Lipid-Lowering Potential of Almond Hulls (Quercetin, Baicalein, and Kaempferol): Insights from Network Pharmacology and Molecular Dynamics
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Extraction of Bioactive Compounds from Almond Hulls
2.3. Network Pharmacology, Molecular Docking, and Molecular Dynamics Simulation
2.3.1. Bioactive Compound Screening
2.3.2. Computational Screening and Prediction of Bioactive Compound–Disease Target Interactions
2.3.3. Retrieval of Overlapping Genes Between Bioactive Compounds and Diseases
2.3.4. Network Construction and Therapeutic Target Identification in Protein–Protein Interaction Analysis
2.3.5. KEGG and GO Enrichment Analysis
2.3.6. Molecular Docking
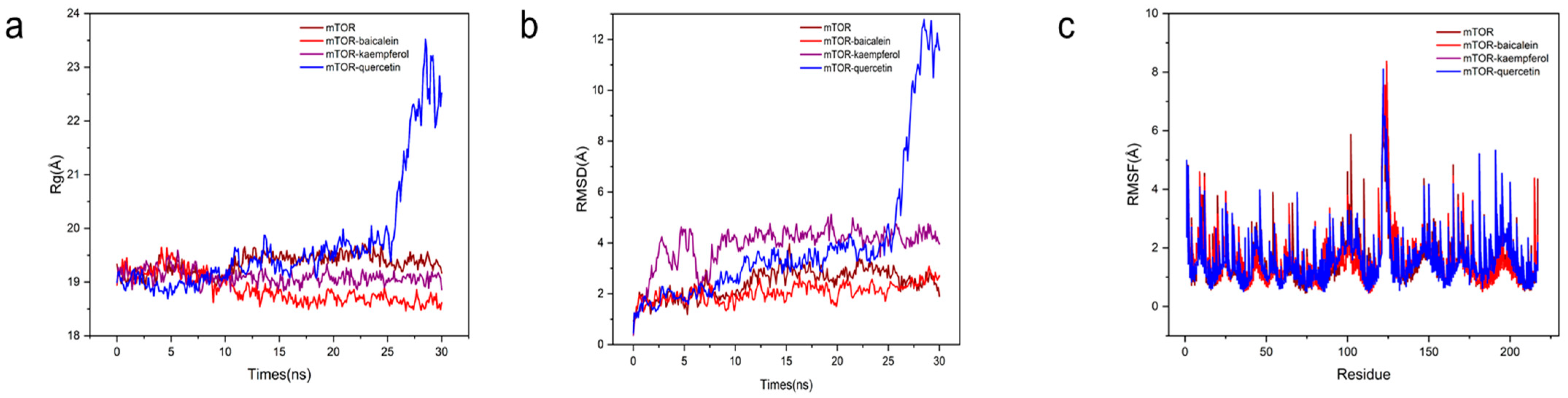
2.3.7. Molecular Dynamics (MD) Simulations
2.4. In Vitro Lipid-Lowering Assay
2.4.1. Pancreatic Lipase Inhibition Assay
2.4.2. Cellular Lipid-Lowering Experiment
Cell Survival Assay
Quantitative Analysis of Cellular Triglycerides, Total Cholesterol, and Antioxidant Biomarkers
Oil Red O Staining
2.5. Statistical Methods
3. Results
3.1. Network Pharmacology Analysis
3.1.1. TCMSP-Driven Screening and Identification of Potential Bioactive Compounds
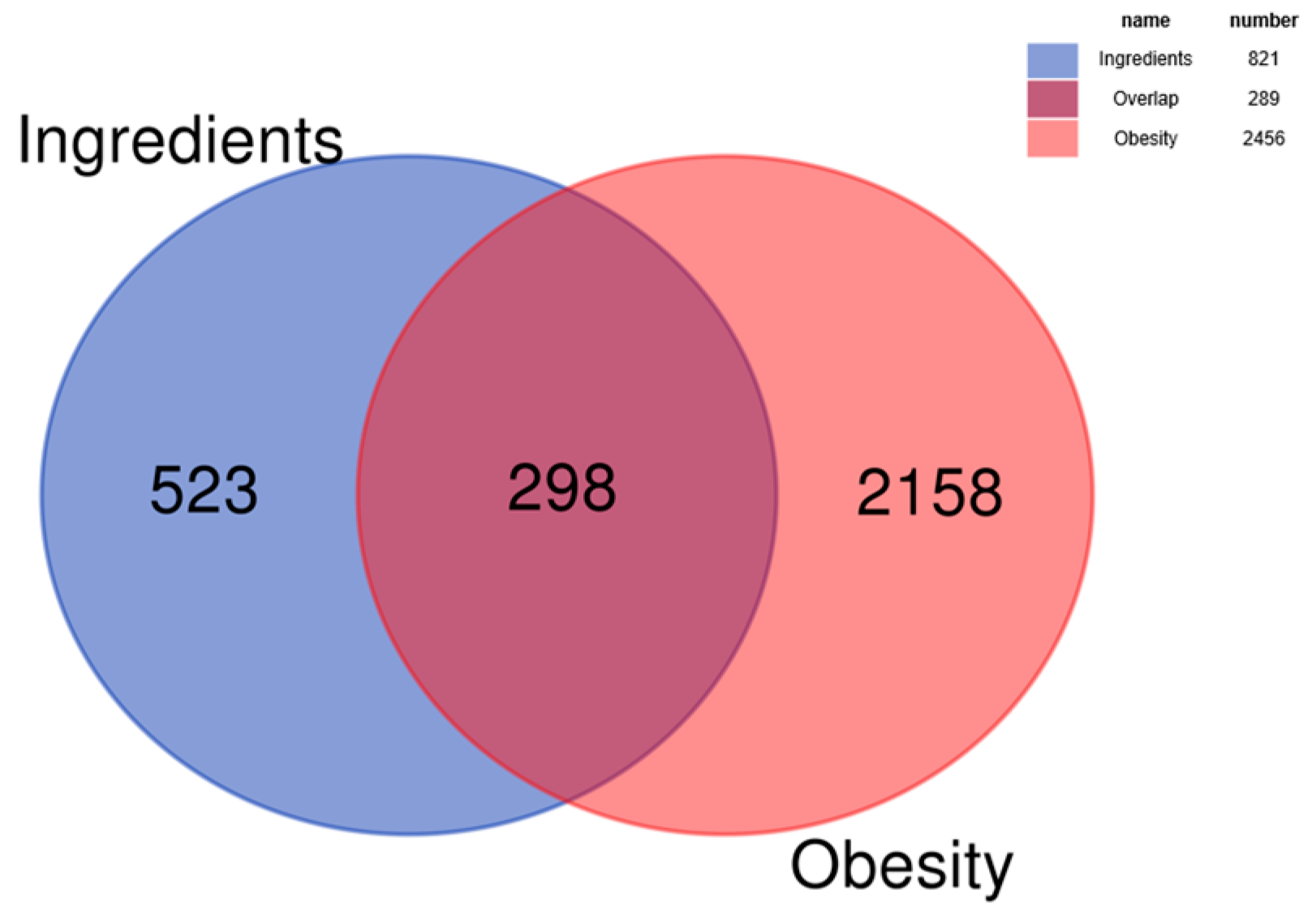
3.1.2. Intersection Analysis of Bioactive Compounds and Disease Targets
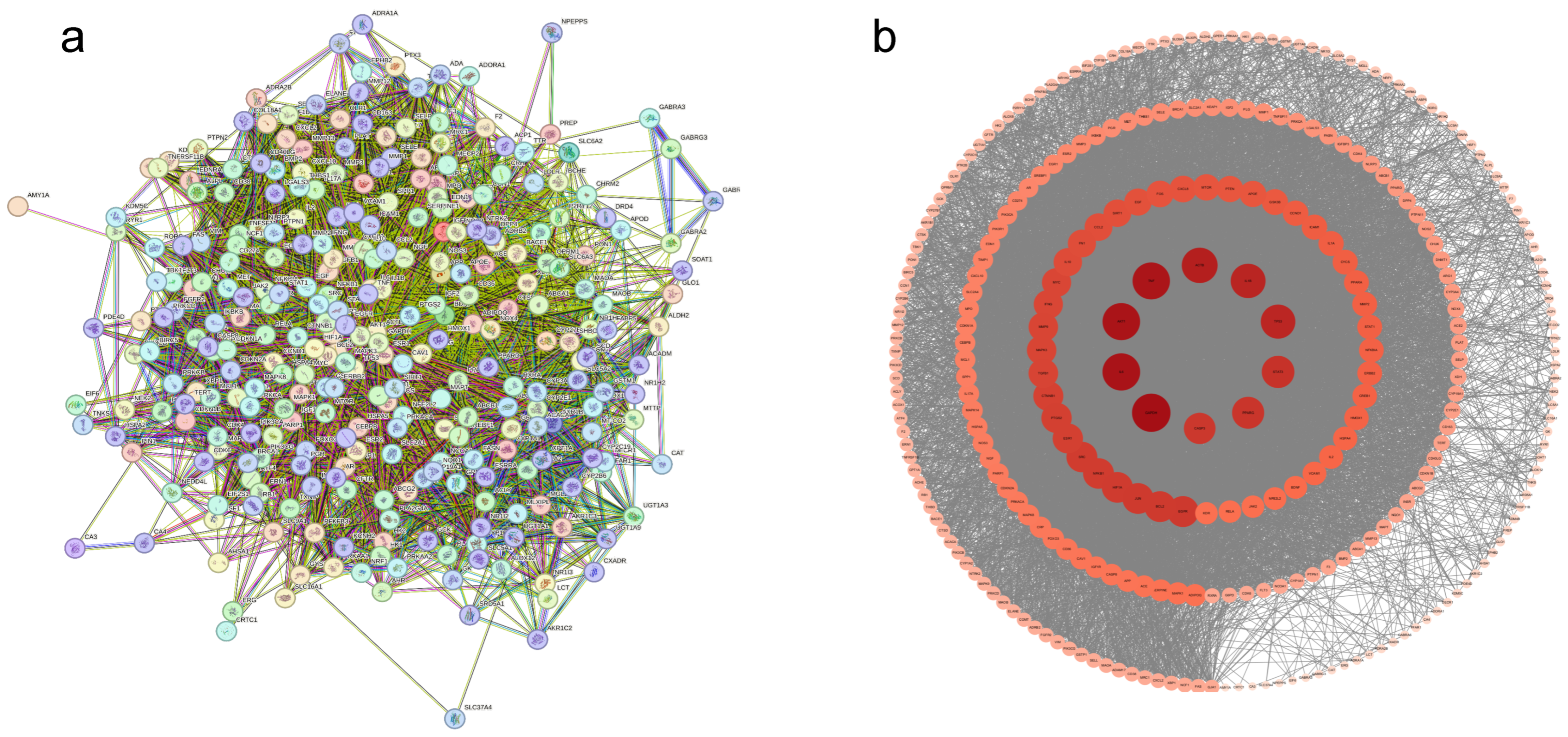
3.1.3. Protein–Protein Interaction Network
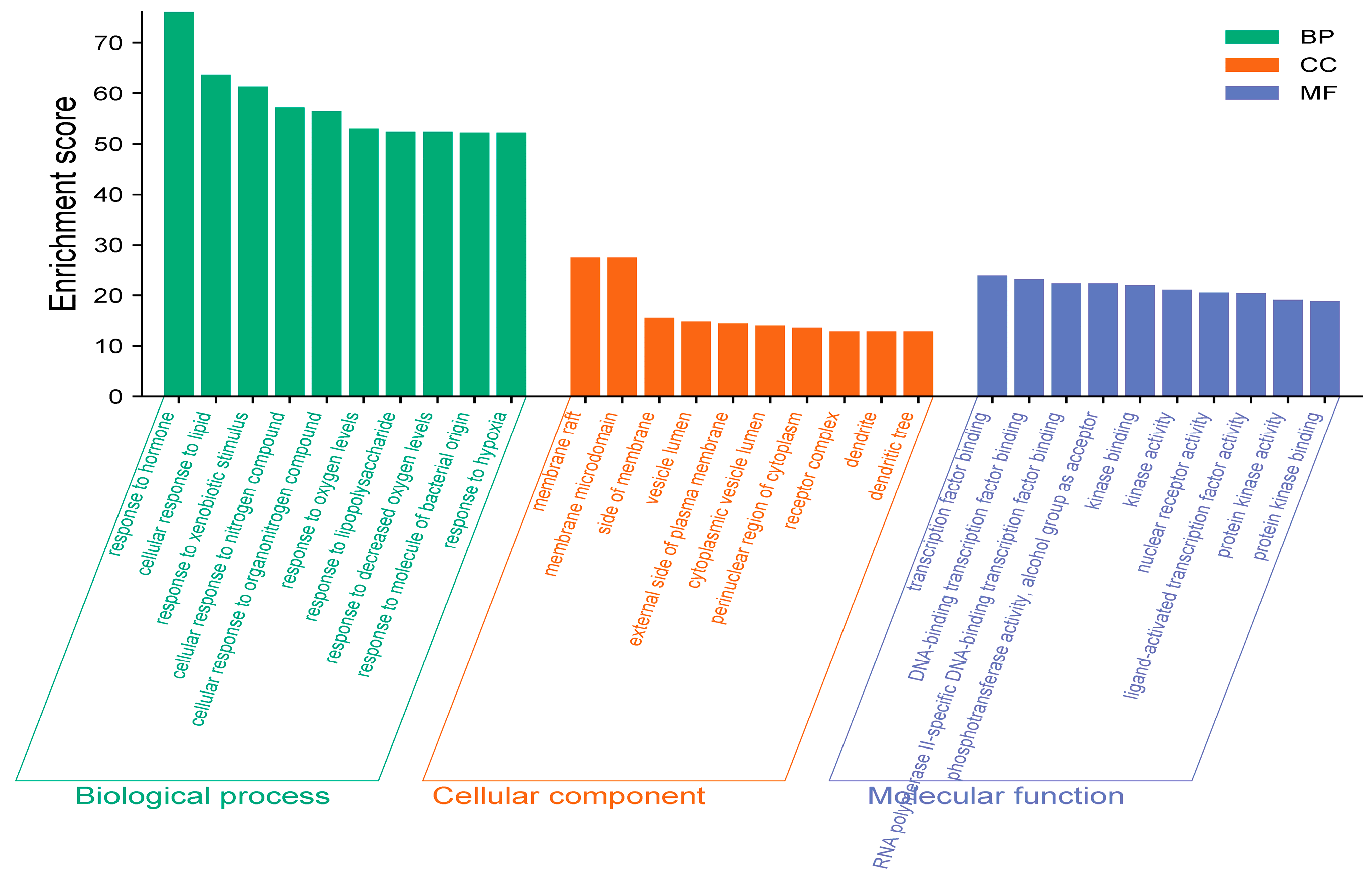
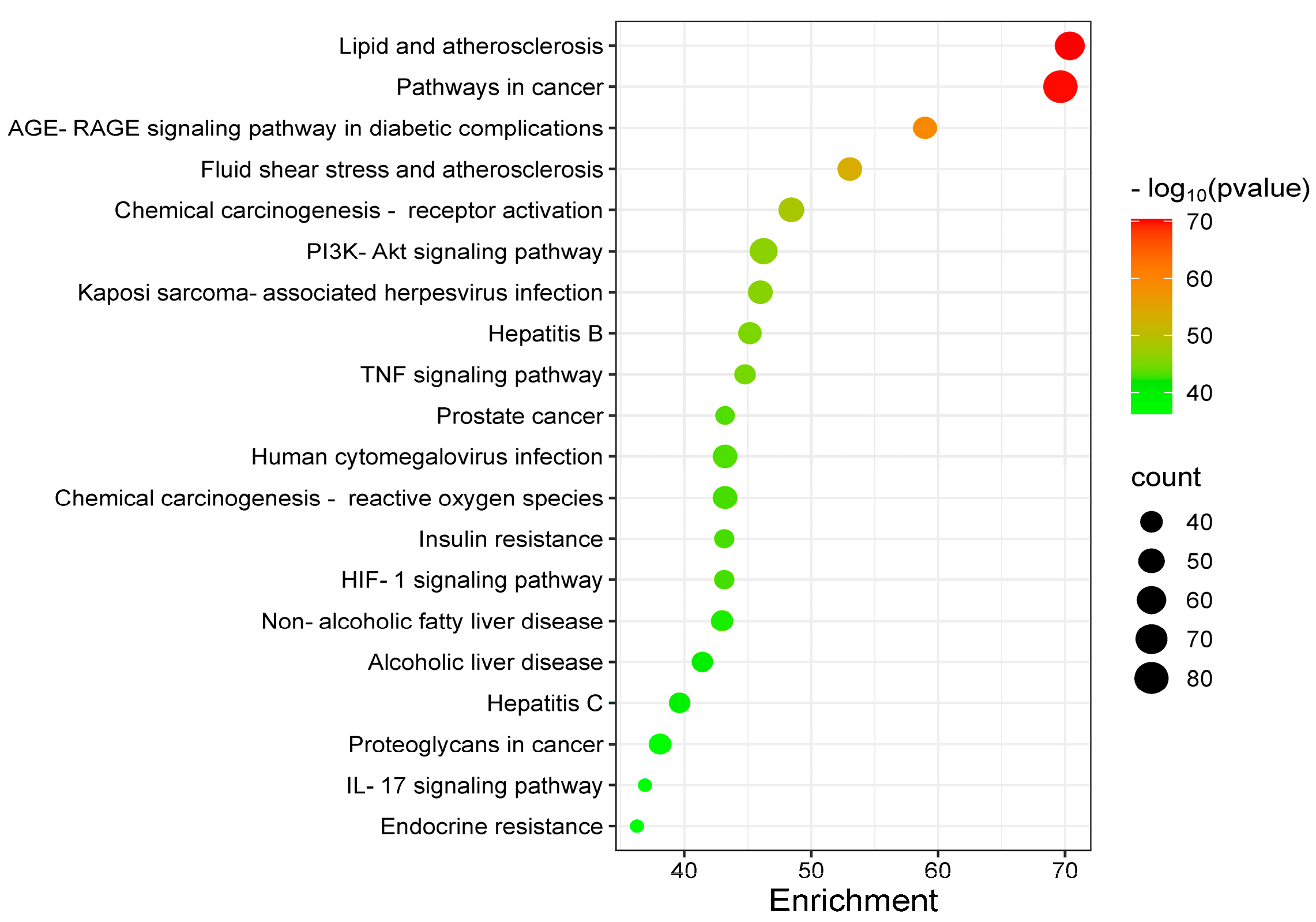
3.1.4. GO and KEGG Analysis
3.1.5. Compound–Target–Disease Network
3.1.6. Molecular Docking and Molecular Dynamic Simulation
3.2. In Vitro Evaluation of Hypolipidemic Efficacy
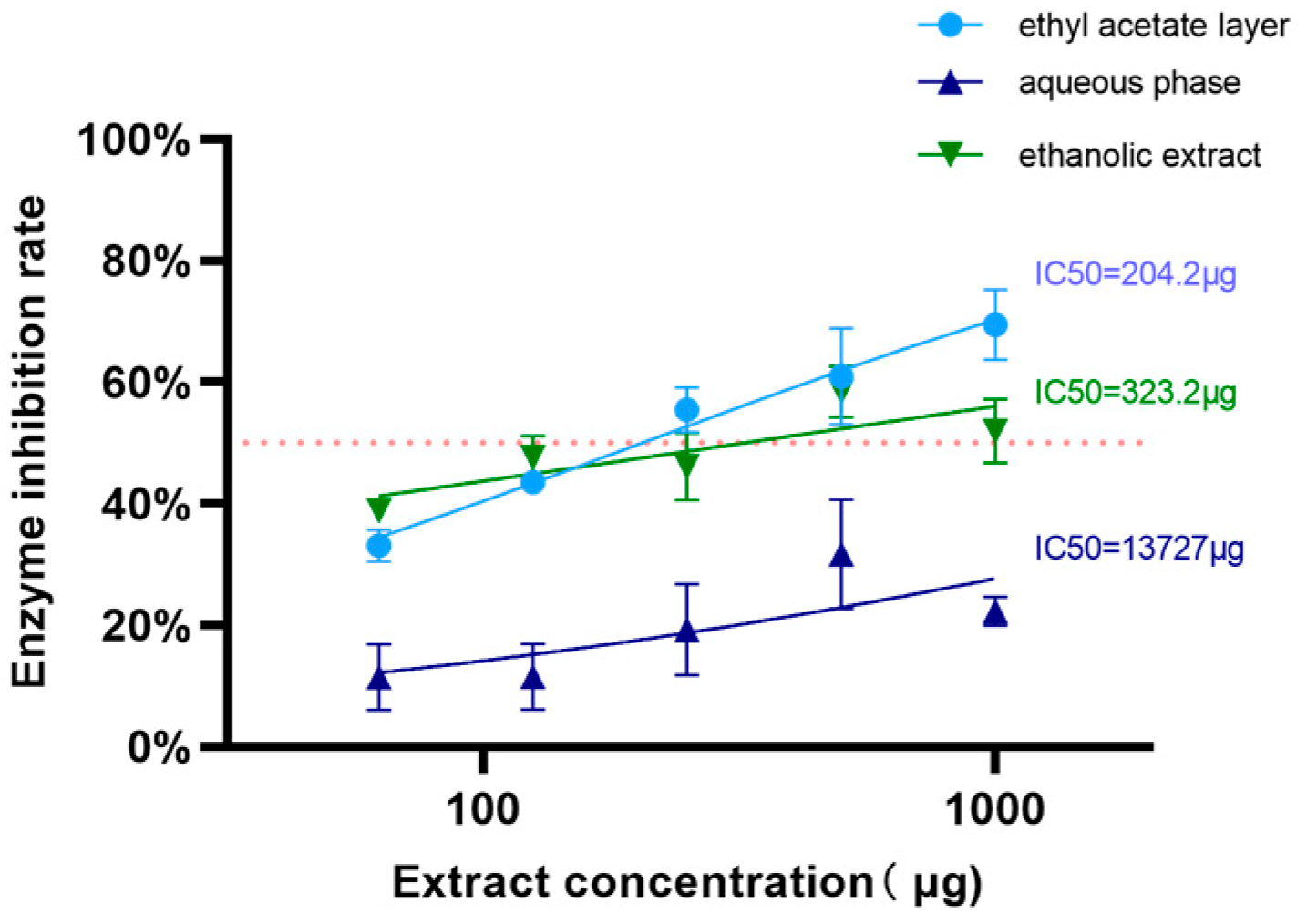
3.2.1. Lipase Inhibitory Effects
3.2.2. Cell-Based Investigation Results
Cell Proliferation Assay Data Interpretation
Intracellular Triglyceride and Total Cholesterol
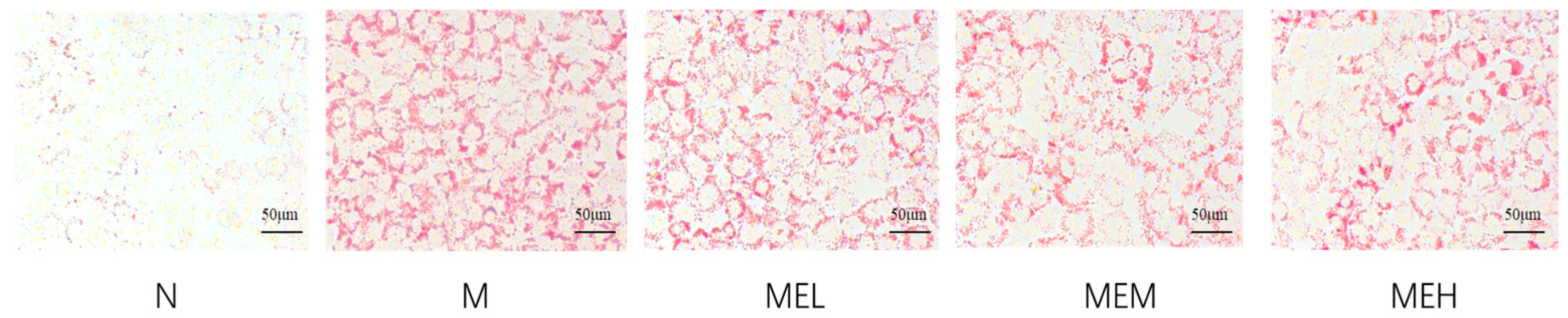
Oil Red O Staining Result
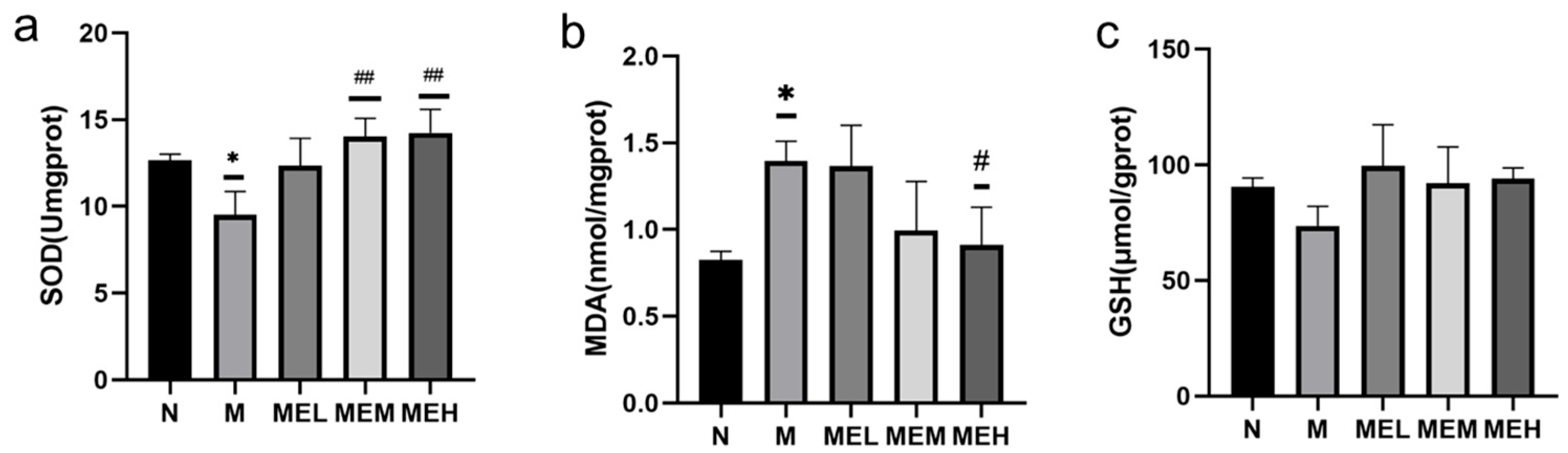
Results of Cellular Antioxidant Indicators
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xu, L.; Yang, Q.; Zhou, J. Mechanisms of Abnormal Lipid Metabolism in the Pathogenesis of Disease. Int. J. Mol. Sci. 2024, 25, 8465. [Google Scholar] [CrossRef] [PubMed]
- Natesan, V.; Kim, S.J. Lipid Metabolism, Disorders and Therapeutic Drugs-Review. Biomol. Ther. 2021, 29, 596–604. [Google Scholar] [CrossRef]
- Younossi, Z.; Anstee, Q.M.; Marietti, M.; Hardy, T.; Henry, L.; Eslam, M.; George, J.; Bugianesi, E. Global burden of NAFLD and NASH: Trends, predictions, risk factors and prevention. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 11–20. [Google Scholar] [CrossRef]
- International Diabetes Federation. IDF Diabetes Atlas, 10th ed.; Brussels, Belgium, 2021; Available online: https://www.diabetesatlas.org (accessed on 10 November 2024).
- World Obesity Federation. Available online: https://www.worldobesity.org/ (accessed on 10 November 2024).
- Liu, Y.; Yang, K.; Jia, Y.; Shi, J.; Tong, Z.; Fang, D.; Yang, B.; Su, C.; Li, R.; Xiao, X.; et al. Gut microbiome alterations in high-fat-diet-fed mice are associated with antibiotic tolerance. Nat. Microbiol. 2021, 6, 874–884. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhao, J.; Tian, C.; Dong, L.; Kang, Z.; Wang, J.; Zhao, S.; Li, M.; Tong, X. Mechanisms of regulation of glycolipid metabolism by natural compounds in plants: Effects on short-chain fatty acids. Nutr. Metab. 2024, 21, 49. [Google Scholar] [CrossRef]
- Chen, C.Y.; Lapsley, K.; Blumberg, J. Perspective A nutrition and health perspective on almonds. J. Sci. Food Agric. 2006, 86, 2245–2250. [Google Scholar] [CrossRef]
- Prgomet, I.; Gonçalves, B.; Domínguez-Perles, R.; Pascual-Seva, N.; Barros, A.I.R.N.A. Valorization Challenges to Almond Residues: Phytochemical Composition and Functional Application. Molecules 2017, 22, 1774. [Google Scholar] [CrossRef]
- Almond Board of California (ABC). Almond Industry Position Report. 2023. Available online: https://www.almonds.com/tools-and-resources/crop-reports/position-reports (accessed on 3 November 2024).
- Esfahlan, A.; Jamei, R.; Esfahlan, R. The importance of almond (Prunus amygdalus L.) and its by-products. Food Chem. 2010, 120, 349–360. [Google Scholar] [CrossRef]
- Wijeratne, S.S.; Abou-Zaid, M.M.; Shahidi, F. Antioxidant polyphenols in almond and its coproducts. J. Sci. Food Agric. 2006, 54, 312–318. [Google Scholar] [CrossRef]
- Li, W.; Wei, J.; Su, Y.; Wu, B.; Zhang, P. Determination of Oleanolic Acid and Ursolic Acid in Almond Green Husk Extract by High Performance Liquid Chromatograph. Food Sci. 2016, 37, 151–157. (In Chinese) [Google Scholar]
- Li, W.; Xi, D.; Tiemur, A.; Su, Y.H.; Wu, B. Content determination of total flavonoids and optimization of the extraction process of extracts from green husk of almonds. Food Sci. 2015, 40, 200–205. (In Chinese) [Google Scholar]
- Sang, S.; Lapsley, K.; Rosen, R.T.; Ho, C.T. New prenylated benzoic acid and other constituents from almond hulls (Prunus amygdalus Batsch). J. Sci. Food Agric. 2002, 50, 607–609. [Google Scholar] [CrossRef] [PubMed]
- Takeoka, G.; Dao, L.; Teranishi, R.; Wong, R.; Flessa, S.; Harden, L.; Edwards, R. Identification of three triterpenoids in almond hulls. J. Sci. Food Agric. 2000, 48, 3437–3439. [Google Scholar] [CrossRef] [PubMed]
- Takeoka, G.; Dao, L. Antioxidant constituents of almond [Prunus dulcis (Mill.) D.A. Webb] hulls. J. Sci. Food Agric. 2003, 51, 496–501. [Google Scholar] [CrossRef]
- Li, W. Analysis of Chemical Constituents and Evaluation of Biological Activities to Almonds Green Husk; Xinjiang University: Ürümqi, China, 2017. (In Chinese) [Google Scholar]
- Harnafi, M.; Bekkouch, O.; Touiss, I.; Khatib, S.; Mokhtari, I.; Milenkovic, D.; Harnafi, H.; Amrani, S. Phenolic-Rich Extract from Almond (Prunus dulcis) Hulls Improves Lipid Metabolism in Triton WR-1339 and High-Fat Diet-Induced Hyperlipidemic Mice and Prevents Lipoprotein Oxidation: A Comparison with Fenofibrate and Butylated Hydroxyanisole. Prev. Nutr. Food Sci. 2020, 25, 254–262. [Google Scholar] [CrossRef] [PubMed]
- Kahlaoui, M.; Bertolino, M.; Barbosa-Pereira, L.; Ben Haj Kbaier, H.; Bouzouita, N.; Zeppa, G. Almond Hull as a Functional Ingredient of Bread: Effects on Physico-Chemical, Nutritional, and Consumer Acceptability Properties. Foods 2022, 11, 777. [Google Scholar] [CrossRef]
- Rocchetti, G.; Gregorio, R.P.; Lorenzo, J.M.; Barba, F.J.; Oliveira, P.G.; Prieto, M.A.; Simal-Gandara, J.; Mosele, J.I.; Motilva, M.J.; Tomas, M.; et al. Functional implications of bound phenolic compounds and phenolics-food interaction:a review. Compr. Rev. Food Sci. Food Saf. 2022, 21, 811–842. [Google Scholar] [CrossRef]
- Gorzynik-Debicka, M.; Przychodzen, P.; Cappello, F.; Kuban-Jankowska, A.; Marino Gammazza, A.; Knap, N.; Wozniak, M.; Gorska-Ponikowska, M. Potential Health Benefits of Olive Oil and Plant Polyphenols. Int. J. Mol. Sci. 2018, 19, 686. [Google Scholar] [CrossRef]
- Caballero, J. The latest automated docking technologies for novel drug discovery. Expert Opin. Drug Discov. 2021, 16, 625–645. [Google Scholar] [CrossRef]
- Gao, S.S.; Sun, J.J.; Wang, X.; Hu, Y.Y.; Feng, Q.; Gou, X.J. Research on the Mechanism of Qushi Huayu Decoction in the Intervention of Nonalcoholic Fatty Liver Disease Based on Network Pharmacology and Molecular Docking Technology. BioMed Res. Int. 2020, 2020, 1704960. [Google Scholar] [CrossRef]
- Li, X.; Miao, F.; Xin, R.; Tai, Z.; Pan, H.; Huang, H.; Yu, J.; Chen, Z.; Zhu, Q. Combining network pharmacology, molecular docking, molecular dynamics simulation, and experimental verification to examine the efficacy and immunoregulation mechanism of FHB granules on vitiligo. Front. Immunol 2023, 14, 1194823. [Google Scholar] [CrossRef] [PubMed]
- Ricchi, M.; Odoardi, M.R.; Carulli, L.; Anzivino, C.; Ballestri, S.; Pinetti, A.; Fantoni, L.I.; Marra, F.; Bertolotti, M.; Banni, S.; et al. Differential effect of oleic and palmitic acid on lipid accumulation and apoptosis in cultured hepatocytes. J. Gastroenterol. Hepatol. 2009, 24, 830–840. [Google Scholar] [CrossRef]
- Luo, W.; Deng, J.; He, J.; Yin, L.; You, R.; Zhang, L.; Shen, J.; Han, Z.; Xie, F.; He, J.; et al. Integration of molecular docking, molecular dynamics and network pharmacology to explore the multi-target pharmacology of fenugreek against diabetes. J. Cell. Mol. Med. 2023, 27, 1959–1974. [Google Scholar] [CrossRef]
- Vekic, J.; Zeljkovic, A.; Stefanovic, A.; Jelic-Ivanovic, Z.; Spasojevic-Kalimanovska, V. Obesity and dyslipidemia. Metabolism 2019, 92, 71–81. [Google Scholar] [CrossRef]
- Munjal, K.; Goel, Y.; Gauttam, V.K.; Chopra, H.; Singla, M.; Smriti; Gupta, S.; Sharma, R. Molecular targets and therapeutic potential of baicalein: A review. Drug Target Insights 2024, 18, 30–46. [Google Scholar] [CrossRef]
- Yao, Y.X.; Yu, Y.J.; Dai, S.; Zhang, C.Y.; Xue, X.Y.; Zhou, M.L.; Yao, C.H.; Li, Y.X. Kaempferol efficacy in metabolic diseases: Molecular mechanisms of action in diabetes mellitus, obesity, non-alcoholic fatty liver disease, steatohepatitis, and atherosclerosis. Biomed. Pharmacother. 2024, 175, 116694. [Google Scholar] [CrossRef] [PubMed]
- Alkandahri, M.Y.; Pamungkas, B.T.; Oktoba, Z.; Shafirany, M.Z.; Sulastri, L.; Arfania, M.; Anggraeny, E.N.; Pratiwi, A.; Astuti, F.D.; Indriyani; et al. Hepatoprotective Effect of Kaempferol: A Review of the Dietary Sources, Bioavailability, Mechanisms of Action, and Safety. Adv. Pharmacol. Pharm. Sci. 2023, 2023, 1387665. [Google Scholar] [CrossRef] [PubMed]
- Tie, F.; Ding, J.; Hu, N.; Dong, Q.; Chen, Z.; Wang, H. Kaempferol and Kaempferide Attenuate Oleic Acid-Induced Lipid Accumulation and Oxidative Stress in HepG2 Cells. Int. J. Mol. Sci. 2021, 22, 8847. [Google Scholar] [CrossRef]
- Hosseini, A.; Razavi, B.M.; Banach, M.; Hosseinzadeh, H. Quercetin and metabolic syndrome: A review. Phytother. Res. 2021, 35, 5352–5364. [Google Scholar] [CrossRef]
- Su, L.; Zeng, Y.; Li, G.; Chen, J.; Chen, X. Quercetin improves high-fat diet-induced obesity by modulating gut microbiota and metabolites in C57BL/6J mice. Phytother. Res. 2022, 36, 4558–4572. [Google Scholar] [CrossRef]
- Yi, R.; Liu, Y.; Zhang, X.; Sun, X.; Wang, N.; Zhang, C.; Deng, H.; Yao, X.; Wang, S.; Yang, G. Unraveling Quercetin’s Potential: A Comprehensive Review of Its Properties and Mechanisms of Action, in Diabetes and Obesity Complications. Phytother. Res. 2024, 38, 5641–5656. [Google Scholar] [CrossRef] [PubMed]
- Briscoe, V.J.; Ertl, A.C.; Tate, D.B.; Davis, S.N. Effects of the selective serotonin reuptake inhibitor fluoxetine on counterregulatory responses to hypoglycemia in individuals with type 1 diabetes. Diabetes 2008, 57, 3315–3322. [Google Scholar] [CrossRef] [PubMed]
- Voellinger, R.; Berney, A.; Baumann, P.; Annoni, J.M.; Bryois, C.; Buclin, T.; Büla, C.; Camus, V.; Christin, L.; Cornuz, J.; et al. Major depressive disorder in the general hospital: Adaptation of clinical practice guidelines. Gen. Hosp. Psychiatry 2003, 25, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Chen, Y.; Deng, F.; Chang, B.; Luo, J.; Dong, L.; Lu, X.; Zhang, Y.; Chen, Z.; Zhou, J. D-Mannose Regulates Hepatocyte Lipid Metabolism via PI3K/Akt/mTOR Signaling Pathway and Ameliorates Hepatic Steatosis in Alcoholic Liver Disease. Front. Immunol. 2022, 13, 877650. [Google Scholar] [CrossRef]
- Knight, Z.A.; Gonzalez, B.; Feldman, M.E.; Zunder, E.R.; Goldenberg, D.D.; Williams, O.; Loewith, R.; Stokoe, D.; Balla, A.; Toth, B.; et al. A pharmacological map of the PI3-K family defines a role for p110alpha in insulin signaling. Cell 2006, 125, 733–747. [Google Scholar] [CrossRef]
- Ersahin, T.; Tuncbag, N.; Cetin-Atalay, R. The PI3K/AKT/mTOR interactive pathway. Mol. Biosyst. 2015, 11, 1946–1954. [Google Scholar] [CrossRef]
- Araiz, C.; Yan, A.; Bettedi, L.; Samuelson, I.; Virtue, S.; McGavigan, A.K.; Dani, C.; Vidal-Puig, A.; Foukas, L.C. Enhanced β-adrenergic signalling underlies an age-dependent beneficial metabolic effect of PI3K p110α inactivation in adipose tissue. Nat. Commun. 2020, 11, 1078. [Google Scholar] [CrossRef]
- Cormerais, Y.; Vučetić, M.; Parks, S.K.; Pouyssegur, J. Amino Acid Transporters Are a Vital Focal Point in the Control of mTORC1 Signaling and Cancer. Int. J. Mol. Sci. 2020, 22, 23. [Google Scholar] [CrossRef]
- Liu, G.Y.; Sabatini, D.M. mTOR at the nexus of nutrition, growth, ageing and disease. Nat. Rev. Mol. Cell Biol. 2020, 21, 183–203. [Google Scholar] [CrossRef]
- Eshaghi, F.S.; Ghazizadeh, H.; Kazami-Nooreini, S.; Timar, A.; Esmaeily, H.; Mehramiz, M.; Avan, A.; Ghayour-Mobarhan, M. Association of a genetic variant in AKT1 gene with features of the metabolic syndrome. Genes Dis. 2019, 6, 290–295. [Google Scholar] [CrossRef]
- Li, J.; Fang, Z.; Dal, E.; Zhang, H.; Yu, K.; Ma, M.; Wang, M.; Sun, R.; Lu, M.; Wang, H.; et al. Transmembrane protein 176B regulates amino acid metabolism through the PI3K-Akt-mTOR signaling pathway and promotes gastric cancer progression. Cancer Cell Int. 2024, 24, 95. [Google Scholar] [CrossRef] [PubMed]
- Musso, G.; Gambino, R.; Cassader, M.; Pagano, G. Meta-analysis: Natural history of non-alcoholic fatty liver disease (NAFLD) and diagnostic accuracy of non-invasive tests for liver disease severity. Ann. Med. 2011, 43, 617–649. [Google Scholar] [CrossRef] [PubMed]
- Bae, J.C.; Cho, Y.K.; Lee, W.Y.; Seo, H.I.; Rhee, E.J.; Park, S.E.; Park, C.Y.; Oh, K.W.; Sung, K.C.; Kim, B.I. Impact of nonalcoholic fatty liver disease on insulin resistance in relation to HbA1c levels in nondiabetic subjects. Am. J. Gastroenterol. 2010, 105, 2389–2395. [Google Scholar] [CrossRef] [PubMed]
- Dongiovanni, P.; Stender, S.; Pietrelli, A.; Mancina, R.M.; Cespiati, A.; Petta, S.; Pelusi, S.; Pingitore, P.; Badiali, S.; Maggioni, M.; et al. Causal relationship of hepatic fat with liver damage and insulin resistance in nonalcoholic fatty liver. J. Intern. Med. 2018, 283, 356–370. [Google Scholar] [CrossRef]
- Manchanayake, J.; Chitturi, S.; Nolan, C.; Farrell, G.C. Postprandial hyperinsulinemia is universal in non-diabetic patients with nonalcoholic fatty liver disease. J. Gastroenterol. Hepatol. 2011, 26, 510–516. [Google Scholar] [CrossRef]
- Bort, A.; Sánchez, B.G.; Mateos-Gómez, P.A.; Díaz-Laviada, I.; Rodríguez-Henche, N. Capsaicin Targets Lipogenesis in HepG2 Cells Through AMPK Activation, AKT Inhibition and PPARs Regulation. Int. J. Mol. Sci. 2019, 20, 1660. [Google Scholar] [CrossRef]
- Bugianesi, E.; Gastaldelli, A.; Vanni, E.; Gambino, R.; Cassader, M.; Baldi, S.; Ponti, V.; Pagano, G.; Ferrannini, E.; Rizzetto, M. Insulin resistance in non-diabetic patients with non-alcoholic fatty liver disease: Sites and mechanisms. Diabetologia 2005, 48, 634–642. [Google Scholar] [CrossRef]
- Seppälä-Lindroos, A.; Vehkavaara, S.; Häkkinen, A.M.; Goto, T.; Westerbacka, J.; Sovijärvi, A.; Halavaara, J.; Yki-Järvinen, H. Fat accumulation in the liver is associated with defects in insulin suppression of glucose production and serum free fatty acids independent of obesity in normal men. J. Clin. Endocrinol. Metab. 2002, 87, 3023–3028. [Google Scholar] [CrossRef]
- Donnelly, K.L.; Smith, C.I.; Schwarzenberg, S.J.; Jessurun, J.; Boldt, M.D.; Parks, E.J. Sources of fatty acids stored in liver and secreted via lipoproteins in patients with nonalcoholic fatty liver disease. J. Clin. Investig. 2005, 115, 1343–1351. [Google Scholar] [CrossRef]
- Ye, J. Role of insulin in the pathogenesis of free fatty acid-induced insulin resistance in skeletal muscle. Endocr. Metab. Immune Disord. Drug Targets 2007, 7, 65–74. [Google Scholar] [CrossRef]
- He, Q.; Gao, Z.; Yin, J.; Zhang, J.; Yun, Z.; Ye, J. Regulation of HIF-1{alpha} activity in adipose tissue by obesity-associated factors: Adipogenesis, insulin, and hypoxia. Am. J. Physiol. Endocrinol. Metab. 2011, 300, E877–E885. [Google Scholar] [CrossRef] [PubMed]
- Heilbronn, L.; Smith, S.R.; Ravussin, E. Failure of fat cell proliferation, mitochondrial function and fat oxidation results in ectopic fat storage, insulin resistance and type II diabetes mellitus. Int. J. Obes. 2004, 28 (Suppl. S4), S12–S21. [Google Scholar] [CrossRef] [PubMed]
- Sabry, K.; Jamshidi, Z.; Emami, S.A.; Sahebka, A. Potential therapeutic effects of baicalin and baicalein. Avicenna J. Phytomed. 2024, 14, 23–49. [Google Scholar] [PubMed]
- Baradaran Rahimi, V.; Askari, V.R.; Hosseinzadeh, H. Promising influences of Scutellaria baicalensis and its two active constituents, baicalin, and baicalein, against metabolic syndrome: A review. Phytother. Res. 2021, 35, 3558–3574. [Google Scholar] [CrossRef]



| Time (min) | Flow Rate (mL/min) | Phase A (%) | Phase B (%) |
|---|---|---|---|
| 0 | 0.35 | 95 | 5 |
| 9 | 0.35 | 5 | 95 |
| 10 | 0.35 | 5 | 95 |
| 11.1 | 0.35 | 95 | 5 |
| 14 | 0.35 | 95 | 5 |
| Mass Spectrometry Conditions | Parameters |
|---|---|
| Source temperature (°C) | 500 |
| Ion spray voltage (positive ion mode) (V) | 5500 |
| Ion spray voltage (negative ion mode) (V) | 4500 |
| Ion source gas I (psi) | 50 |
| Ion source gas II (psi) | 60 |
| Curtain gas (psi) | 25 |
| Collision gas | medium |
| Collision-activated dissociation | high |
| Reagents | Sample Group (µL) | Blank Group (µL) | Sample Control Group (µL) | Blank Control Group (µL) |
|---|---|---|---|---|
| Enzyme solution | 50 | 50 | 50 | 50 |
| Sample solution | 20 | 0 | 20 | 0 |
| Sample solvent | 0 | 20 | 0 | 20 |
| Buffer solution | 110 | |||
| PNPB | 20 | |||
| Compounds | OB (%) | DL |
|---|---|---|
| Catechin | 49.67 | 0.24 |
| Skimmin | 38.35 | 0.32 |
| Sesamin | 56.55 | 0.83 |
| Tamarixetin | 32.86 | 0.31 |
| Isorhamnetin | 49.60 | 0.31 |
| Baicalein | 33.52 | 0.21 |
| Fraxin | 36.76 | 0.42 |
| 5-O-Coumaroylquinic acid | 37.63 | 0.29 |
| Kaempferol | 41.88 | 0.24 |
| Wogonin | 30.68 | 0.23 |
| Nobiletin | 61.67 | 0.52 |
| Arctiin | 34.45 | 0.84 |
| Gossypetin | 35.00 | 0.31 |
| Laricitrin | 35.38 | 0.34 |
| Azaleatin | 54.28 | 0.30 |
| Syringetin | 36.82 | 0.37 |
| Herbacetin | 36.07 | 0.27 |
| Queretin | 46.43 | 0.28 |
| Morin | 46.23 | 0.27 |
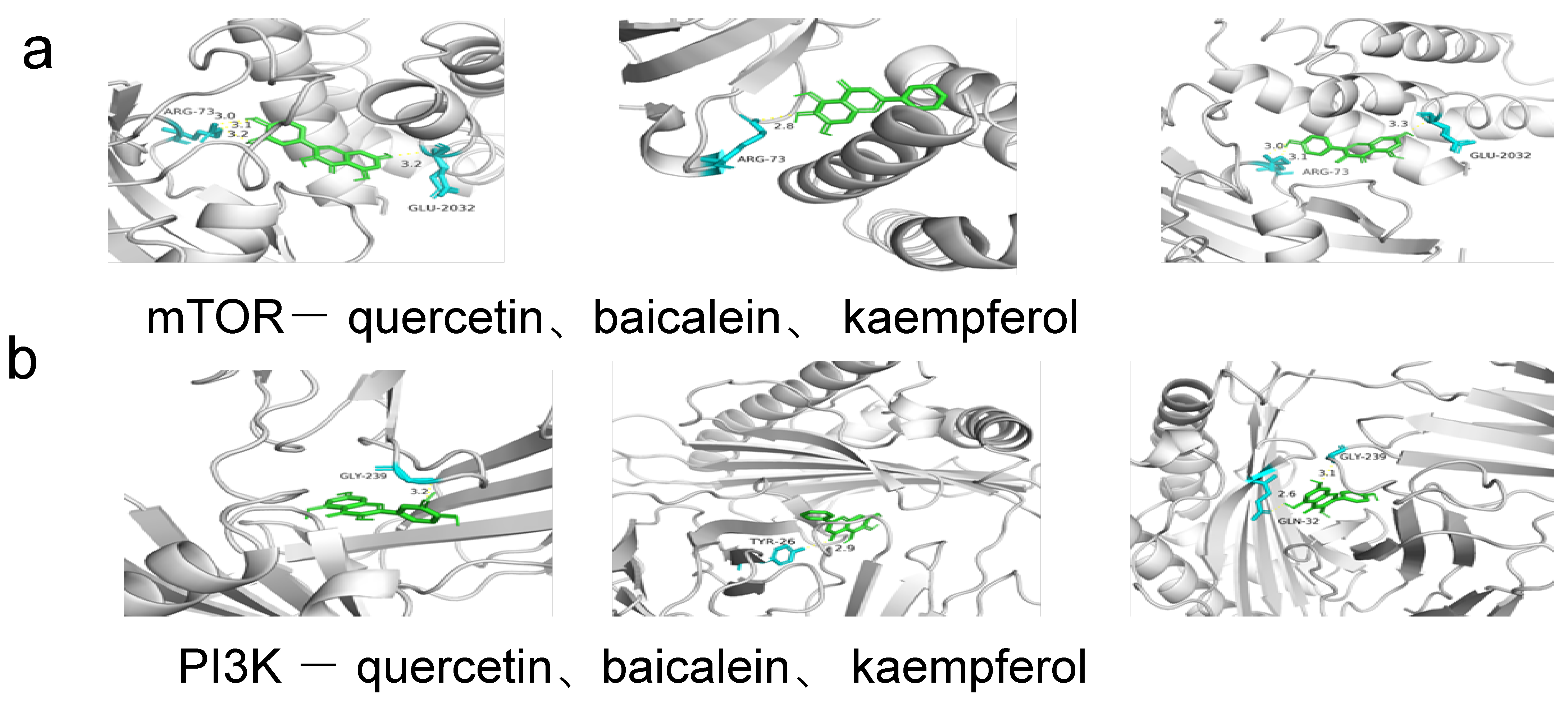
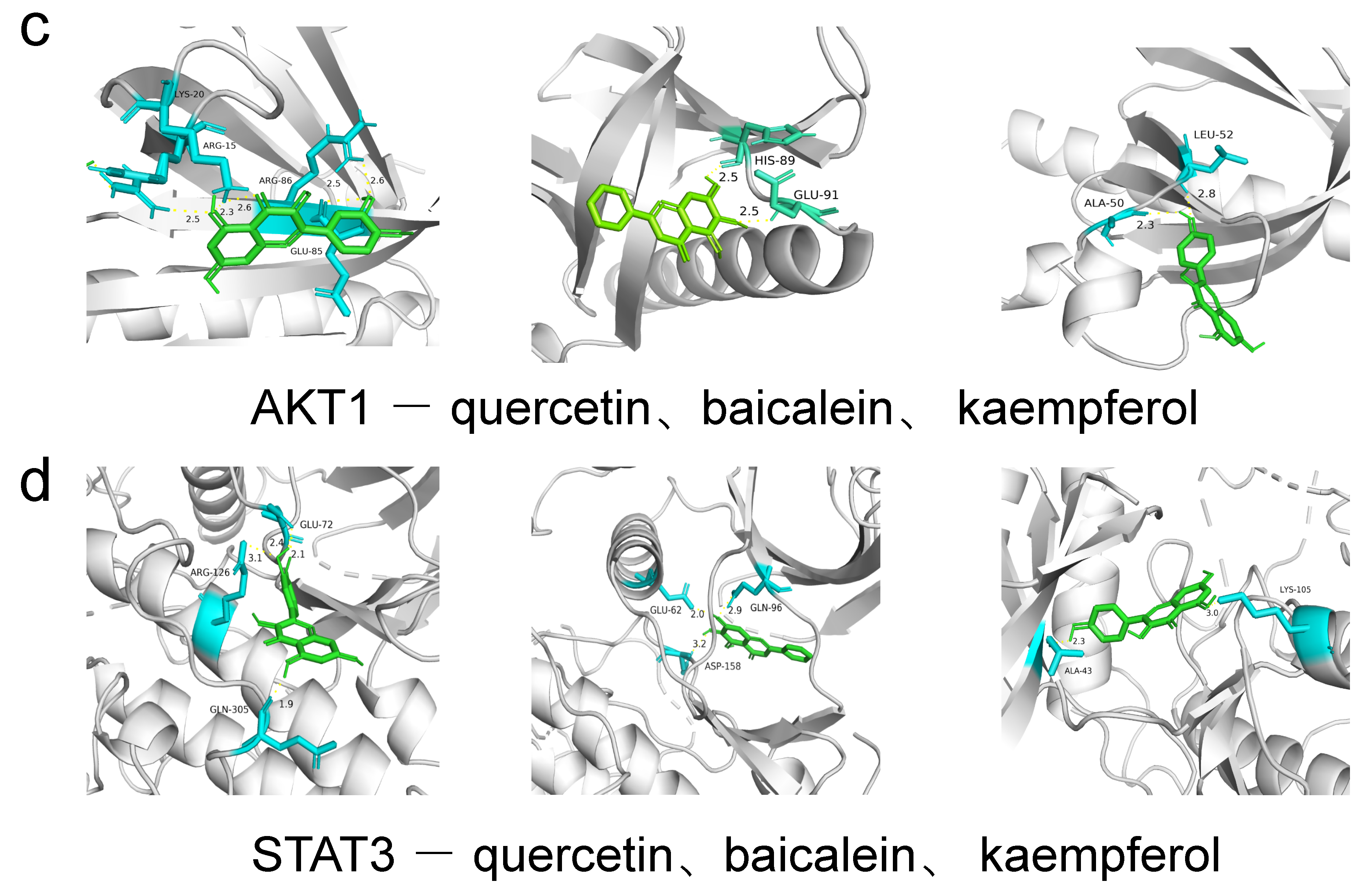
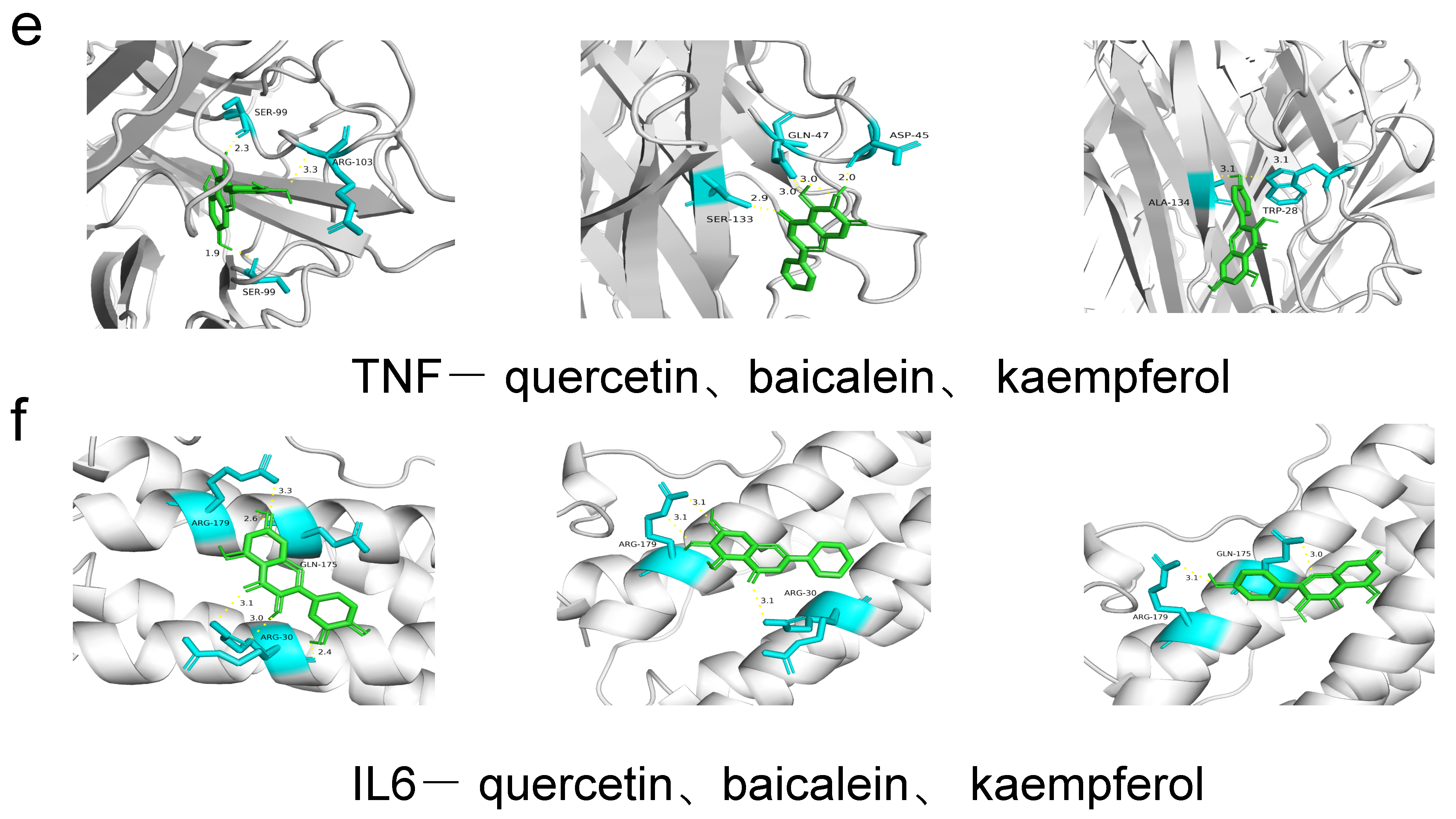
| Ingredient | Receptor | PDB ID | Binding Energy (KJ/mol) | Binding Site |
|---|---|---|---|---|
| quercetin | PI3K | 7I1c | −8.0 | GLY-239 |
| quercetin | AKT1 | 1h10 | −6.3 | ARG-86, GLU-85, LYS-20 |
| quercetin | IL6 | 1a1u | −7.1 | ARG-179, GLN-175, ARG-30 |
| quercetin | TNF | 1a8m | −8.8 | SER-99, ARG-103 |
| quercetin | STAT3 | 5ax3 | −8.5 | GLN-305, ARG-126, GLU-72 |
| quercetin | mTOR | 4dri | −9.2 | GLU-2032, ARG-73 |
| baicalein | PI3K | 7I1c | −8.0 | TYR-26 |
| baicalein | AKT1 | 1h10 | −6.7 | HIS-89, GLU-91 |
| baicalein | IL6 | 1a1u | −6.9 | ARG-179, ARG-30 |
| baicalein | TNF | 1a8m | −6.5 | SER-133, GLN-47, ASP-45 |
| baicalein | STAT3 | 5ax3 | −8.3 | ASP-158, GLU-62, GLN-32 |
| baicalein | mTOR | 4dri | −9.5 | ARG-73 |
| kaempferol | PI3K | 7I1c | −7.9 | GLY-239, GLN-32 |
| kaempferol | AKT1 | 1h10 | −6.1 | ALA-50, LEU-52 |
| kaempferol | IL6 | 1a1u | −6.7 | ARG-179, GLN-175 |
| kaempferol | TNF | 1a8m | −6.9 | ALA-134, TRP-28 |
| kaempferol | STAT3 | 5ax3 | −8.1 | LYS-105, ALA-43 |
| kaempferol | mTOR | 4dri | −9.3 | ARG-73, GLU-2032 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miao, Q.; Sun, L.; Wu, J.; Zhu, X.; Liu, J.; Ruan, R.; Huang, G.; Mi, S.; Cheng, Y. Lipid-Lowering Potential of Almond Hulls (Quercetin, Baicalein, and Kaempferol): Insights from Network Pharmacology and Molecular Dynamics. Curr. Issues Mol. Biol. 2025, 47, 450. https://doi.org/10.3390/cimb47060450
Miao Q, Sun L, Wu J, Zhu X, Liu J, Ruan R, Huang G, Mi S, Cheng Y. Lipid-Lowering Potential of Almond Hulls (Quercetin, Baicalein, and Kaempferol): Insights from Network Pharmacology and Molecular Dynamics. Current Issues in Molecular Biology. 2025; 47(6):450. https://doi.org/10.3390/cimb47060450
Chicago/Turabian StyleMiao, Qiming, Lu Sun, Jiayuan Wu, Xinyue Zhu, Juer Liu, Roger Ruan, Guangwei Huang, Shengquan Mi, and Yanling Cheng. 2025. "Lipid-Lowering Potential of Almond Hulls (Quercetin, Baicalein, and Kaempferol): Insights from Network Pharmacology and Molecular Dynamics" Current Issues in Molecular Biology 47, no. 6: 450. https://doi.org/10.3390/cimb47060450
APA StyleMiao, Q., Sun, L., Wu, J., Zhu, X., Liu, J., Ruan, R., Huang, G., Mi, S., & Cheng, Y. (2025). Lipid-Lowering Potential of Almond Hulls (Quercetin, Baicalein, and Kaempferol): Insights from Network Pharmacology and Molecular Dynamics. Current Issues in Molecular Biology, 47(6), 450. https://doi.org/10.3390/cimb47060450






