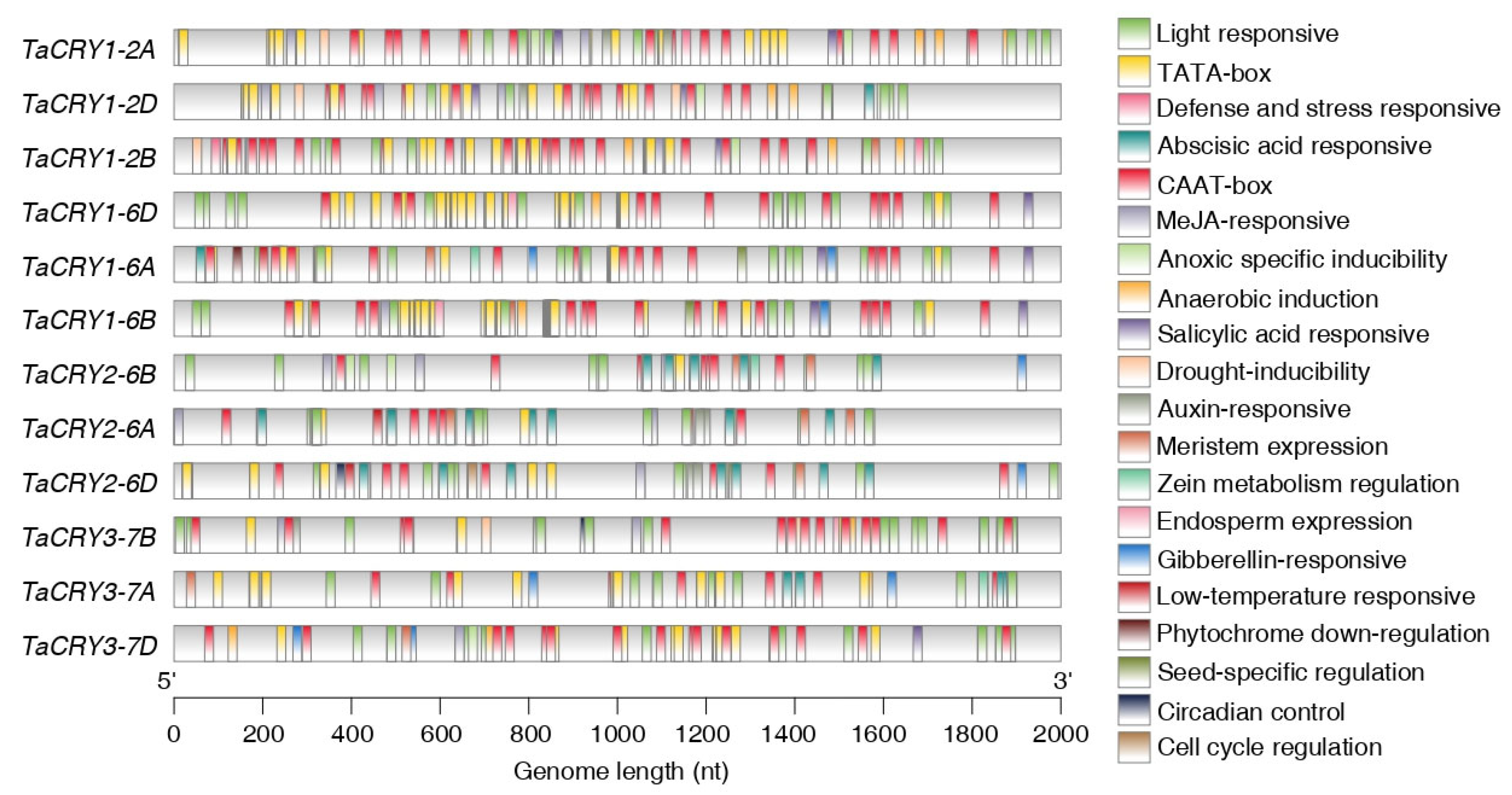
3.6. Genomic Landscape of Cis-Acting Elements in TaCRY Members
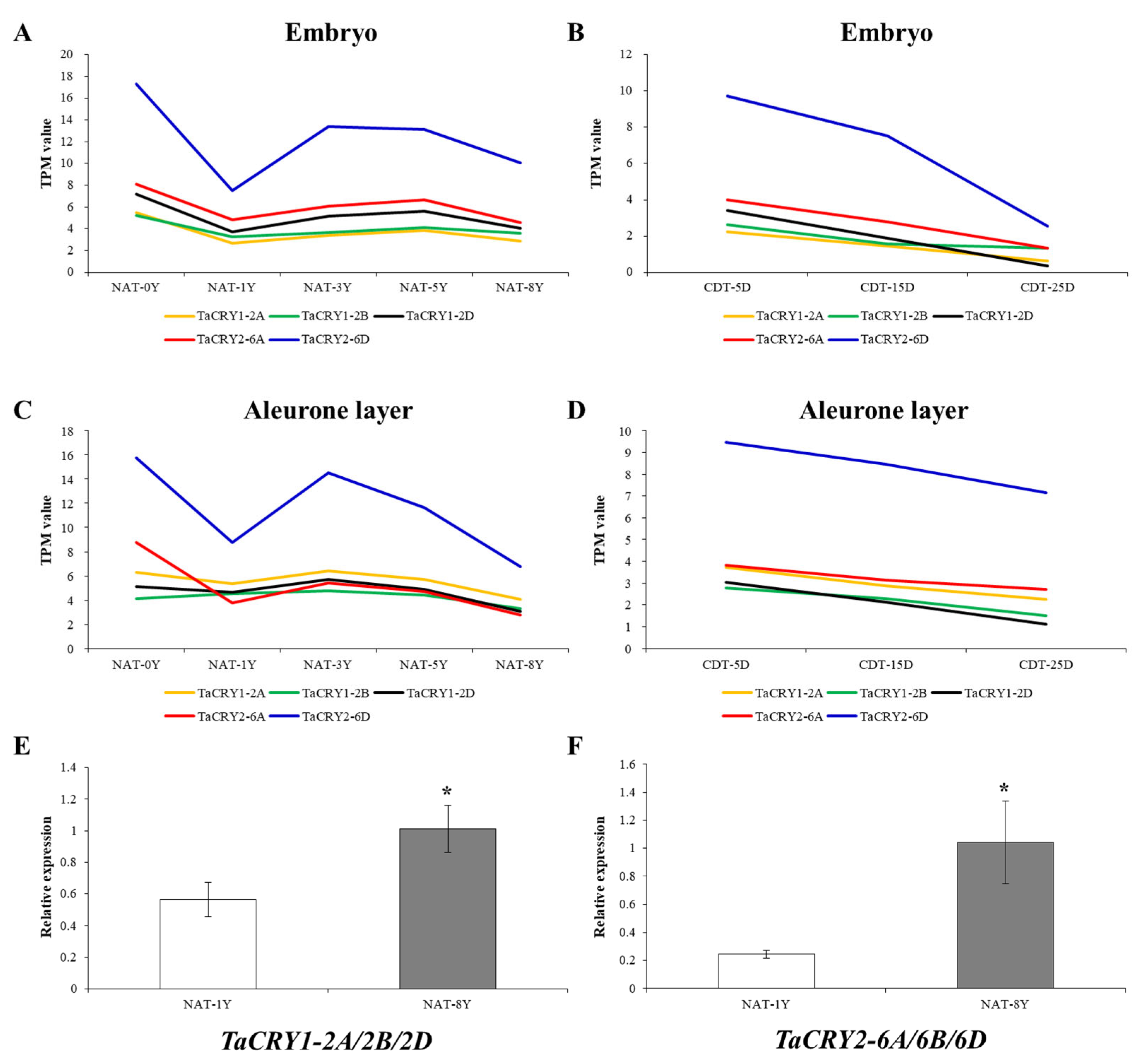
3.7. Expression Analysis of TaCRY Members Related to Seed Vigor and Candidate Gene Identification
4. Discussion
4.1. Evolutionary Expansion and Functional Diversification of the TaCRY Members
4.2. Structural Conservation and Subfamily-Specific Functional Modules
4.3. Functional Implications of the TaCRY Members for Wheat Seed Vigor
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, Q.; Lin, C.T. Mechanisms of cryptochrome-mediated photoresponses in plants. Annu. Rev. Plant Biol. 2020, 71, 103–129. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lin, C.T. The two action mechanisms of plant cryptochromes. Trends Plant Sci. 2025, 30, 775–791. [Google Scholar] [CrossRef]
- Ahmad, M.; Cashmore, A.R. HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 1993, 366, 162–166. [Google Scholar] [CrossRef] [PubMed]
- Sang, Y.; Li, Q.H.; Rubio, V.; Zhang, Y.C.; Mao, J.; Deng, X.W.; Yang, H.Q. N-terminal domain-mediated homodimerization is required for photoreceptor activity of Arabidopsis cryptochrome 1. Plant Cell 2005, 17, 1569–1584. [Google Scholar] [CrossRef]
- Guo, H.; Yang, H.; Mockler, T.C.; Lin, C.T. Regulation of flowering time by Arabidopsis photoreceptors. Science 1998, 279, 1360–1363. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.T.; Yu, X.H.; Li, K.W.; Klejnot, J.; Yang, H.Y.; Lisiero, D.; Lin, C.T. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 2008, 322, 1535–1539. [Google Scholar] [CrossRef]
- Huang, Y.H.; Baxter, R.; Smith, B.S.; Partch, C.L.; Colbert, C.L.; Deisenhofer, J. Crystal structure of cryptochrome 3 from Arabidopsis thaliana and its implications for photolyase activity. Proc. Natl. Acad. Sci. USA 2006, 103, 17701–17706. [Google Scholar] [CrossRef]
- Selby, C.P.; Sancar, A. A cryptochrome/photolyase class of enzymes with single-stranded DNA-specific photolyase activity. Proc. Natl. Acad. Sci. USA 2006, 103, 17696–17700. [Google Scholar] [CrossRef]
- Ma, L.; Wang, X.; Guan, Z.Y.; Wang, L.X.; Wang, Y.D.; Zheng, L.; Gong, Z.; Shen, C.C.; Wang, J.; Zhang, D.L.; et al. Structural insights into BIC-mediated inactivation of Arabidopsis cryptochrome 2. Nat. Struct. Mol. Biol. 2020, 27, 472–479. [Google Scholar] [CrossRef]
- Shao, K.; Zhang, X.; Li, X.; Hao, Y.H.; Huang, X.W.; Ma, M.L.; Zhang, M.H.; Yu, F.; Liu, H.T.; Zhang, P. The oligomeric structures of plant cryptochromes. Nat. Struct. Mol. Biol. 2020, 27, 480–488. [Google Scholar] [CrossRef]
- Qu, G.P.; Jiang, B.C.; Lin, C.T. The dual-action mechanism of Arabidopsis cryptochromes. J. Integr. Plant Biol. 2024, 66, 883–896. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Lin, C.T. A structural view of plant CRY2 photoactivation and inactivation. Nat. Struct. Mol. Biol. 2020, 27, 401–403. [Google Scholar] [CrossRef]
- Zeng, D.S.; Lv, J.Q.; Li, X.; Liu, H.T. The Arabidopsis blue-light photoreceptor CRY2 is active in darkness to inhibit root growth. Cell 2025, 188, 60–76.e20. [Google Scholar] [CrossRef] [PubMed]
- Chaves, I.; Pokorny, R.; Byrdin, M.; Hoang, N.; Ritz, T.; Brettel, K.; Essen, L.O.; van der Horst, G.T.J.; Batschauer, A.; Ahmad, M. The cryptochromes: Blue light photoreceptors in plants and animals. Annu. Rev. Plant Biol. 2011, 62, 335–364. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Guan, Z.Y.; Wang, Q.; Yan, X.H.; Wang, J.; Wang, Z.Z.; Cao, J.B.; Zhang, D.L.; Gong, X.; Yin, P. Structural insights into the photoactivation of Arabidopsis CRY2. Nat. Plants 2020, 6, 1432–1438. [Google Scholar] [CrossRef]
- Hao, Y.H.; Zhang, X.; Liu, Y.Q.; Ma, M.L.; Huang, X.W.; Liu, H.T.; Zhang, P. Cryo–EM structure of the CRY2 and CIB1 fragment complex provides insights into CIB1-mediated photosignaling. Plant Commun. 2023, 4, 100475. [Google Scholar] [CrossRef]
- Wang, Q.; Zuo, Z.C.; Wang, X.; Gu, L.F.; Yoshizumi, T.; Yang, Z.H.; Yang, L.; Liu, Q.; Liu, W.; Han, Y.J.; et al. Photoactivation and inactivation of Arabidopsis cryptochrome 2. Science 2016, 354, 343–347. [Google Scholar] [CrossRef]
- Zhao, Z.W.; Dent, C.; Liang, H.F.; Lv, J.Q.; Shang, G.D.; Liu, Y.W.; Feng, F.; Wang, F.; Pang, J.H.; Li, X.; et al. CRY2 interacts with CIS1 to regulate thermosensory flowering via FLM alternative splicing. Nat. Commun. 2022, 13, 7045. [Google Scholar] [CrossRef]
- Ponnu, J.; Riedel, T.; Penner, E.; Schrader, A.; Hoecker, U. Cryptochrome 2 competes with COP1 substrates to repress COP1 ubiquitin ligase activity during Arabidopsis photomorphogenesis. Proc. Natl. Acad. Sci. USA 2019, 116, 27133–27141. [Google Scholar] [CrossRef]
- Pedmale, U.V.; Huang, S.C.; Zander, M.; Cole, B.J.; Hetzel, J.; Ljung, K.; Reis, P.A.B.; Sridevi, P.; Nito, K.; Nery, J.R.; et al. Cryptochromes interact directly with PIFs to control plant growth in limiting blue light. Cell 2016, 164, 233–245. [Google Scholar] [CrossRef]
- Ma, D.B.; Li, X.; Guo, Y.X.; Chu, J.F.; Fang, S.; Yan, C.Y.; Noel, J.P.; Liu, H.T. Cryptochrome 1 interacts with PIF4 to regulate high temperature-mediated hypocotyl elongation in response to blue light. Proc. Natl. Acad. Sci. USA 2016, 113, 224–229. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Wang, Q.; Deng, W.X.; Wang, X.; Piao, M.X.; Cai, D.W.; Li, Y.X.; Barshop, W.D.; Yu, X.L.; Zhou, T.T.; et al. Molecular basis for blue light-dependent phosphorylation of Arabidopsis cryptochrome 2. Nat. Commun. 2017, 8, 15234. [Google Scholar] [CrossRef] [PubMed]
- Sajeev, N.; Koornneef, M.; Bentsink, L. A commitment for life: Decades of unraveling the molecular mechanisms behind seed dormancy and germination. Plant Cell 2024, 36, 1358–1376. [Google Scholar] [CrossRef]
- Yang, L.W.; Liu, S.R.; Lin, R.C. The role of light in regulating seed dormancy and germination. J. Integr. Plant Biol. 2020, 62, 1310–1326. [Google Scholar] [CrossRef]
- Tognacca, R.S.; Botto, J.F. Post-transcriptional regulation of seed dormancy and germination: Current understanding and future directions. Plant Commun. 2021, 2, 100169. [Google Scholar] [CrossRef] [PubMed]
- Oh, E.; Kim, J.; Park, E.; Kim, J.I.; Kang, C.; Choi, G. PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell 2004, 16, 3045–3058. [Google Scholar] [CrossRef]
- Arana, M.V.; Sanchez-Lamas, M.; Strasser, B.; Ibarra, S.E.; Cerdan, P.D.; Botto, J.F.; Sanchez, R.A. Functional diversity of phytochrome family in the control of light and gibberellin-mediated germination in Arabidopsis. Plant Cell Environ. 2014, 37, 2014–2023. [Google Scholar] [CrossRef]
- Oh, E.; Yamaguchi, S.; Kamiya, Y.; Bae, G.; Chung, W.I.; Choi, G. Light activates the degradation of PIL5 protein to promote seed germination through gibberellin in Arabidopsis. Plant J. 2006, 47, 124–139. [Google Scholar] [CrossRef]
- Ali, F.; Qanmber, G.; Li, F.G.; Wang, Z. Updated role of ABA in seed maturation, dormancy, and germination. J. Adv. Res. 2021, 35, 199–214. [Google Scholar] [CrossRef]
- Gubler, F.; Hughes, T.; Waterhouse, P.; Jacobsen, J. Regulation of dormancy in barley by blue light and afterripening: Effects on abscisic acid and gibberellin metabolism. Plant Physiol. 2008, 147, 886–896. [Google Scholar] [CrossRef]
- Barrero, J.M.; Jacobsen, J.V.; Talbot, M.J.; White, R.G.; Swain, S.M.; Garvin, D.F.; Gubler, F. Grain dormancy and light quality effects on germination in the model grass Brachypodium distachyon. New Phytol. 2012, 193, 376–386. [Google Scholar] [CrossRef] [PubMed]
- Barrero, J.M.; Downie, A.B.; Xu, Q.; Gubler, F. A role for barley CRYPTOCHROME1 in light regulation of grain dormancy and germination. Plant Cell 2014, 26, 1094–1104. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, 200–204. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bo, Y.; Han, L.Y.; He, J.E.; Lanczycki, C.J.; Lu, S.N.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45, 200–203. [Google Scholar] [CrossRef]
- Lu, S.N.; Wang, J.Y.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, 265–268. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.Y.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; Gwadz, M.; Lu, S.N.; Marchler, G.H.; Song, J.S.; Thanki, N.; Yamashita, R.A.; et al. The conserved domain database in 2023. Nucleic Acids Res. 2023, 51, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.M.; Song, W.J.; Xie, X.M.; Wang, Z.H.; Guan, P.F.; Peng, H.R.; Jiao, Y.N.; Ni, Z.F.; Sun, Q.X.; Guo, W.L. A collinearity-incorporating homology inference strategy for connecting emerging assemblies in the triticeae tribe as a pilot practice in the plant pangenomic era. Mol. Plant 2020, 13, 1694–1708. [Google Scholar] [CrossRef]
- Savojardo, C.; Martelli, P.L.; Fariselli, P.; Profiti, G.; Casadio, R. BUSCA: An integrative web server to predict subcellular localization of proteins. Nucleic Acids Res. 2018, 46, 459–466. [Google Scholar] [CrossRef]
- Chou, K.C.; Shen, H. Cell-PLoc: A package of Web servers for predicting subcellular localization of proteins in various organisms. Nat. Protoc. 2008, 3, 153–162. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, 78–82. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, 39–49. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Liang, W.Z.; Dong, H.X.; Guo, X.J.; Rodríguez, V.; Cheng, M.P.; Li, M.L.; Benech-Arnold, R.; Pu, Z.E.; Wang, J.R. Identification of long-lived and stable mRNAs in the aged seeds of wheat. Seed Biol. 2023, 2, 14. [Google Scholar] [CrossRef]
- Heberle, H.; Meirelles, G.V.; da Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinform. 2015, 16, 169. [Google Scholar] [CrossRef]
- Chen, L.T.; Sun, A.Q.; Yang, M.; Chen, L.L.; Ma, X.L.; Li, M.L.; Yin, Y.P. Seed vigor evaluation based on adversity resistance index of wheat seed germination under stress conditions. Chin. J. Appl. Ecol. 2016, 27, 2968–2974. [Google Scholar]
- Zeng, Z.K.; Guo, C.; Yan, X.F.; Song, J.Q.; Wang, C.P.; Xu, X.T.; Hao, Y.F. QTL mapping and KASP marker development for seed vigor related traits in common wheat. Front. Plant Sci. 2022, 13, 994973. [Google Scholar] [CrossRef]
- Hofmann, N. Cryptochromes and seed dormancy: The molecular mechanism of blue light inhibition of grain germination. Plant Cell 2014, 26, 846. [Google Scholar] [CrossRef]
- Liao, J.K.; Deng, B.; Yang, Q.X.; Li, Y.; Zhang, Y.X.; Cong, J.J.; Wang, X.Q.; Kohnen, M.V.; Liu, Z.J.; Lu, M.Z.; et al. Insights into cryptochrome modulation of ABA signaling to mediate dormancy regulation in Marchantia polymorpha. New Phytol. 2023, 238, 1479–1497. [Google Scholar] [CrossRef]
- Stawska, M.; Oracz, K. PhyB and HY5 are involved in the blue light-mediated alleviation of dormancy of Arabidopsis seeds possibly via the modulation of expression of genes related to light, GA, and ABA. Int. J. Mol. Sci. 2019, 20, 5882. [Google Scholar] [CrossRef] [PubMed]
- Richter, A.S.; Nägele, T.; Grimm, B.; Kaufmann, K.; Schroda, M.; Leister, D.; Kleine, T. Retrograde signaling in plants: A critical review focusing on the GUN pathway and beyond. Plant Commun. 2023, 4, 100511. [Google Scholar] [CrossRef] [PubMed]
- Cackett, L.; Luginbuehl, L.H.; Schreier, T.B.; Lopez-Juez, E.; Hibberd, J.M. Chloroplast development in green plant tissues: The interplay between light, hormone, and transcriptional regulation. New Phytol. 2022, 233, 2000–2016. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.K.; Sun, X.Y.; Peng, J.; Li, F.G.; Ali, F.; Wang, Z. Regulation of seed germination: ROS, epigenetic, and hormonal aspects. J. Adv. Res. 2025, 71, 107–125. [Google Scholar] [CrossRef]
- Castro, B.; Citterico, M.; Kimura, S.; Stevens, D.M.; Wrzaczek, M.; Coaker, G. Stress-induced reactive oxygen species compartmentalization, perception and signalling. Nat. Plants 2021, 7, 403–412. [Google Scholar] [CrossRef]
- Kachroo, P.; Burch-Smith, T.M.; Grant, M. An emerging role for chloroplasts in disease and defense. Annu. Rev. Phytopathol. 2021, 59, 423–445. [Google Scholar] [CrossRef]
- Pirredda, M.; Fañanás-Pueyo, I.; Oñate-Sánchez, L.; Mira, S. Seed longevity and aging: A review on physiological and genetic factors with an emphasis on hormonal regulation. Plants 2023, 13, 41. [Google Scholar] [CrossRef]
| Gene Name | AA (aa) | MW (Da) | pI | Instability Index | Aliphatic Index | GRAVY | Subcellular Localization |
|---|---|---|---|---|---|---|---|
| TaCRY1-2A | 712 | 80,626.7 | 5.54 | 53.09 | 76.03 | −0.428 | nucleus |
| TaCRY1-2B | 712 | 80,497.49 | 5.41 | 54.06 | 76.43 | −0.41 | nucleus |
| TaCRY1-2D | 696 | 78,841.74 | 5.58 | 52.95 | 77.08 | −0.412 | nucleus |
| TaCRY1-6A | 698 | 79,033.49 | 5.34 | 51.53 | 73.8 | −0.475 | nucleus |
| TaCRY2-6A | 653 | 73,450.14 | 5.31 | 43.87 | 85.25 | −0.328 | nucleus |
| TaCRY1-6B | 684 | 77,644.21 | 5.55 | 51.61 | 74.61 | −0.465 | nucleus |
| TaCRY2-6B | 650 | 73,258.94 | 5.62 | 41.17 | 85.49 | −0.363 | nucleus |
| TaCRY1-6D | 698 | 79,049.55 | 5.52 | 52.02 | 74.63 | −0.491 | nucleus |
| TaCRY2-6D | 653 | 73,643.45 | 5.44 | 44.96 | 83.91 | −0.351 | nucleus |
| TaCRY3-7A | 593 | 65,929.05 | 9.44 | 44.01 | 77.69 | −0.389 | chloroplast |
| TaCRY3-7B | 598 | 66,333.77 | 9.56 | 43.98 | 79.65 | −0.346 | chloroplast |
| TaCRY3-7D | 597 | 65,522.85 | 9.62 | 41.73 | 76.68 | −0.376 | chloroplast |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, G.; Cui, X.; Wang, J.; Lei, M.; Liu, X.; Wang, Y.; Wang, H.; Liu, L.; Mu, Z.; Xin, X. Genome-Wide Identification and Characterization of TaCRY Gene Family and Its Expression in Seed Aging Process of Wheat. Curr. Issues Mol. Biol. 2025, 47, 522. https://doi.org/10.3390/cimb47070522
Cui G, Cui X, Wang J, Lei M, Liu X, Wang Y, Wang H, Liu L, Mu Z, Xin X. Genome-Wide Identification and Characterization of TaCRY Gene Family and Its Expression in Seed Aging Process of Wheat. Current Issues in Molecular Biology. 2025; 47(7):522. https://doi.org/10.3390/cimb47070522
Chicago/Turabian StyleCui, Guoqing, Xiuyan Cui, Junjie Wang, Menglin Lei, Xia Liu, Yanzhen Wang, Haigang Wang, Longlong Liu, Zhixin Mu, and Xia Xin. 2025. "Genome-Wide Identification and Characterization of TaCRY Gene Family and Its Expression in Seed Aging Process of Wheat" Current Issues in Molecular Biology 47, no. 7: 522. https://doi.org/10.3390/cimb47070522
APA StyleCui, G., Cui, X., Wang, J., Lei, M., Liu, X., Wang, Y., Wang, H., Liu, L., Mu, Z., & Xin, X. (2025). Genome-Wide Identification and Characterization of TaCRY Gene Family and Its Expression in Seed Aging Process of Wheat. Current Issues in Molecular Biology, 47(7), 522. https://doi.org/10.3390/cimb47070522






