PAICS-Driven Purine Biosynthesis and Its Prognostic Implications in Lung Adenocarcinoma: A Novel Risk Stratification Model and Therapeutic Insights
Abstract
1. Introduction
2. Materials and Methods
2.1. Public Datasets Acquisition
2.2. CHCAMS Cohort
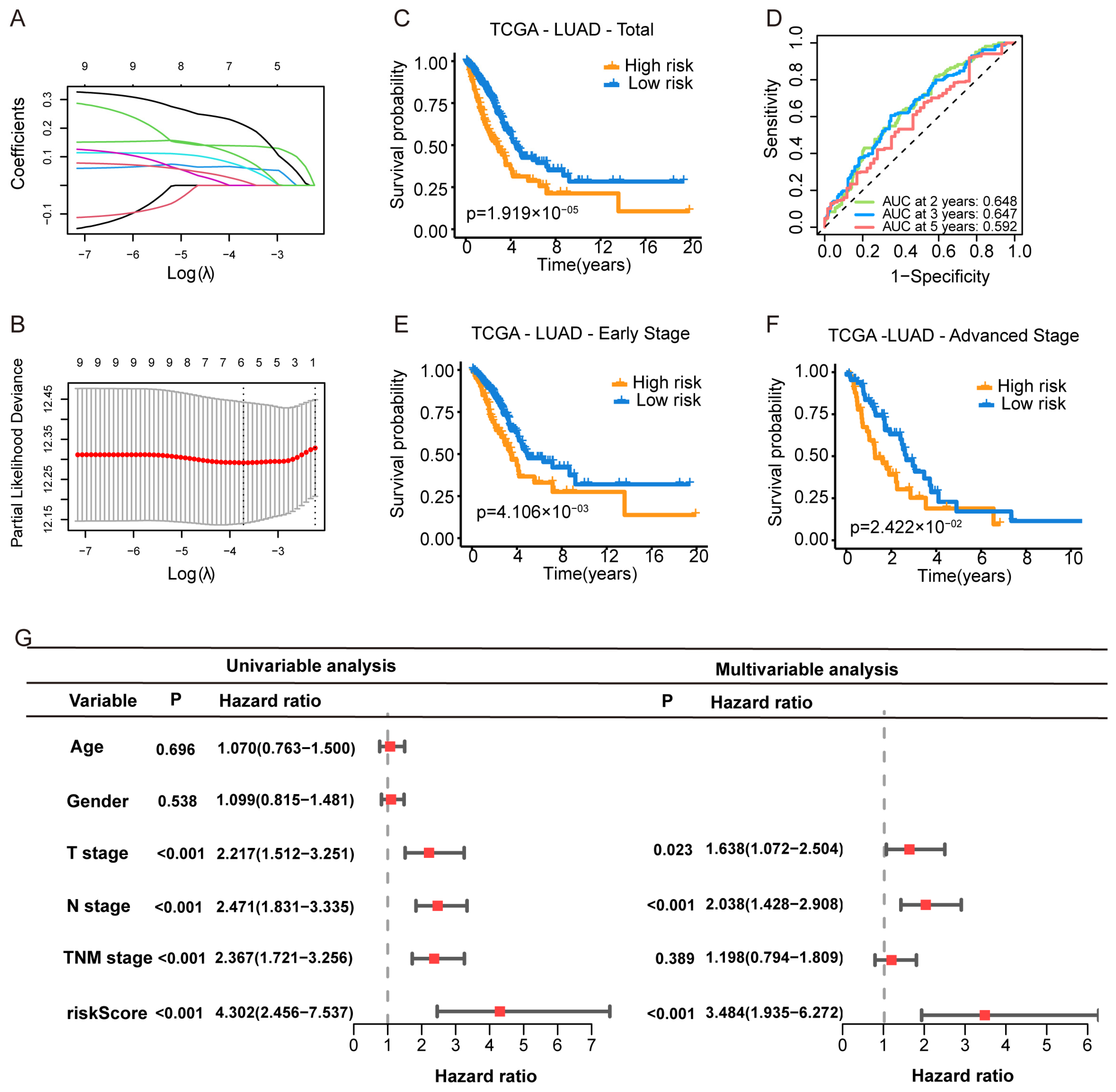
2.3. Construction and Validation of the PSRS
2.4. Exploration of Risk Score Correlationshanisms Related to the PBRS-Based Stratification in LUAD
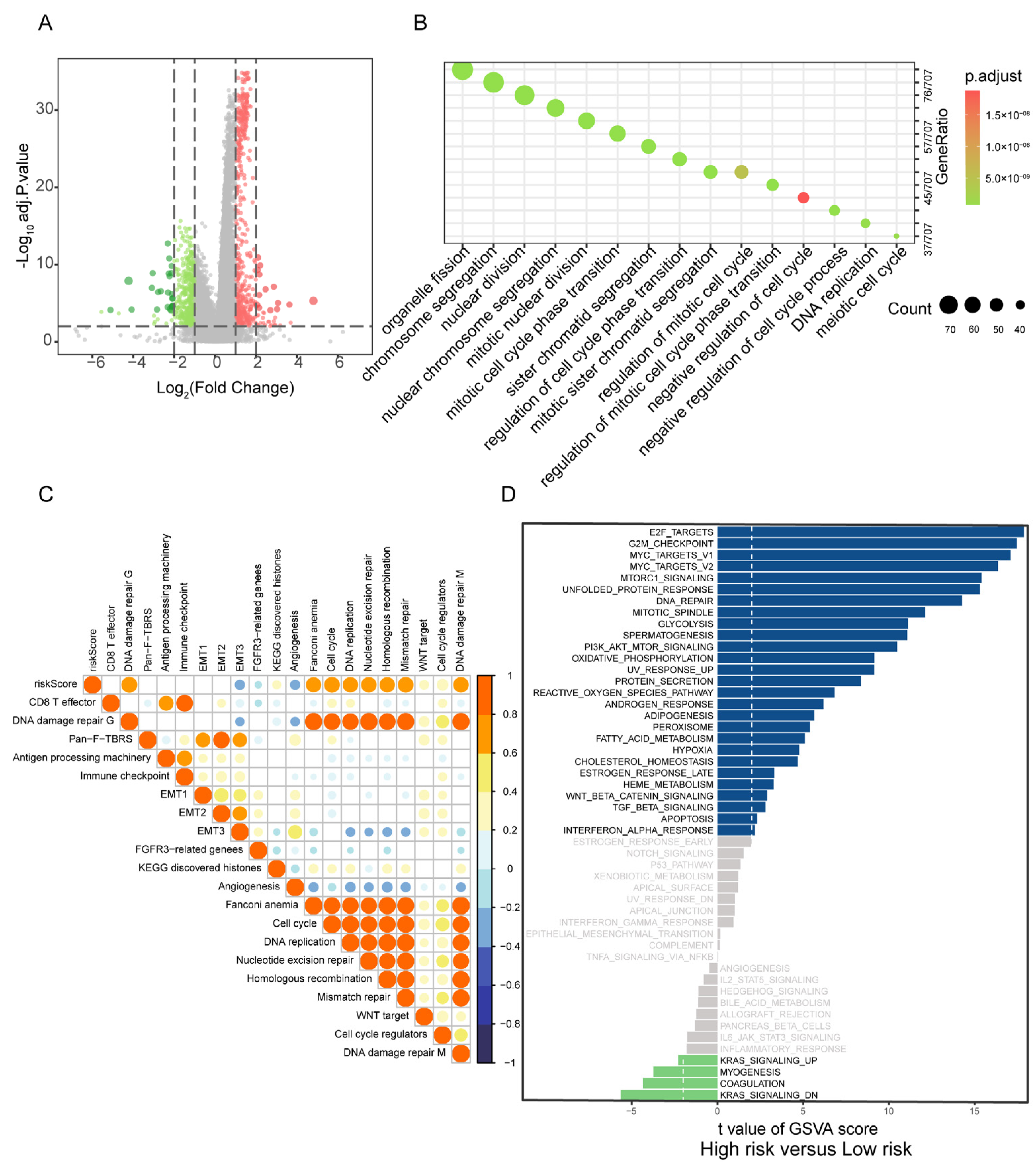
2.4.1. Differential Biological Processes
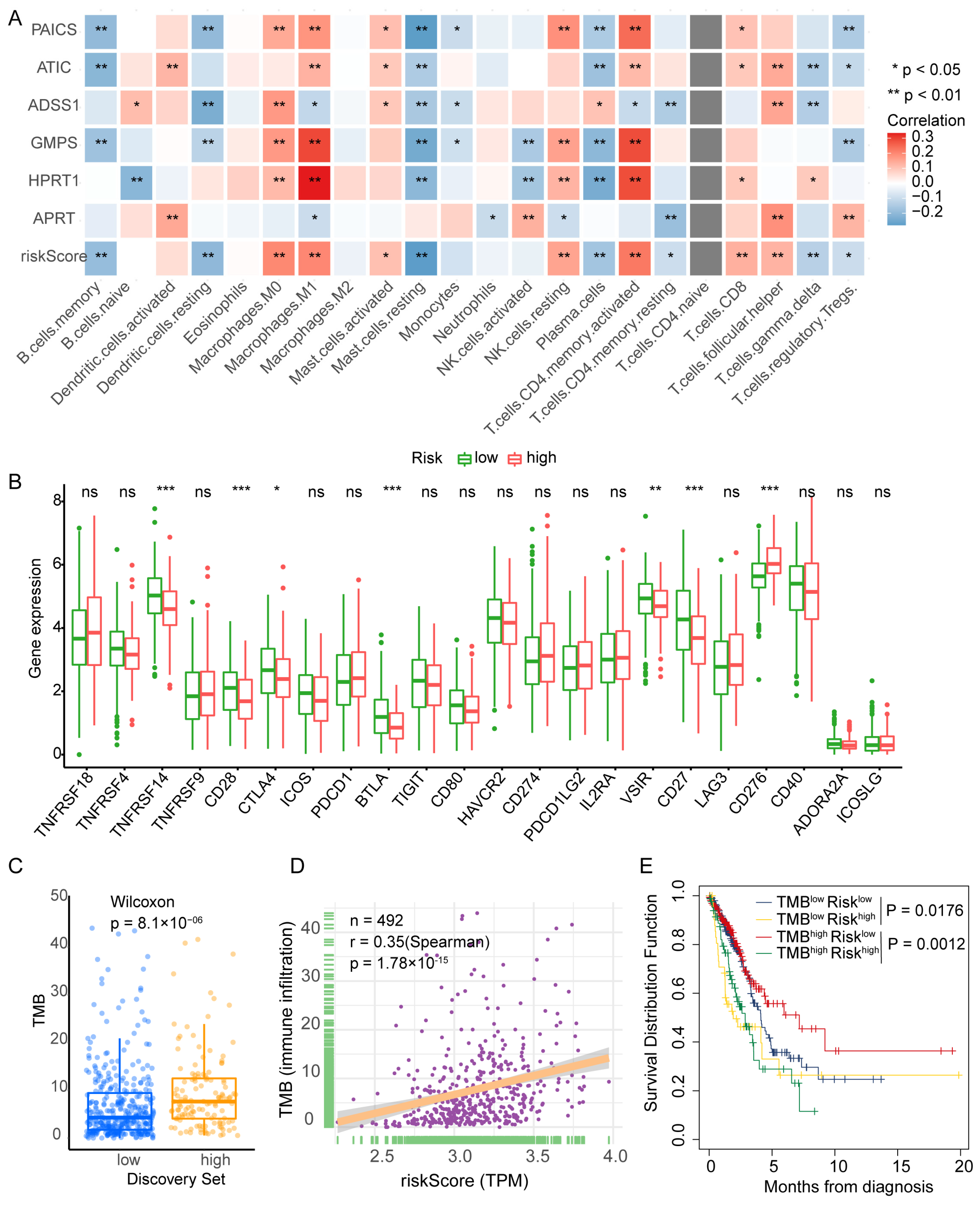
2.4.2. Immune Correlation Analysis
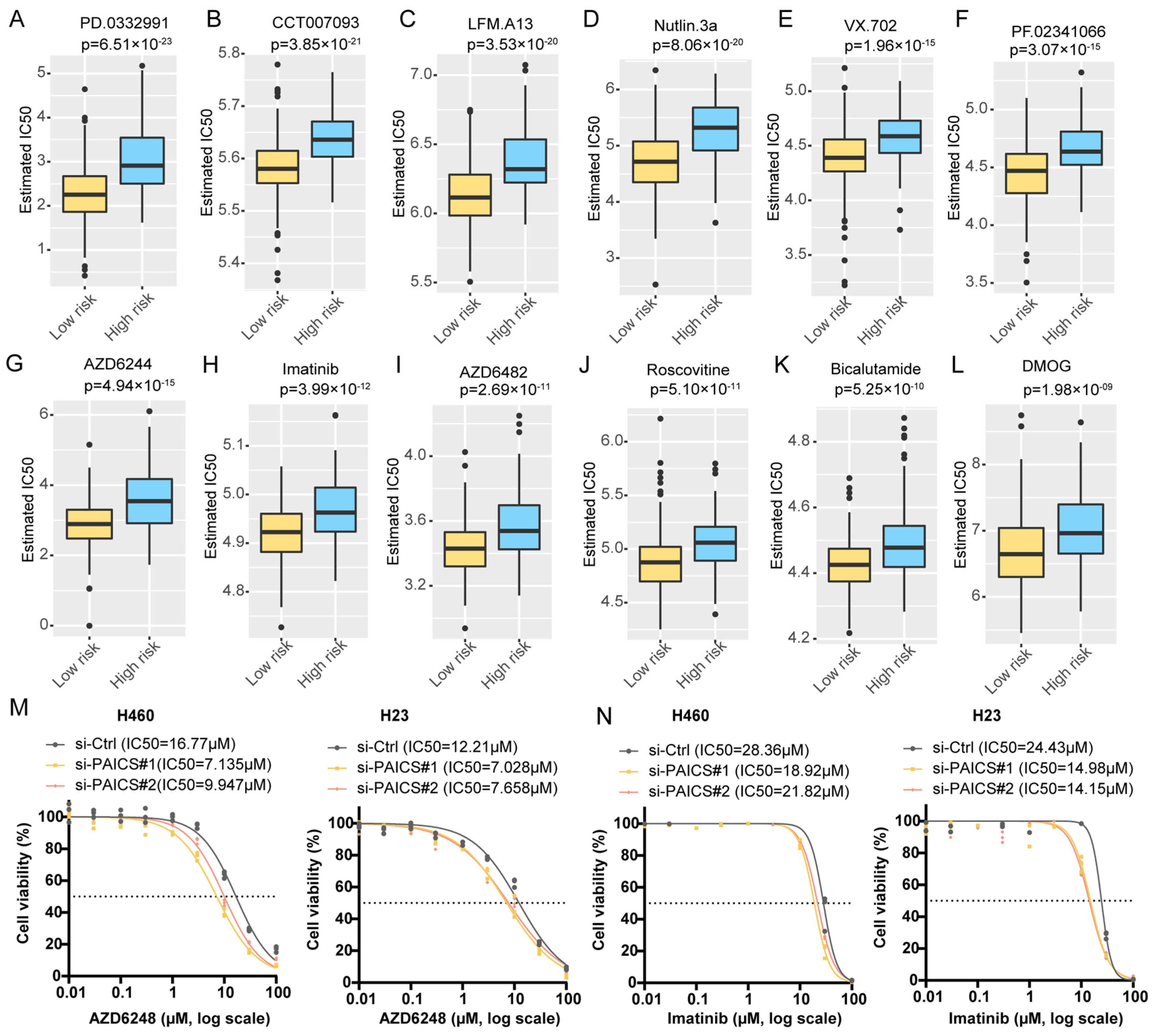
2.4.3. Drug Sensitivity Prediction
3. Results
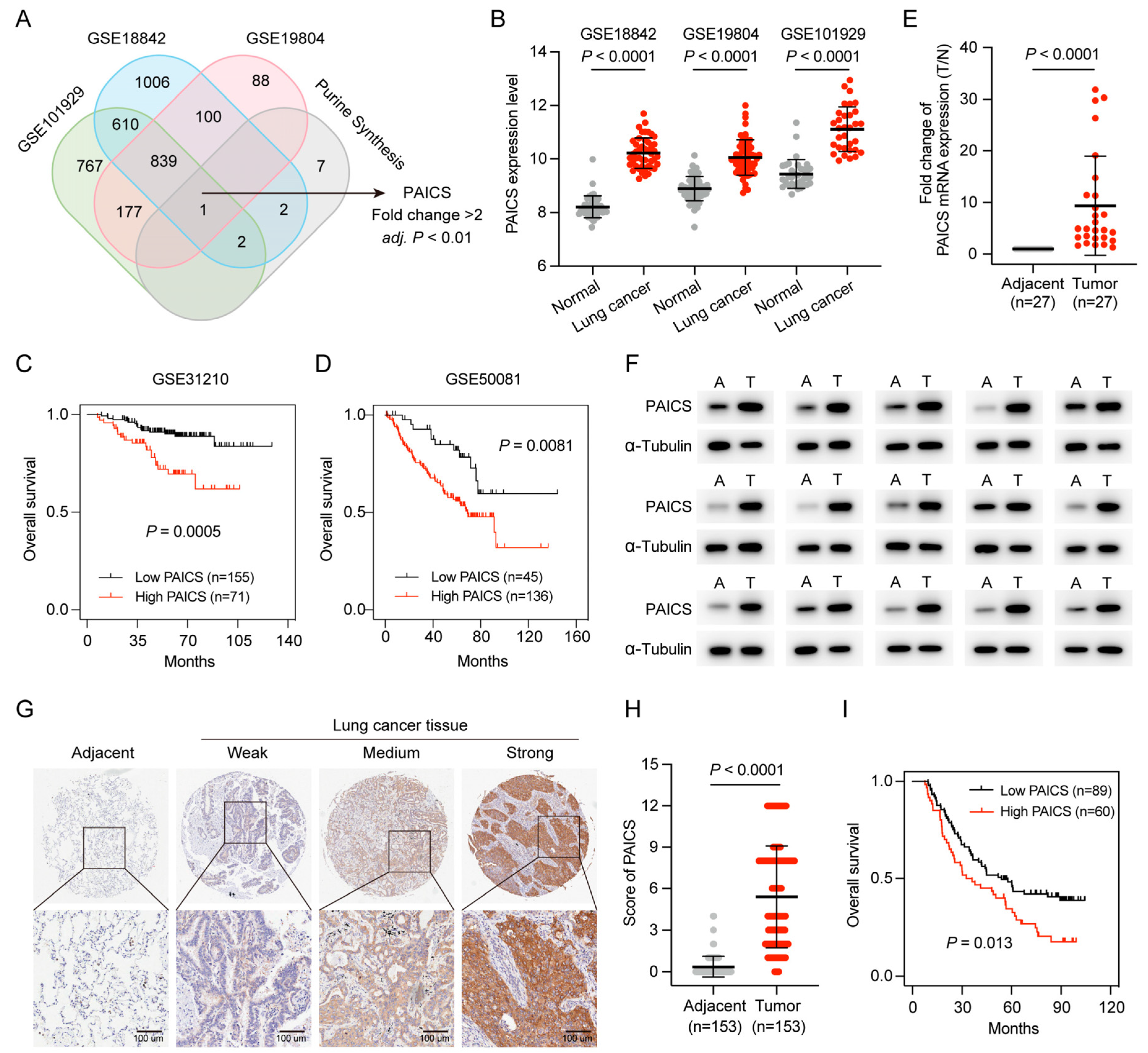
3.1. Purine Metabolism Enrichment in LUAD and Its Association with Poor Prognosis
3.2. Construction of the Purine Biosynthesis-Related Score (PBRS) System for Overall Survival in LUAD Patients
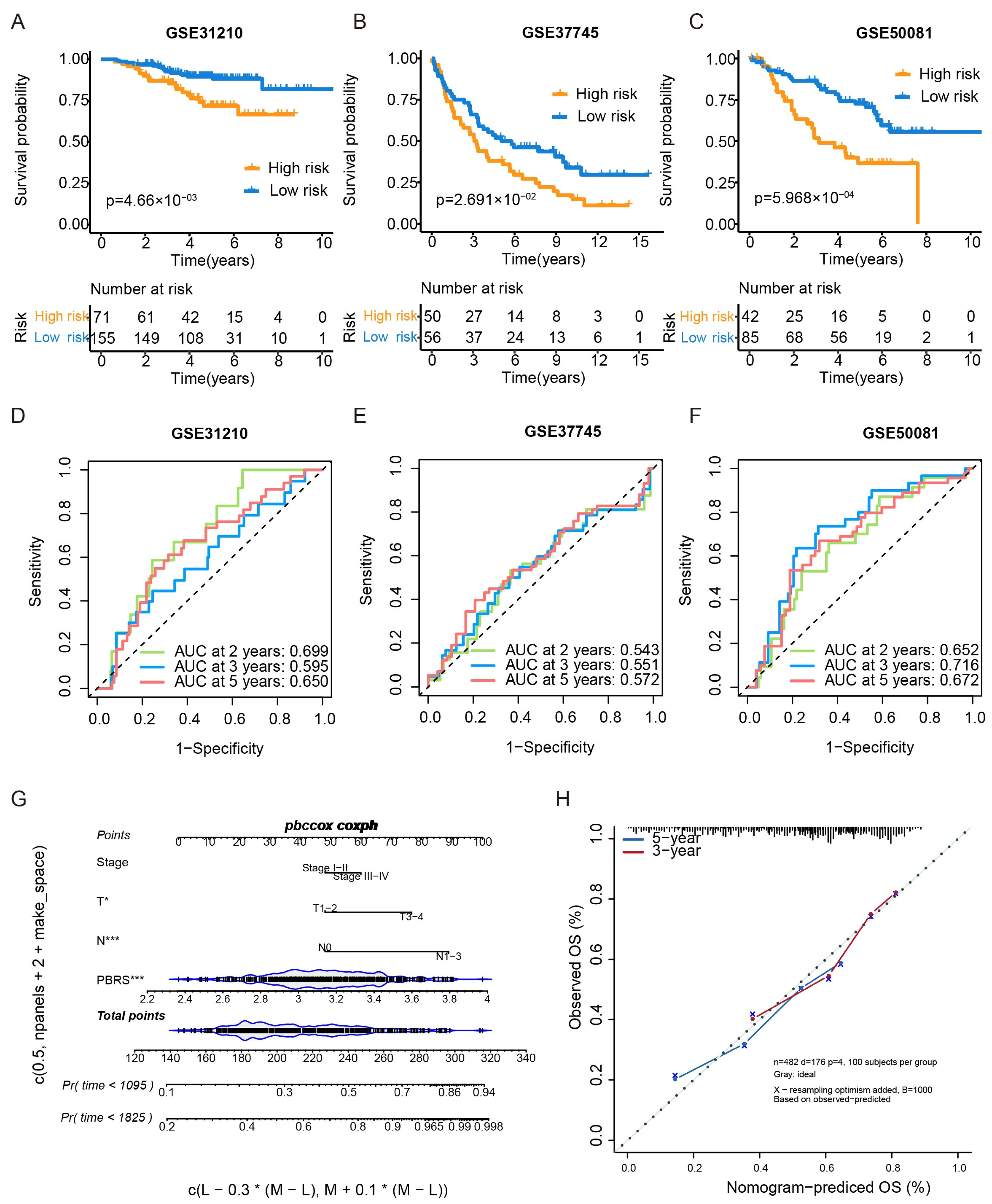
3.3. Validation of the PBRS
3.4. Correlation of PBRS with Biological Processes
3.5. PBRS Stratifies Immune Infiltration and TMB Heterogeneity in LUAD
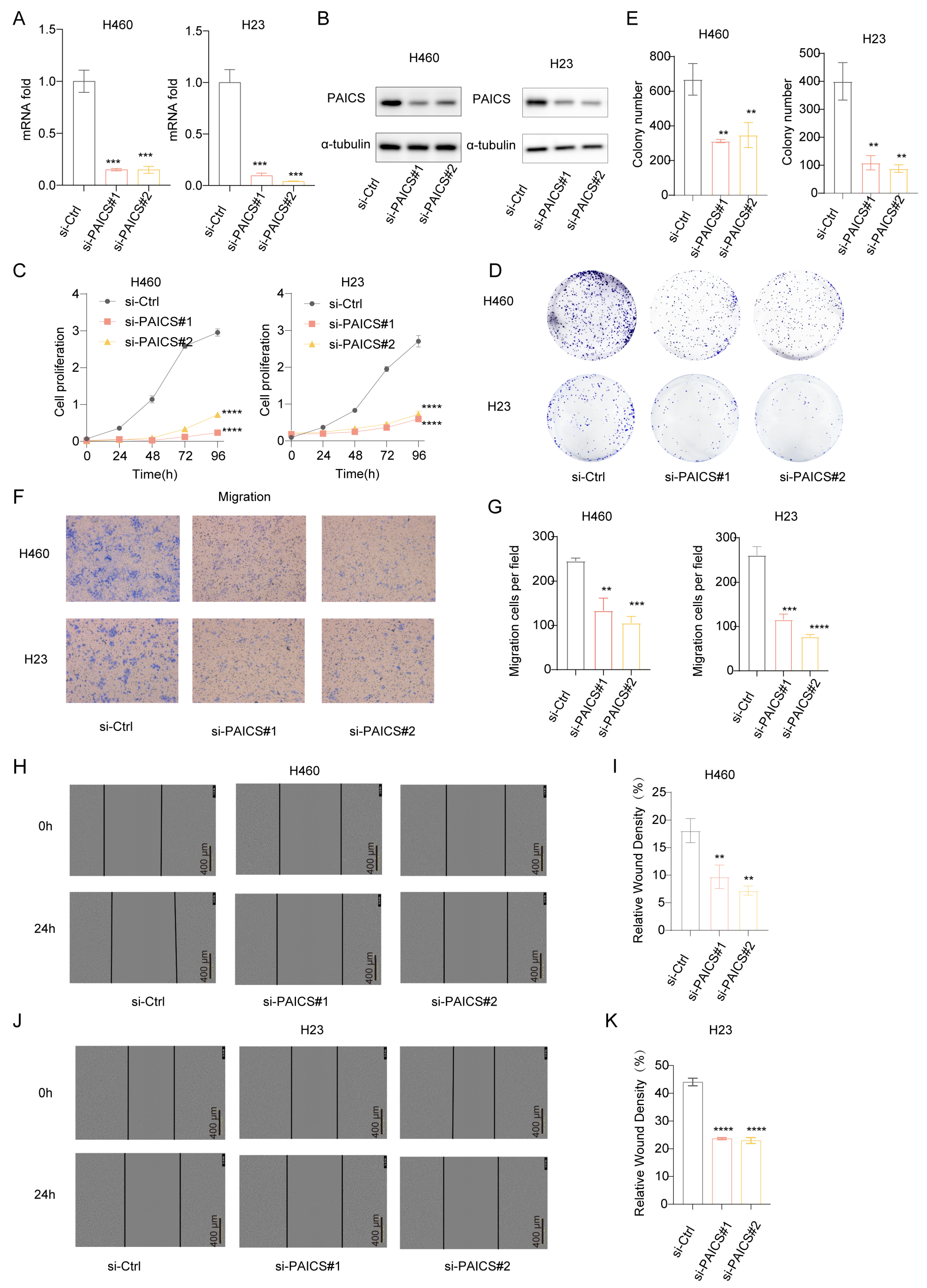
3.6. PAICS as a Key Driver of LUAD Malignancy
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global Cancer Statistics 2022: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef]
- Mazzilli, S.A.; Rahal, Z.; Rouhani, M.J.; Janes, S.M.; Kadara, H.; Dubinett, S.M.; Spira, A.E. Translating Premalignant Biology to Accelerate Non-Small-Cell Lung Cancer Interception. Nat. Rev. Cancer 2025, 25, 379–392. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhuang, X.; Pan, C.-H.; Yan, Y.; Thummalapalli, R.; Hallin, J.; Torborg, S.; Singhal, A.; Chang, J.C.; Manchado, E.; et al. Alveolar Differentiation Drives Resistance to KRAS Inhibition in Lung Adenocarcinoma. Cancer Discov. 2024, 14, 308–325. [Google Scholar] [CrossRef] [PubMed]
- Basil, M.C.; Katzen, J.; Engler, A.E.; Guo, M.; Herriges, M.J.; Kathiriya, J.J.; Windmueller, R.; Ysasi, A.B.; Zacharias, W.J.; Chapman, H.A.; et al. The Cellular and Physiological Basis for Lung Repair and Regeneration: Past, Present, and Future. Cell Stem Cell 2020, 26, 482–502. [Google Scholar] [CrossRef]
- Antonia, S.J.; Villegas, A.; Daniel, D.; Vicente, D.; Murakami, S.; Hui, R.; Kurata, T.; Chiappori, A.; Lee, K.H.; De Wit, M.; et al. Overall Survival with Durvalumab after Chemoradiotherapy in Stage III NSCLC. N. Engl. J. Med. 2018, 379, 2342–2350. [Google Scholar] [CrossRef]
- Spigel, D.R.; Faivre-Finn, C.; Gray, J.E.; Vicente, D.; Planchard, D.; Paz-Ares, L.; Vansteenkiste, J.F.; Garassino, M.C.; Hui, R.; Quantin, X.; et al. Five-Year Survival Outcomes from the PACIFIC Trial: Durvalumab After Chemoradiotherapy in Stage III Non-Small-Cell Lung Cancer. J. Clin. Oncol. 2022, 40, 1301–1311. [Google Scholar] [CrossRef]
- Camidge, D.R.; Pao, W.; Sequist, L.V. Acquired Resistance to TKIs in Solid Tumours: Learning from Lung Cancer. Nat. Rev. Clin. Oncol. 2014, 11, 473–481. [Google Scholar] [CrossRef]
- Qian, X.; Guo, X.; Li, T.; Hu, W.; Zhang, L.; Wu, C.; Ye, F. Efficacy of Immune Checkpoint Inhibitors in EGFR-Mutant NSCLC Patients with EGFR-TKI Resistance: A Systematic Review and Meta-Analysis. Front. Pharmacol. 2022, 13, 926890. [Google Scholar] [CrossRef]
- Hastings, K.; Yu, H.A.; Wei, W.; Sanchez-Vega, F.; DeVeaux, M.; Choi, J.; Rizvi, H.; Lisberg, A.; Truini, A.; Lydon, C.A.; et al. EGFR Mutation Subtypes and Response to Immune Checkpoint Blockade Treatment in Non-Small-Cell Lung Cancer. Ann. Oncol. 2019, 30, 1311–1320. [Google Scholar] [CrossRef]
- Tang, Y.; Fang, W.; Zhang, Y.; Hong, S.; Kang, S.; Yan, Y.; Chen, N.; Zhan, J.; He, X.; Qin, T.; et al. The Association between PD-L1 and EGFR Status and the Prognostic Value of PD-L1 in Advanced Non-Small Cell Lung Cancer Patients Treated with EGFR-TKIs. Oncotarget 2015, 6, 14209–14219. [Google Scholar] [CrossRef]
- Yin, J.; Ren, W.; Huang, X.; Deng, J.; Li, T.; Yin, Y. Potential Mechanisms Connecting Purine Metabolism and Cancer Therapy. Front. Immunol. 2018, 9, 1697. [Google Scholar] [CrossRef] [PubMed]
- Mullen, N.J.; Singh, P.K. Nucleotide Metabolism: A Pan-Cancer Metabolic Dependency. Nat. Rev. Cancer 2023, 23, 275–294. [Google Scholar] [CrossRef]
- Tran, D.H.; Kim, D.; Kesavan, R.; Brown, H.; Dey, T.; Soflaee, M.H.; Vu, H.S.; Tasdogan, A.; Guo, J.; Bezwada, D.; et al. De Novo and Salvage Purine Synthesis Pathways across Tissues and Tumors. Cell 2024, 187, 3602–3618.e20. [Google Scholar] [CrossRef]
- French, J.B.; Jones, S.A.; Deng, H.; Pedley, A.M.; Kim, D.; Chan, C.Y.; Hu, H.; Pugh, R.J.; Zhao, H.; Zhang, Y.; et al. Spatial Colocalization and Functional Link of Purinosomes with Mitochondria. Science 2016, 351, 733–737. [Google Scholar] [CrossRef] [PubMed]
- Ben-Sahra, I.; Hoxhaj, G.; Ricoult, S.J.H.; Asara, J.M.; Manning, B.D. mTORC1 Induces Purine Synthesis through Control of the Mitochondrial Tetrahydrofolate Cycle. Science 2016, 351, 728–733. [Google Scholar] [CrossRef]
- Wu, Z.; Bezwada, D.; Cai, F.; Harris, R.C.; Ko, B.; Sondhi, V.; Pan, C.; Vu, H.S.; Nguyen, P.T.; Faubert, B.; et al. Electron Transport Chain Inhibition Increases Cellular Dependence on Purine Transport and Salvage. Cell Metab. 2024, 36, 1504–1520. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, T.; Nakata, A.; Chen, X.; Nishi, K.; Meguro-Horike, M.; Sasaki, S.; Kita, K.; Horike, S.; Saitoh, K.; Kato, K.; et al. Cancer Stem-like Properties and Gefitinib Resistance Are Dependent on Purine Synthetic Metabolism Mediated by the Mitochondrial Enzyme MTHFD2. Oncogene 2019, 38, 2464–2481. [Google Scholar] [CrossRef]
- Geng, P.; Ye, F.; Dou, P.; Hu, C.; He, J.; Zhao, J.; Li, Q.; Bao, M.; Li, X.; Liu, X.; et al. HIF-1α-HPRT1 Axis Promotes Tumorigenesis and Gefitinib Resistance by Enhancing Purine Metabolism in EGFR-Mutant Lung Adenocarcinoma. J. Exp. Clin. Canc. Res. 2024, 43, 269. [Google Scholar] [CrossRef]
- Yang, L.; Li, A.; Yu, W.; Wang, H.; Zhang, L.; Wang, D.; Wang, Y.; Zhang, R.; Lei, Q.; Liu, Z.; et al. Blockade of Purine Metabolism Reverses Macrophage Immunosuppression and Enhances Anti-Tumor Immunity in Non-Small Cell Lung Cancer. Drug Resist. Updates 2025, 78, 101175. [Google Scholar] [CrossRef]
- Huo, A.; Xiong, X. PAICS as a Potential Target for Cancer Therapy Linking Purine Biosynthesis to Cancer Progression. Life Sci. 2023, 331, 122070. [Google Scholar] [CrossRef]
- Chakravarthi, B.V.S.K.; Goswami, M.T.; Pathi, S.S.; Dodson, M.; Chandrashekar, D.S.; Agarwal, S.; Nepal, S.; Hodigere Balasubramanya, S.A.; Siddiqui, J.; Lonigro, R.J.; et al. Expression and Role of PAICS, a De Novo Purine Biosynthetic Gene in Prostate Cancer. Prostate 2017, 77, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Hany, D.; Vafeiadou, V.; Picard, D. CRISPR-Cas9 Screen Reveals a Role of Purine Synthesis for Estrogen Receptor α Activity and Tamoxifen Resistance of Breast Cancer Cells. Sci. Adv. 2023, 9, eadd3685. [Google Scholar] [CrossRef]
- Meng, M.; Chen, Y.; Jia, J.; Li, L.; Yang, S. Knockdown of PAICS Inhibits Malignant Proliferation of Human Breast Cancer Cell Lines. Biol. Res. 2018, 51, 24. [Google Scholar] [CrossRef] [PubMed]
- Du, B.; Zhang, Z.; Di, W.; Xu, W.; Yang, L.; Zhang, S.; He, G.; Yang, R.; Wang, M. PAICS Is Related to Glioma Grade and Can Promote Glioma Growth and Migration. J. Cell. Mol. Med. 2021, 25, 7720–7733. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhu, L.; Mao, J.; Zheng, H.; Hu, Z.; Yang, S.; Mao, T.; Zhou, T.; Cao, P.; Wu, H.; et al. Genome-Scale CRISPR-Cas9 Screen Identifies PAICS as a Therapeutic Target for EGFR Wild-Type Non-Small Cell Lung Cancer. MedComm 2024, 5, e483. [Google Scholar] [CrossRef]
- Goswami, M.T.; Chen, G.; Chakravarthi, B.V.S.K.; Pathi, S.S.; Anand, S.K.; Carskadon, S.L.; Giordano, T.J.; Chinnaiyan, A.M.; Thomas, D.G.; Palanisamy, N.; et al. Role and Regulation of Coordinately Expressed de Novo Purine Biosynthetic Enzymes PPAT and PAICS in Lung Cancer. Oncotarget 2015, 6, 23445–23461. [Google Scholar] [CrossRef]
- Okayama, H.; Kohno, T.; Ishii, Y.; Shimada, Y.; Shiraishi, K.; Iwakawa, R.; Furuta, K.; Tsuta, K.; Shibata, T.; Yamamoto, S.; et al. Identification of Genes Upregulated in ALK-Positive and EGFR/KRAS/ALK-Negative Lung Adenocarcinomas. Cancer Res. 2012, 72, 100–111. [Google Scholar] [CrossRef]
- Botling, J.; Edlund, K.; Lohr, M.; Hellwig, B.; Holmberg, L.; Lambe, M.; Berglund, A.; Ekman, S.; Bergqvist, M.; Pontén, F.; et al. Biomarker Discovery in Non-Small Cell Lung Cancer: Integrating Gene Expression Profiling, Meta-Analysis, and Tissue Microarray Validation. Clin. Cancer Res. 2013, 19, 194–204. [Google Scholar] [CrossRef]
- Der, S.D.; Sykes, J.; Pintilie, M.; Zhu, C.-Q.; Strumpf, D.; Liu, N.; Jurisica, I.; Shepherd, F.A.; Tsao, M.-S. Validation of a Histology-Independent Prognostic Gene Signature for Early-Stage, Non-Small-Cell Lung Cancer Including Stage IA Patients. J. Thorac. Oncol. 2014, 9, 59–64. [Google Scholar] [CrossRef]
- Heagerty, P.J.; Lumley, T.; Pepe, M.S. Time-Dependent ROC Curves for Censored Survival Data and a Diagnostic Marker. Biometrics 2000, 56, 337–344. [Google Scholar] [CrossRef]
- The Gene Ontology Consortium. The Gene Ontology Resource: 20 Years and Still GOing Strong. Nucleic Acids Res. 2019, 47, D330–D338. [Google Scholar] [CrossRef] [PubMed]
- Mariathasan, S.; Turley, S.J.; Nickles, D.; Castiglioni, A.; Yuen, K.; Wang, Y.; Kadel , E.E., III; Koeppen, H.; Astarita, J.L.; Cubas, R.; et al. TGFβ Attenuates Tumour Response to PD-L1 Blockade by Contributing to Exclusion of T Cells. Nature 2018, 554, 544–548. [Google Scholar] [CrossRef]
- Hänzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene Set Variation Analysis for Microarray and RNA-Seq Data. BMC Bioinf. 2013, 14, 7. [Google Scholar] [CrossRef] [PubMed]
- Maeser, D.; Gruener, R.F.; Huang, R.S. oncoPredict: An R Package for Predicting in Vivo or Cancer Patient Drug Response and Biomarkers from Cell Line Screening Data. Brief. Bioinform. 2021, 22, bbab260. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Qin, Y.; Ji, S.; Shi, X.; Dai, W.; Fan, G.; Li, S.; Xu, W.; Liu, W.; Liu, M.; et al. MTAP Deficiency-Induced Metabolic Reprogramming Creates a Vulnerability to Cotargeting De Novo Purine Synthesis and Glycolysis in Pancreatic Cancer. Cancer Res. 2021, 81, 4964–4980. [Google Scholar] [CrossRef]
- Hansen, L.J.; Sun, R.; Yang, R.; Singh, S.X.; Chen, L.H.; Pirozzi, C.J.; Moure, C.J.; Hemphill, C.; Carpenter, A.B.; Healy, P.; et al. MTAP Loss Promotes Stemness in Glioblastoma and Confers Unique Susceptibility to Purine Starvation. Cancer Res. 2019, 79, 3383–3394. [Google Scholar] [CrossRef]
- Lv, Y.; Wang, X.; Li, X.; Xu, G.; Bai, Y.; Wu, J.; Piao, Y.; Shi, Y.; Xiang, R.; Wang, L. Nucleotide de Novo Synthesis Increases Breast Cancer Stemness and Metastasis via cGMP-PKG-MAPK Signaling Pathway. PLOS Biol. 2020, 18, e3000872. [Google Scholar] [CrossRef]
- Tabata, S.; Umemura, S.; Narita, M.; Udagawa, H.; Ishikawa, T.; Tsuboi, M.; Goto, K.; Ishii, G.; Tsuchihara, K.; Ochiai, A.; et al. Metabolic Hallmarks for Purine Nucleotide Biosynthesis in Small Cell Lung Carcinoma. Mol. Cancer Res. 2024, 22, 82–93. [Google Scholar] [CrossRef]
- Xu, X.; Wang, L.; Zang, Q.; Li, S.; Li, L.; Wang, Z.; He, J.; Qiang, B.; Han, W.; Zhang, R.; et al. Rewiring of Purine Metabolism in Response to Acidosis Stress in Glioma Stem Cells. Cell Death Dis. 2021, 12, 277. [Google Scholar] [CrossRef]
- Wang, Q.; Guan, Y.F.; Hancock, S.E.; Wahi, K.; Van Geldermalsen, M.; Zhang, B.K.; Pang, A.; Nagarajah, R.; Mak, B.; Freidman, N.; et al. Inhibition of Guanosine Monophosphate Synthetase (GMPS) Blocks Glutamine Metabolism and Prostate Cancer Growth. J. Pathol. 2021, 254, 135–146. [Google Scholar] [CrossRef]
- Huang, Z.; Xie, N.; Illes, P.; Di Virgilio, F.; Ulrich, H.; Semyanov, A.; Verkhratsky, A.; Sperlagh, B.; Yu, S.-G.; Huang, C.; et al. From Purines to Purinergic Signalling: Molecular Functions and Human Diseases. Signal Transduct. Target. Ther. 2021, 6, 162. [Google Scholar] [CrossRef]
- Keshet, R.; Lee, J.S.; Adler, L.; Iraqi, M.; Ariav, Y.; Lim, L.Q.J.; Lerner, S.; Rabinovich, S.; Oren, R.; Katzir, R.; et al. Targeting Purine Synthesis in ASS1-Expressing Tumors Enhances the Response to Immune Checkpoint Inhibitors. Nat. Cancer 2020, 1, 894–908. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Paila, U.D.; Teraoka, S.N.; Wright, J.A.; Huang, X.; Quinlan, A.R.; Gatti, R.A.; Concannon, P. Identification of ATIC as a Novel Target for Chemoradiosensitization. Int. J. Radiat. Oncol.*Biol.*Phys. 2018, 100, 162–173. [Google Scholar] [CrossRef]
- Karanika, S.; Karantanos, T.; Li, L.; Corn, P.G.; Thompson, T.C. DNA Damage Response and Prostate Cancer: Defects, Regulation and Therapeutic Implications. Oncogene 2015, 34, 2815–2822. [Google Scholar] [CrossRef]
- Huang, N.; Xu, C.; Deng, L.; Li, X.; Bian, Z.; Zhang, Y.; Long, S.; Chen, Y.; Zhen, N.; Li, G.; et al. PAICS Contributes to Gastric Carcinogenesis and Participates in DNA Damage Response by Interacting with Histone Deacetylase 1/2. Cell Death Dis. 2020, 11, 507. [Google Scholar] [CrossRef] [PubMed]
- Fok, J.H.L.; Ramos-Montoya, A.; Vazquez-Chantada, M.; Wijnhoven, P.W.G.; Follia, V.; James, N.; Farrington, P.M.; Karmokar, A.; Willis, S.E.; Cairns, J.; et al. AZD7648 Is a Potent and Selective DNA-PK Inhibitor that Enhances Radiation, Chemotherapy and Olaparib Activity. Nat. Commun. 2019, 10, 5065. [Google Scholar] [CrossRef]
- Wang, M.; Chen, S.; Wei, Y.; Wei, X. DNA-PK Inhibition by M3814 Enhances Chemosensitivity in Non-Small Cell Lung Cancer. Acta Pharm. Sin. B 2021, 11, 3935–3949. [Google Scholar] [CrossRef]
- Sen, T.; Rodriguez, B.L.; Chen, L.; Corte, C.M.D.; Morikawa, N.; Fujimoto, J.; Cristea, S.; Nguyen, T.; Diao, L.; Li, L.; et al. Targeting DNA Damage Response Promotes Anti-Tumor Immunity through STING-Mediated T-Cell Activation in Small Cell Lung Cancer. Cancer Discov. 2019, 9, 646–661. [Google Scholar] [CrossRef]
- Lang, L.; Tao, J.; Yang, C.; Li, W. Tumor Suppressive Role of microRNA-4731-5p in Breast Cancer through Reduction of PAICS-Induced FAK Phosphorylation. Cell Death Discov. 2022, 8, 154. [Google Scholar] [CrossRef]
- Hosea, R.; Hillary, S.; Naqvi, S.; Wu, S.; Kasim, V. The Two Sides of Chromosomal Instability: Drivers and Brakes in Cancer. Signal Transduct. Target. Ther. 2024, 9, 75. [Google Scholar] [CrossRef]
- Agarwal, S.; Chakravarthi, B.V.S.K.; Behring, M.; Kim, H.-G.; Chandrashekar, D.S.; Gupta, N.; Bajpai, P.; Elkholy, A.; Balasubramanya, S.A.H.; Hardy, C.; et al. PAICS, a Purine Nucleotide Metabolic Enzyme, Is Involved in Tumor Growth and the Metastasis of Colorectal Cancer. Cancers 2020, 12, 772. [Google Scholar] [CrossRef]
- Barfeld, S.J.; Fazli, L.; Persson, M.; Marjavaara, L.; Urbanucci, A.; Kaukoniemi, K.M.; Rennie, P.S.; Ceder, Y.; Chabes, A.; Visakorpi, T.; et al. Myc-Dependent Purine Biosynthesis Affects Nucleolar Stress and Therapy Response in Prostate Cancer. Oncotarget 2015, 6, 12587–12602. [Google Scholar] [CrossRef] [PubMed]
- Ren, B.; Cam, H.; Takahashi, Y.; Volkert, T.; Terragni, J.; Young, R.A.; Dynlacht, B.D. E2F Integrates Cell Cycle Progression with DNA Repair, Replication, and G2/M Checkpoints. Genes Dev. 2002, 16, 245–256. [Google Scholar] [CrossRef]
- Bansal, P.; Osman, D.; Gan, G.N.; Simon, G.R.; Boumber, Y. Recent Advances in Immunotherapy in Metastatic NSCLC. Front. Oncol. 2016, 6, 239. [Google Scholar] [CrossRef]
- Yang, Y.; Yang, F.; Huang, Z.; Li, Y.; Shi, H.; Sun, Q.; Ma, Y.; Wang, Y.; Zhang, Y.; Yang, S.; et al. T Cells, NK Cells, and Tumor-Associated Macrophages in Cancer Immunotherapy and the Current State of the Art of Drug Delivery Systems. Front. Immunol. 2023, 14, 1199173. [Google Scholar] [CrossRef]
- Palermo, B.; Franzese, O.; Frisullo, G.; D’Ambrosio, L.; Panetta, M.; Campo, G.; D’Andrea, D.; Sperduti, I.; De Nicola, F.; Goeman, F.; et al. CD28/PD1 Co-Expression: Dual Impact on CD8+ T Cells in Peripheral Blood and Tumor Tissue, and Its Significance in NSCLC Patients’ Survival and ICB Response. J. Exp. Clin. Cancer Res. 2023, 42, 287. [Google Scholar] [CrossRef] [PubMed]
- Getu, A.A.; Tigabu, A.; Zhou, M.; Lu, J.; Fodstad, Ø.; Tan, M. New Frontiers in Immune Checkpoint B7-H3 (CD276) Research and Drug Development. Mol. Cancer 2023, 22, 43. [Google Scholar] [CrossRef]
- Ren, S.; Tian, Q.; Amar, N.; Yu, H.; Rivard, C.J.; Caldwell, C.; Ng, T.L.; Tu, M.; Liu, Y.; Gao, D.; et al. The Immune Checkpoint, HVEM May Contribute to Immune Escape in Non-Small Cell Lung Cancer Lacking PD-L1 Expression. Lung Cancer 2018, 125, 115–120. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Chen, Z.; Bartman, C.R.; Xing, X.; Olszewski, K.; Rabinowitz, J.D. One-Carbon Unit Supplementation Fuels Purine Synthesis in Tumor-Infiltrating T Cells and Augments Checkpoint Blockade. Cell Chem. Biol. 2024, 31, 932–943.e8. [Google Scholar] [CrossRef]
- Li, S.; Yu, J.; Huber, A.; Kryczek, I.; Wang, Z.; Jiang, L.; Li, X.; Du, W.; Li, G.; Wei, S.; et al. Metabolism Drives Macrophage Heterogeneity in the Tumor Microenvironment. Cell Rep. 2022, 39, 110609. [Google Scholar] [CrossRef]
- Rowe, J.H.; Elia, I.; Shahid, O.; Gaudiano, E.F.; Sifnugel, N.E.; Johnson, S.; Reynolds, A.G.; Fung, M.E.; Joshi, S.; LaFleur, M.W.; et al. Formate Supplementation Enhances Anti-Tumor CD8+ T Cell Fitness and Efficacy of PD-1 Blockade. Cancer Discov. 2023, 13, 2566–2583. [Google Scholar] [CrossRef]
- Ni, J.; Liu, Q.; Xie, S.; Carlson, C.; Von, T.; Vogel, K.; Riddle, S.; Benes, C.; Eck, M.; Roberts, T.; et al. Functional Characterization of an Isoform-Selective Inhibitor of PI3K-P110β as a Potential Anticancer Agent. Cancer Discov. 2012, 2, 425–433. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Chen, D.; Liu, G.; Wu, Q.; Zhao, H.; Guo, D.; Jiang, X.; Li, M.; Meng, Y.; Yin, Y.; et al. PI3Kβ Functions as a Protein Kinase to Promote Cellular Protein O-GlcNAcylation and Acetyl-CoA Production for Tumor Growth. Mol. Cell 2025, 85, 1411–1425.e8. [Google Scholar] [CrossRef] [PubMed]
- Macioszek, S.; Dudzik, D.; Bartoszewski, R.; Stokowy, T.; Lambrechts, D.; Boeckx, B.; Wozniak, A.; Schöffski, P.; Markuszewski, M.J. Metabolomic and Transcriptomic Response to Imatinib Treatment of Gastrointestinal Stromal Tumour in Xenograft-Bearing Mice. Transl. Oncol. 2023, 30, 101632. [Google Scholar] [CrossRef]
- Zarou, M.M.; Rattigan, K.M.; Sarnello, D.; Shokry, E.; Dawson, A.; Ianniciello, A.; Dunn, K.; Copland, M.; Sumpton, D.; Vazquez, A.; et al. Inhibition of Mitochondrial Folate Metabolism Drives Differentiation through mTORC1 Mediated Purine Sensing. Nat. Commun. 2024, 15, 1931. [Google Scholar] [CrossRef]
- Chou, M.-C.; Wang, Y.-H.; Chen, F.-Y.; Kung, C.-Y.; Wu, K.-P.; Kuo, J.-C.; Chan, S.-J.; Cheng, M.-L.; Lin, C.-Y.; Chou, Y.-C.; et al. PAICS Ubiquitination Recruits UBAP2 to Trigger Phase Separation for Purinosome Assembly. Mol. Cell 2023, 83, 4123–4140.e12. [Google Scholar] [CrossRef]
- Niu, N.; Zeng, J.; Ke, X.; Zheng, W.; Fu, C.; Lv, S.; Fu, J.; Yu, Y. ATIC Facilitates Cell Growth and Migration by Upregulating Myc Expression in Lung Adenocarcinoma. Oncol. Lett. 2022, 23, 131. [Google Scholar] [CrossRef]
- Miller, J.C.; Blake, D.C.; Herzog, C.R. Adenylosuccinate Synthetase 1 Gene Is a Novel Target of Deletion in Lung Adenocarcinoma. Mol. Carcinog. 2009, 48, 1116–1122. [Google Scholar] [CrossRef]
- Su, X.; Feng, C.; Wang, S.; Shi, L.; Gu, Q.; Zhang, H.; Lan, X.; Zhao, Y.; Qiang, W.; Ji, M.; et al. The Noncoding RNAs SNORD50A and SNORD50B-Mediated TRIM21-GMPS Interaction Promotes the Growth of P53 Wild-Type Breast Cancers by Degrading P53. Cell Death Differ. 2021, 28, 2450–2464. [Google Scholar] [CrossRef] [PubMed]
- Pey, J.; San José-Eneriz, E.; Ochoa, M.C.; Apaolaza, I.; De Atauri, P.; Rubio, A.; Cendoya, X.; Miranda, E.; Garate, L.; Cascante, M.; et al. In-Silico Gene Essentiality Analysis of Polyamine Biosynthesis Reveals APRT as a Potential Target in Cancer. Sci. Rep. 2017, 7, 14358. [Google Scholar] [CrossRef]
- Wang, L.; Wang, Y.; Han, N.; Wang, X.; Ruan, M. HPRT Promotes Proliferation and Metastasis in Head and Neck Squamous Cell Carcinoma through Direct Interaction with STAT3. Exp. Cell Res. 2021, 399, 112424. [Google Scholar] [CrossRef]
- Ali, E.S.; Sahu, U.; Villa, E.; O’Hara, B.P.; Gao, P.; Beaudet, C.; Wood, A.W.; Asara, J.M.; Ben-Sahra, I. ERK2 Phosphorylates PFAS to Mediate Posttranslational Control of De Novo Purine Synthesis. Mol. Cell 2020, 78, 1178–1191.e6. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, J.; Chen, Q.-A.; Yang, Y.; Zhang, L.; Lin, W.; Hong, Y.; Gao, Y. PAICS-Driven Purine Biosynthesis and Its Prognostic Implications in Lung Adenocarcinoma: A Novel Risk Stratification Model and Therapeutic Insights. Curr. Issues Mol. Biol. 2025, 47, 366. https://doi.org/10.3390/cimb47050366
Liu J, Chen Q-A, Yang Y, Zhang L, Lin W, Hong Y, Gao Y. PAICS-Driven Purine Biosynthesis and Its Prognostic Implications in Lung Adenocarcinoma: A Novel Risk Stratification Model and Therapeutic Insights. Current Issues in Molecular Biology. 2025; 47(5):366. https://doi.org/10.3390/cimb47050366
Chicago/Turabian StyleLiu, Jinhui, Qi-An Chen, Yannan Yang, Lin Zhang, Weihao Lin, Yuheng Hong, and Yibo Gao. 2025. "PAICS-Driven Purine Biosynthesis and Its Prognostic Implications in Lung Adenocarcinoma: A Novel Risk Stratification Model and Therapeutic Insights" Current Issues in Molecular Biology 47, no. 5: 366. https://doi.org/10.3390/cimb47050366
APA StyleLiu, J., Chen, Q.-A., Yang, Y., Zhang, L., Lin, W., Hong, Y., & Gao, Y. (2025). PAICS-Driven Purine Biosynthesis and Its Prognostic Implications in Lung Adenocarcinoma: A Novel Risk Stratification Model and Therapeutic Insights. Current Issues in Molecular Biology, 47(5), 366. https://doi.org/10.3390/cimb47050366





