HLA-DRB1 and DQB1 Allelic Polymorphism and Multiple Sclerosis in a Moroccan Population
Abstract
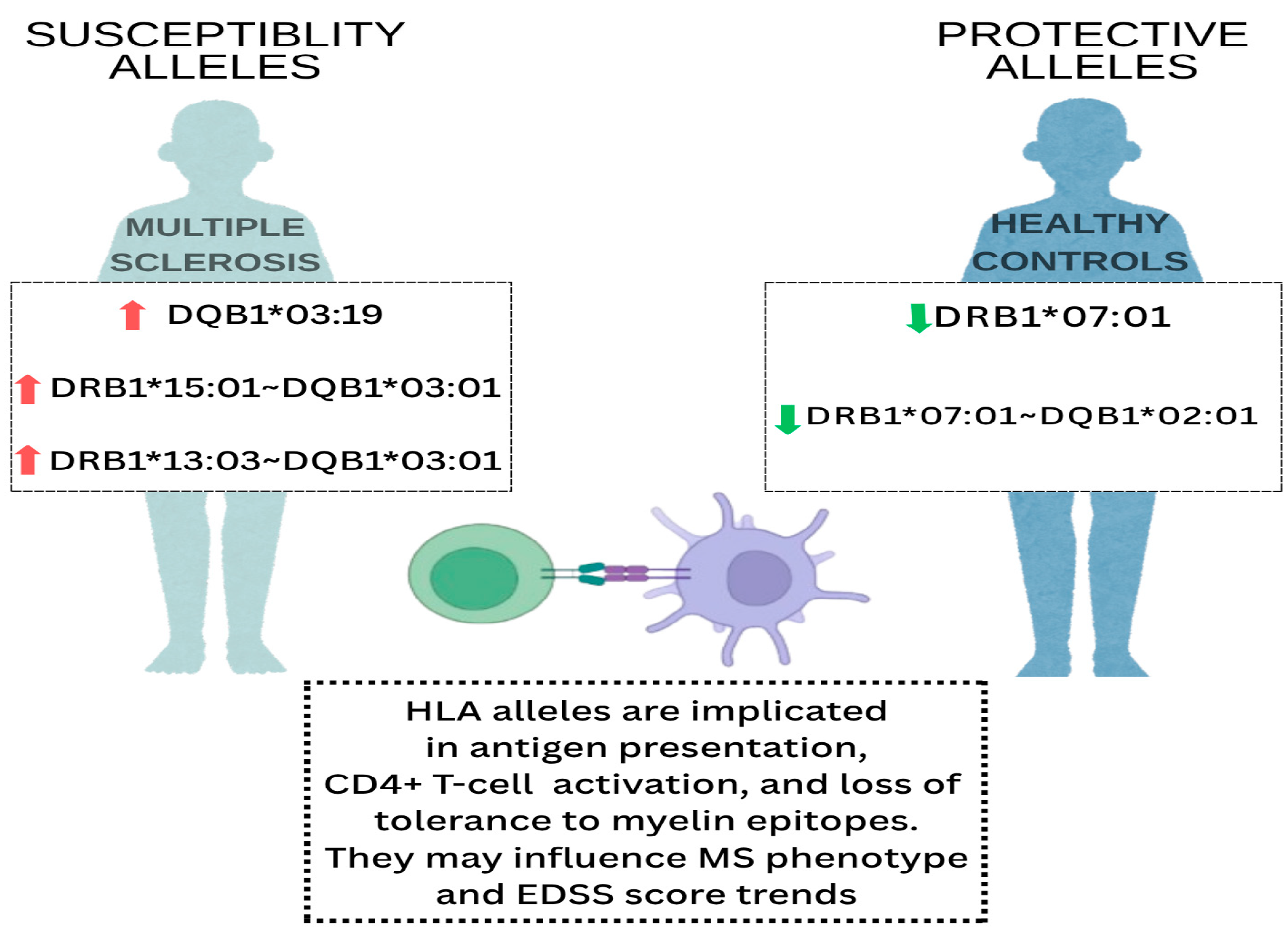
1. Introduction
2. Materials and Methods
2.1. Study Population Selection
2.2. Sample Collection
2.3. DNA Extraction and HLA Typing
2.4. Statistical Analysis
2.5. Ethical Consideration
3. Results
3.1. Descriptive Analysis of Patients with MS
3.2. Distribution of HLA-DRB1 Alleles in Patients with MS and Controls
3.3. Distribution of HLA-DQB1 Alleles in Patients with MS and Controls
3.4. Association of HLA DRB1 and DQB1 Alleles in Patients with MS and Controls
3.5. HLA-DRB1 and DQB1 Allele Polymorphisms and Clinical Forms of MS
3.5.1. Association of DRB1 and DQB1 Alleles with MS Subtypes
3.5.2. DRB1 and DQB1 Allele Polymorphisms and EDSS Score in MS
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MS | Multiple Sclerosis |
| HLA | Human Leukocyte Antigen |
| CNS | Central Nervous System |
| SSO | Sequence-Specific Oligonucleotide |
| MBP | Myelin Basic Protein |
| RRMS | Relapsing–Remitting Multiple Sclerosis |
| PPMS | Primary Progressive Multiple Sclerosis |
| SPMS | Secondary Progressive Multiple Sclerosis |
| EDTA | Ethylene Diamine Tetraacetic Acid |
| DNA | Deoxyribonucleic Acid |
| PCR | Polymerase Chain Reaction |
| PBMCs | Peripheral Blood Mononuclear Cells |
| ORs | Odds Ratios |
| CIs | Confidence Intervals |
| FDR | False Discovery Rate |
| EDSS | Expanded Disability Status Scale |
| MENA | Middle East and North Africa |
References
- Maghbooli, Z.; Sahraian, M.A.; Naser Moghadasi, A. Multiple sclerosis and human leukocyte antigen genotypes: Focus on the Middle East and North Africa region. Mult. Scler. J. Exp. Transl. Clin. 2020, 6, 2055217319881775. [Google Scholar] [CrossRef]
- Baranzini, S.E.; Oksenberg, J.R. The Genetics of Multiple Sclerosis: From 0 to 200 in 50 Years. Trends Genet. 2017, 33, 960–970. [Google Scholar] [CrossRef]
- Olsson, T.; Barcellos, L.F.; Alfredsson, L. Interactions between genetic, lifestyle and environmental risk factors for multiple sclerosis. Nat. Rev. Neurol. 2017, 13, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Sawcer, S.; Franklin, R.J.; Ban, M. Multiple sclerosis genetics. Lancet Neurol. 2014, 13, 700–709. [Google Scholar] [CrossRef] [PubMed]
- Alcina, A.; Abad-Grau Mdel, M.; Fedetz, M.; Izquierdo, G.; Lucas, M.; Fernández, O.; Ndagire, D.; Catalá-Rabasa, A.; Ruiz, A.; Gayán, J.; et al. Multiple sclerosis risk variant HLA-DRB1*1501 associates with high expression of DRB1 gene in different human populations. PLoS ONE. 2012, 7, e29819. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hollenbach, J.A.; Oksenberg, J.R. The immunogenetics of multiple sclerosis: A comprehensive review. J. Autoimmun. 2015, 64, 13–25. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- De Silvestri, A.; Capittini, C.; Mallucci, G.; Bergamaschi, R.; Rebuffi, C.; Pasi, A.; Martinetti, M.; Tinelli, C. The Involvement of HLA Class II Alleles in Multiple Sclerosis: A Systematic Review with Meta-analysis. Dis Markers. 2019, 2019, 1409069. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Aljumah, M.A.; Saadah, O.; Alharbi, T. HLA-DRB1 association with multiple sclerosis in Saudi Arabia. Neurosciences 2014, 19, 206–210. [Google Scholar] [PubMed]
- Messadi, A.; Najiba, F.M.; Ouerhani, S.; Zaweli, J.; Louatti, I.; Layouni, S.; Nciri, B.; Bouaicha, G.; Kouki, W.; Yedeas, M.; et al. HLA class II alleles and multiple sclerosis in Tunisian patients. Clin. Neurol. Neurosurg. 2010, 112, 849–852. [Google Scholar] [CrossRef]
- Fadhlaoui-Zid, K.; Martinez-Cruz, B.; Khodjet-el-khil, H.; Mendizabal, I.; Benammar-Elgaaied, A.; Comas, D. Genetic structure of Tunisian ethnic groups revealed by paternal lineages. Am. J. Phys. Anthropol. 2011, 146, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Lotfi, R.; El Kardoudi, A.; Chigr, F. Multiple sclerosis in Morocco: Epidemiological, clinical, and therapeutic profile. Mult. Scler. Relat. Disord. 2024, 81, 105347. [Google Scholar] [CrossRef] [PubMed]
- Fguirouche, A.; Ouahmani, F.; Brahim, I.; Hazime, R.; Louhab, N.; Kissani, N.; Chraa, M.; Admou, B. Distribution of Major HLA-A, -B, -DR, and -DQ Loci Potentially Associated with Multiple Sclerosis in a Healthy Population from Southern Morocco. Clin. Pract. 2025, 15, 10. [Google Scholar] [CrossRef] [PubMed]
- Bove, R.; Chitnis, T. The role of gender and sex hormones in determining the onset and outcome of multiple sclerosis. Mult. Scler. 2014, 20, 520–526. [Google Scholar] [CrossRef] [PubMed]
- Baroncini, D.; Annovazzi, P.O.; De Rossi, N.; Mallucci, G.; Torri Clerici, V.; Tonietti, S.; Mantero, V.; Ferrò, M.T.; Messina, M.J.; Barcella, V.; et al. Impact of natural menopause on multiple sclerosis: A multicentre study. J. Neurol. Neurosurg. Psychiatry 2019, 90, 1201–1206. [Google Scholar] [CrossRef] [PubMed]
- Hensiek, A.E.; Sawcer, S.J.; Feakes, R.; Deans, J.; Mander, A.; Akesson, E.; Roxburgh, R.; Coraddu, F.; Smith, S.; Compston, D.A.S. HLA-DR15 is associated with female sec and younger age at diagnostic in multiple sclerosis. Lournal Neurol. Neurosurg. Psychiatry 2002, 72, 184–187. [Google Scholar] [CrossRef]
- Michelis, D.; Brunetti, N.; Solaro, C.; Mancardi, G.L.; Uccelli, A.; Inglese, M.; Laroni, A. Aging with multiple sclerosis: Clinical characterization of an elderly population, a cross-sectional study. Mult. Scler. Relat. Disord. 2023, 69, 104464. [Google Scholar] [CrossRef] [PubMed]
- Rojas, O.L.; Rojas-Villarraga, A.; Cruz-Tapias, P.; Sánchez, J.L.; Suárez-Escudero, J.C.; Patarroyo, M.A.; Anaya, J.M. HLA class II polymorphism in Latin American patients with multiple sclerosis. Autoimmun. Rev. 2010, 9, 407–413. [Google Scholar] [CrossRef] [PubMed]
- Qiu, W.; James, I.; Carroll, W.M.; Mastaglia, F.L.; Kermode, A.G. HLA-DR allele polymorphism and multiple sclerosis in Chinese populations: A meta-analysis. Mult Scler. 2011, 17, 382–388. [Google Scholar] [CrossRef] [PubMed]
- Ouadghiri, S.; El Alaoui Toussi, K.; Brick, C.; Ait Benhaddou, E.H.; Benseffaj, N.; Benomar, A.; El Yahyaoui, M.; Essakalli, M. Genetic factors and multiple sclerosis in the Moroccan population: A role for HLA class II. Pathol. Biol. 2013, 61, 259–263. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Lin, C.Y.; Dong, Q.; Wang, J.; Wang, W. Relationship between HLA-DRB1 polymorphism and susceptibility or resistance to multiple sclerosis in Caucasians: A meta-analysis of non-family-based studies. Autoimmun. Rev. 2011, 10, 474–481. [Google Scholar] [CrossRef] [PubMed]
- Hedström, A.K.; Hillert, J.; Brenner, N.; Butt, J.; Waterboer, T.; Strid, P.; Kockum, I.; Olsson, T.; Alfredsson, L. DRB1-environment interactions in multiple sclerosis etiology: Results from two Swedish case-control studies. J. Neurol. Neurosurg. Psychiatry 2021, 92, 717–722. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tsai, S.; Santamaria, P. MHC Class II Polymorphisms, Autoreactive T-Cells, and Autoimmunity. Front Immunol. 2013, 4, 321. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Schmidt, H.; Williamson, D.; Ashley-Koch, A. HLA-DR15 haplotype and multiple sclerosis: A HuGE review. Am. J. Epidemiol. 2007, 165, 1097–1109. [Google Scholar]
- Stürner, K.H.; Siembab, I.; Schön, G.; Stellmann, J.P.; Heidari, N.; Fehse, B.; Heesen, C.; Eiermann, T.H.; Martin, R.; Binder, T.M. Is multiple sclerosis progression associated with the HLA-DR15 haplotype? Mult. Scler. J. Exp. Transl. Clin. 2019, 5, 2055217319894615. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sønderstrup, G.; McDevitt, H.O. DR, DQ, and you: MHC alleles and autoimmunity. J. Clin. Invest. 2001, 107, 795–796. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
| Demographic Characteristics | MS Patients | Controls | p-Value |
|---|---|---|---|
| Mean age (years) | 40 ± 1.67 | 32 ± 1.88 | 0.012 |
| Male (%) | 35% | 50% | 0.108 |
| Female (%) | 65% | 50% | 0.108 |
| Clinical subtypes | Nb of patients (n) | Percentage (%) | |
| Relapsing–Remitting MS (RRMS) | 19 | 47.5% | |
| Secondary Progressive MS (SPMS) | 12 | 30.0% | |
| Primary Progressive MS (PPMS) | 9 | 22.5% |
| HLA DRB1 Alleles | Frequency in Patients, n = 40 (%) | Frequency in Controls, n = 100 (%) | Odds Ratio | 95% CI | p-Value | c-p* |
|---|---|---|---|---|---|---|
| DRB1*15:01 | 8.8 | 4.08 | 2.18 | 0.79–6.04 | 0.12 | 0.36 |
| DRB1*15:02 | 5 | 2 | 2.58 | 0.35–18.97 | 0.32 | 1 |
| DRB1*03:01 | 9.2 | 8.43 | 1.15 | 0.61–2.14 | 0.67 | 0.89 |
| DRB1*13:01 | 4 | 29 | 0.13 | 0.03–0.57 | 0.03 | 0.25 |
| DRB1*13:03 | 9 | 14.5 | 0.32 | 0.07–1.49 | 0.15 | 1 |
| DRB1*07:01 | 10 | 17 | 0.41 | 0.14–1.38 | 0.004 | 0.027 |
| DRB1*04:01 | 7.0 | 5.0 | 0.69 | 0.22–2.14 | 0.45 | 1 |
| DRB1*11:01 | 1 | 17 | 0.12 | 0.03–1.00 | 0.03 | 0.25 |
| DRB1*01:01 | 1 | 9 | 0.0 | 0.01–2.09 | 0.05 | 0.34 |
| HLA DQB1 Alleles | Frequency in Patients, n = 40 (%) | Frequency in Controls, n = 100 (%) | Odds Ratio | 95% CI | p-Value | c-p* |
|---|---|---|---|---|---|---|
| DQB1*06:02 | 10 | 6 | 1.74 | 0.47–6.41 | 0.10 | 0.30 |
| DQB1*02:01 | 40 | 16 | 0.48 | 0.23–1.02 | 0.06 | 0.39 |
| DQB1*03:19 | 3.7 | 0 | ∞ | 0.22–∞ | 0.0004 | 0.0011 |
| DQB1*03:01 | 12 | 30 | 0.42 | 0.20–0.95 | 0.03 | 0.11 |
| DQB1*05:01 | 4.0 | 3 | 1.33 | 0.36–4.90 | 0.72 | 0.69 |
| DQB1*04:01 | 2.0 | 1.5 | 1.35 | 0.31–5.88 | 0.49 | 0.56 |
| HLA DR–DQ Associations | Frequency in Patients (%) | Frequency in Controls (%) | Odds Ratio | 95% CI | p-Value | c-p* |
|---|---|---|---|---|---|---|
| DRB1*15:01–DQB1*06:02 | 9 | 5 | 1.8 | 1.5–5.4 | 0.004 | 0.009 |
| DRB1*13:03–DQB1*03:01 | 6.2 | 0.0 | ∞ | 1.42–∞ | 0.0004 | 0.0011 |
| DRB1*07:01–DQB1*02:01 | 1.3 | 13 | 0.10 | 0.02–0.42 | 0.001 | 0.044 |
| DRB1*03:01–DQB1*02:01 | 10.4 | 3 | 0.11 | 0.04–0.85 | 0.01 | 0.49 |
| DRB1*04:01–DQB1*03:01 | 5.0 | 7.0 | 0.69 | 0.22–2.14 | 0.45 | 1 |
| DRB1*01:01–DQB1*05:01 | 4.0 | 3.05 | 1.33 | 0.36–4.90 | 0.69 | 1 |
| DR and DQ Alleles | PPMS % | RRMS % | SPMS % | p-Value * |
|---|---|---|---|---|
| DRB1*15:01 | 3 | 11.1 | 8.3 | 0.50 |
| DRB1*03:01 | 31 | 33.6 | 25 | 0.66 |
| DRB1*13:01 | 0 | 10.5 | 8.3 | 0.66 |
| DRB1*13:03 | 0 | 6 | 0 | 0.003 |
| DRB1*07:01 | 10 | 10.5 | 8.3 | 0.83 |
| DRB1*04:01 | 0 | 5.3 | 16.7 | 0.48 |
| DRB1*11:01 | 11 | 0 | 0 | 0.02 |
| DQB1*02:01 | 2 | 42.1 | 41.7 | 0.001 |
| DQB1*06:02 | 1 | 10.5 | 25 | 0.71 |
| DQB1*03:19 | 0 | 5 | 2.5 | 0.15 |
| DQB1*03:01 | 10.5 | 44.5 | 8.3 | 0.009 |
| DQB1*05:01 | 0 | 5.3 | 8.3 | 0.76 |
| DQB1*04:01 | 0 | 8.3 | 0 | 0.06 |
| DR and DQ Alleles | Mean EDSS Score | Trend | p-Value * |
|---|---|---|---|
| DRB1*15:01 | 5 | Higher disability | 0.548 |
| DRB1*03:01 | 3.4 | Moderate disability | 0.548 |
| DRB1*13:01 | 5.8 | Higher disability | 0.548 |
| DRB1*13:03 | 4 | Moderate disability | 0.548 |
| DRB1*07:01 | 4.4 | Moderate disability | 0.548 |
| DRB1*04:01 | 2.5 | Low disability | 0.548 |
| DRB1*11:01 | 2 | Moderate disability | 0.548 |
| DQB1*02:01 | 3.5 | Moderate disability | 0.763 |
| DQB1*06:02 | 6 | Higher disability | 0.763 |
| DQB1*03:19 | 4.3 | Moderate disability | 0.763 |
| DQB1*03:01 | 4 | Moderate disability | 0.763 |
| DQB1*05:01 | 3 | Moderate disability | 0.763 |
| DQB1*04:01 | 3 | Moderate disability | 0.763 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fguirouche, A.; Naji, Y.; Guennouni, M.; Hazime, R.; Zahlane, S.; Chraa, M.; Kissani, N.; Louhab, N.; Admou, B. HLA-DRB1 and DQB1 Allelic Polymorphism and Multiple Sclerosis in a Moroccan Population. Curr. Issues Mol. Biol. 2025, 47, 458. https://doi.org/10.3390/cimb47060458
Fguirouche A, Naji Y, Guennouni M, Hazime R, Zahlane S, Chraa M, Kissani N, Louhab N, Admou B. HLA-DRB1 and DQB1 Allelic Polymorphism and Multiple Sclerosis in a Moroccan Population. Current Issues in Molecular Biology. 2025; 47(6):458. https://doi.org/10.3390/cimb47060458
Chicago/Turabian StyleFguirouche, Abir, Yahya Naji, Morad Guennouni, Raja Hazime, Safa Zahlane, Mohamed Chraa, Najib Kissani, Nissrine Louhab, and Brahim Admou. 2025. "HLA-DRB1 and DQB1 Allelic Polymorphism and Multiple Sclerosis in a Moroccan Population" Current Issues in Molecular Biology 47, no. 6: 458. https://doi.org/10.3390/cimb47060458
APA StyleFguirouche, A., Naji, Y., Guennouni, M., Hazime, R., Zahlane, S., Chraa, M., Kissani, N., Louhab, N., & Admou, B. (2025). HLA-DRB1 and DQB1 Allelic Polymorphism and Multiple Sclerosis in a Moroccan Population. Current Issues in Molecular Biology, 47(6), 458. https://doi.org/10.3390/cimb47060458







