Krüppel-like Factor 4-Deficient Cells Are Sensitive to Etoposide-Induced DNA Damage
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Drug Treatment
2.2. Plasmid Transfection
2.3. Western Blot Analysis
2.4. Immunostaining
2.5. Trypan Blue Assay
2.6. Statistical Analysis
3. Results
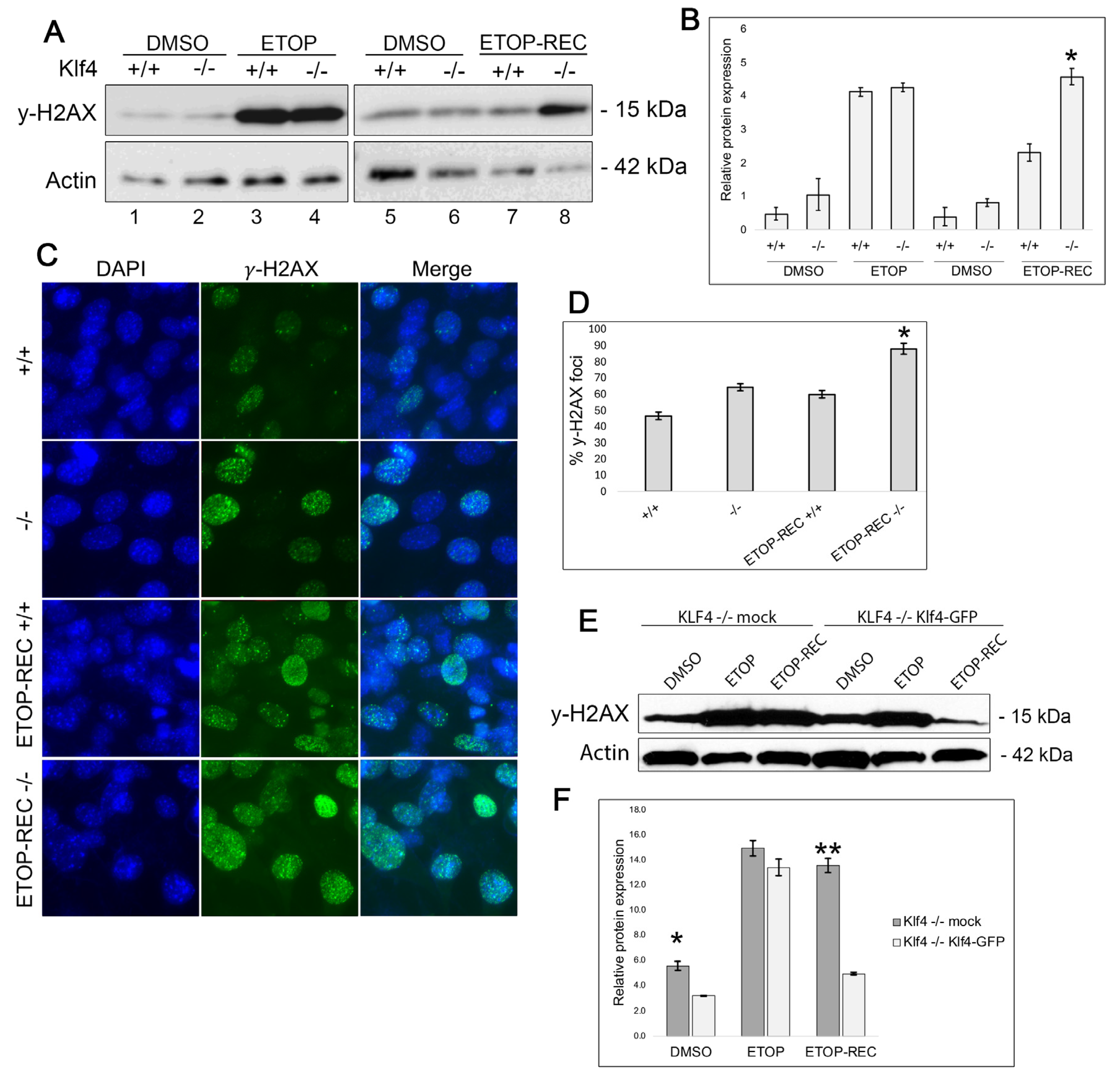
3.1. Klf4+/+ MEFs Display Less DNA Damage After Etoposide Treatment
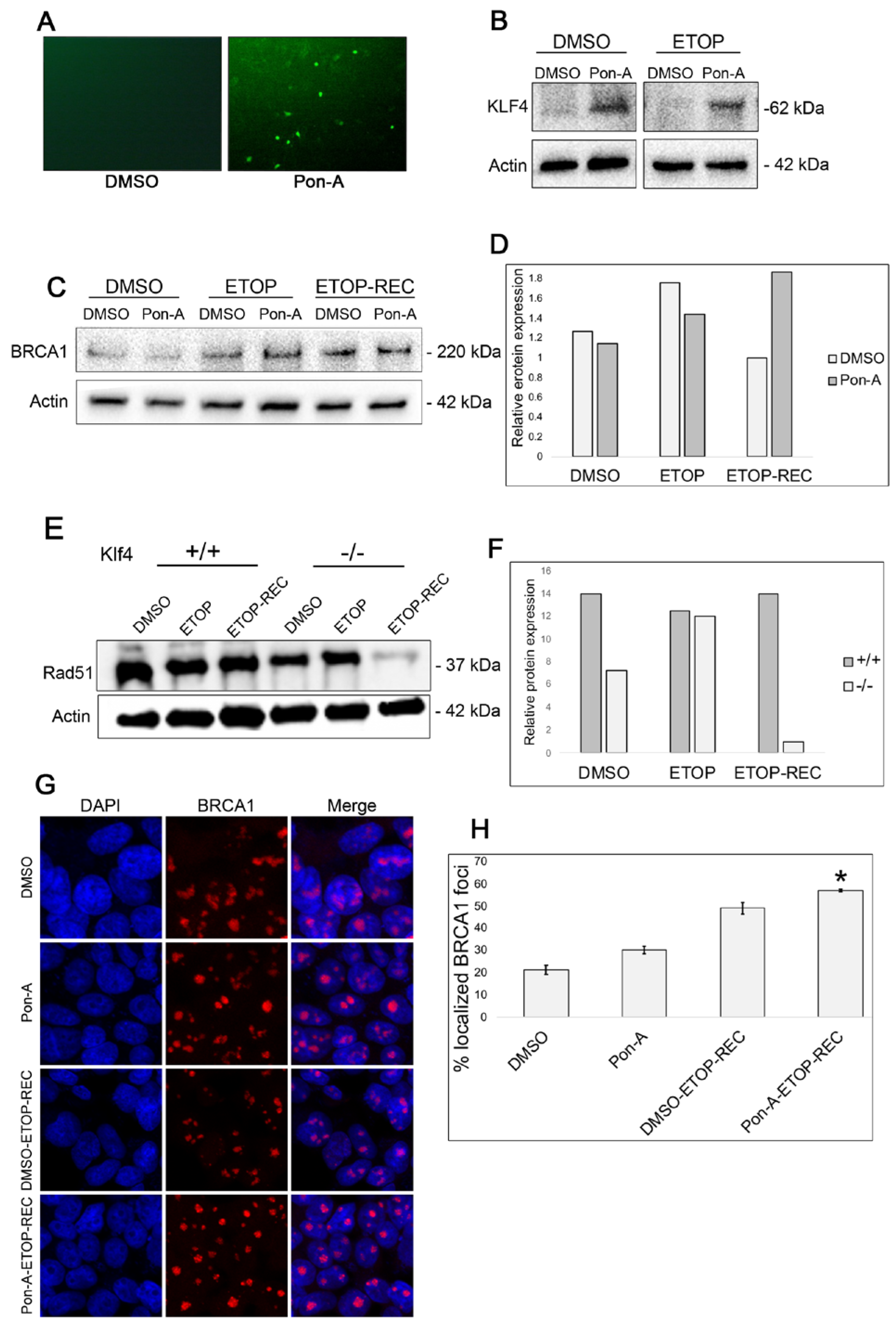
3.2. RKO Cells Expressing KLF4 Show Increased BRCA1 24 h After Etoposide-Induced DNA Damage
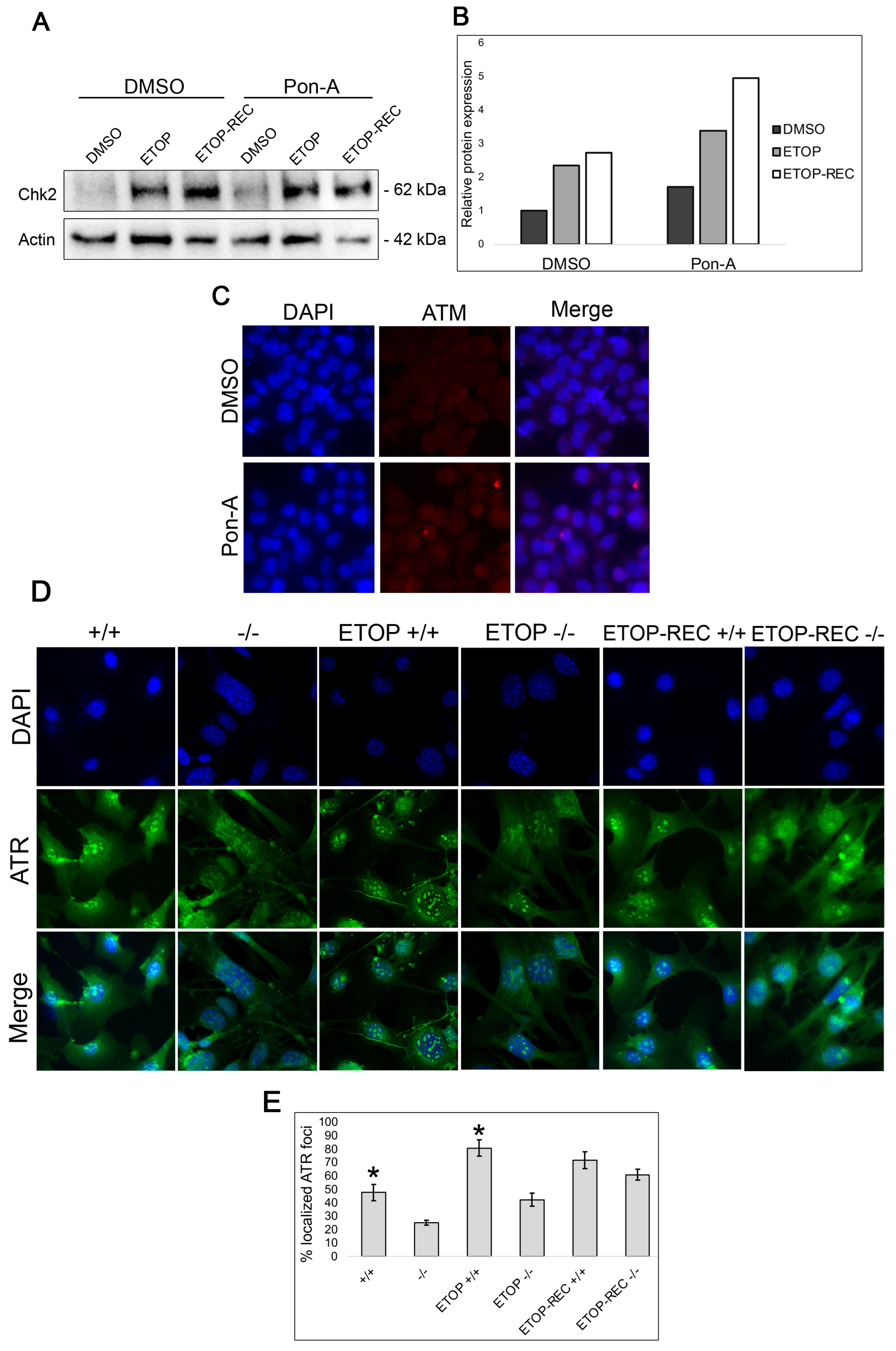
3.3. Genes Involved in DNA Repair Are Upregulated in Klf4 Expressing Cells
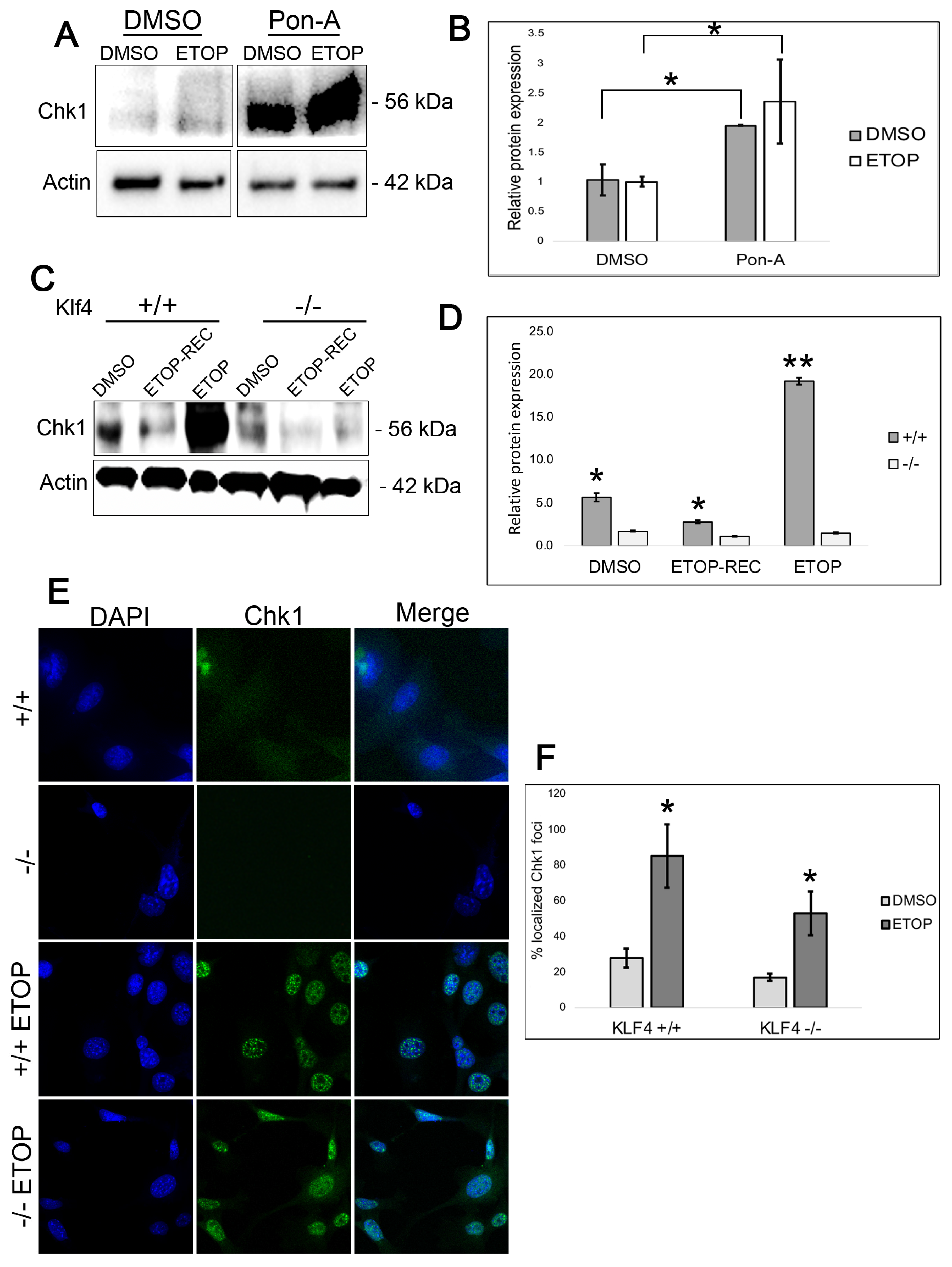
3.4. Chk1 Expression Is Higher at the Basal Level and Immediately After Etoposide DNA Damage in KLF4-Expressing Cells
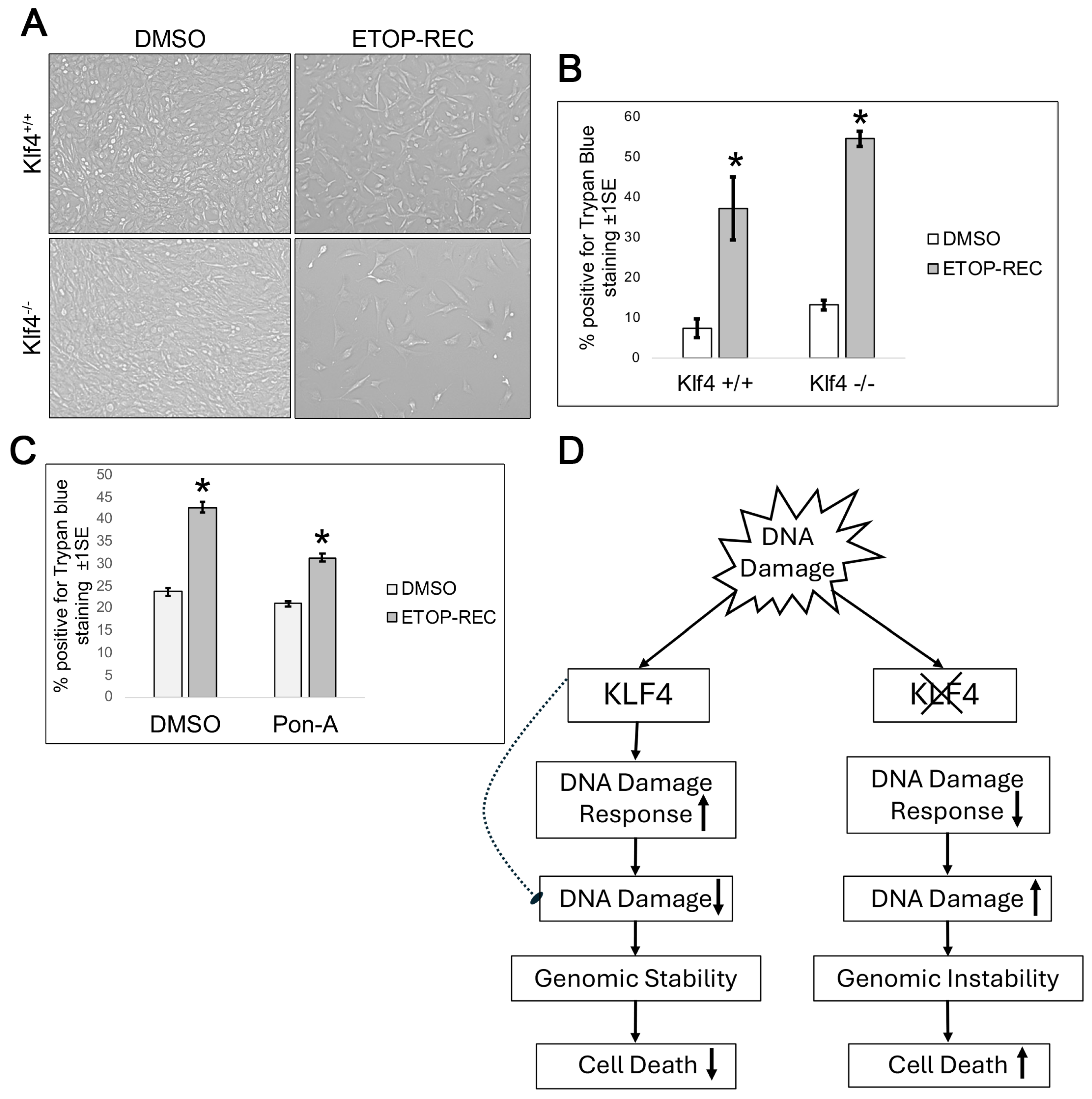
3.5. Cells Wild Type for Klf4 Experience Less Cell Death than Cells Null for Klf4
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, C.; DeRoo, E.P.; Stecyk, C.; Wolsey, M.; Szuchnicki, M.; Hagos, E.G. Impaired autophagy in mouse embryonic fibroblasts null for Krüppel-like factor 4 promotes DNA damage and increases apoptosis upon serum starvation. Mol. Cancer 2015, 14, 101. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; La Rosa, S.; Hagos, E.G. Oxidative DNA damage causes premature senescence in mouse embryonic fibroblasts deficient for Kruppel-like factor 4. Mol. Carcinog. 2014, 54, 889–899. [Google Scholar] [CrossRef] [PubMed]
- Rosencrans, W.; Walsh, Z.; Houerbi, N.; Blum, A.; Belew, A.; Chernak, B.; Liu, C.; Trazo, A.; Olson, A.; Brauer, P.; et al. Cells deficient for Krüppel-Like Factor 4 exhibit mitochondrial dysfunction and impaired mitophagy. Eur. J. Cell Biol. 2020, 99, 151061. [Google Scholar] [CrossRef]
- Li, Z.-Y.; Zhu, Y.-X.; Chen, J.-R.; Chang, X.; Xie, Z.-Z. The role of KLF transcription factor in the regulation of cancer progression. Biomed. Pharmacother. 2023, 162, 114661. [Google Scholar] [CrossRef] [PubMed]
- Ghaleb, A.M.; Elkarim, E.A.; Bialkowska, A.B.; Yang, V.W. KLF4 suppresses tumor formation in genetic and pharmacological mouse models of colonic tumorigenesis. Mol. Cancer Res. 2016, 14, 385–396. [Google Scholar] [CrossRef]
- Yang, V.W.; Liu, Y.; Kim, J.; Shroyer, K.R.; Bialkowska, A.B. Increased genetic instability and accelerated progression of colitis-associated colorectal cancer through intestinal epithelium-specific deletion of Klf4. Mol. Cancer Res. 2019, 17, 165–176. [Google Scholar] [CrossRef]
- Zhai, F.; Cao, C.; Zhang, L.; Zhang, J. miR-543 promotes colorectal cancer proliferation and metastasis by targeting Klf4. Oncotarget 2017, 8, 59246–59256. [Google Scholar] [CrossRef]
- He, Z.; He, J.; Xie, K. KLF4 transcription factor in tumorigenesis. Cell Death Discov. 2023, 9, 118. [Google Scholar] [CrossRef]
- Dang, D.T.; Chen, X.; Feng, J.; Torbenson, M.; Dang, L.H.; Yang, V.W. Overexpression of Krüppel-like factor 4 in the human colon cancer cell line RKO leads to reduced tumorigenicity. Oncogene 2003, 22, 3424–3430. [Google Scholar] [CrossRef]
- El-Karim, E.A.; Hagos, E.G.; Ghaleb, A.M.; Yu, B.; Yang, V.W. Krüppel-like factor 4 regulates genetic stability in mouse embryonic fibroblasts. Mol. Cancer 2013, 12, 1. [Google Scholar] [CrossRef]
- Yoon, H.S.; Chen, X.; Yang, V.W. Krüppel-like factor 4 mediates p53-dependent G1/S cell cycle arrest in response to DNA damage. J. Biol. Chem. 2003, 278, 2101–2105. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Hisamuddin, I.M.; Nandan, M.O.; Babbin, B.A.; Lamb, N.E.; Yang, V.W. Identification of Krüppel-like factor 4 as a potential tumor suppressor gene in colorectal cancer. Oncogene 2004, 23, 395–402. [Google Scholar] [CrossRef]
- Hagos, E.G.; Ghaleb, A.M.; Dalton, W.B.; Bialkowska, A.B.; Yang, V.W. Mouse embryonic fibroblasts null for the Krüppel-like factor 4 gene are genetically unstable. Oncogene 2009, 28, 1197–1205. [Google Scholar] [CrossRef][Green Version]
- Mah, L.J.; El-Osta, A.; Karagiannis, T.C. gammaH2AX: A sensitive molecular marker of DNA damage and repair. Leukemia 2010, 24, 679–686. [Google Scholar] [CrossRef] [PubMed]
- Thielhelm, T.P.; Goncalves, S.; Welford, S.; Mellon, E.A.; Bracho, O.; Estivill, M.; Brown, C.; Morcos, J.; Ivan, M.E.; Telischi, F.; et al. Primary vestibular schwannoma cells activate P21 and RAD51-associated DNA repair following radiation-induced DNA damage. Otol. Neurotol. 2021, 42, 1600–1608. [Google Scholar] [CrossRef]
- Fan, Z.; Luo, H.; Zhou, J.; Wang, F.; Zhang, W.; Wang, J.; Li, S.; Lai, Q.; Xu, Y.; Wang, G.; et al. Checkpoint kinase-1 inhibition and etoposide exhibit a strong synergistic anticancer effect on chronic myeloid leukemia cell line K562 by impairing homologous recombination DNA damage repair. Oncol. Rep. 2020, 44, 2152–2164. [Google Scholar] [CrossRef] [PubMed]
- Bartek, J.; Lukas, J. Chk1 and CHK2 kinases in checkpoint control and cancer. Cancer Cell 2003, 3, 421–429. [Google Scholar] [CrossRef]
- Marín, B.G.; Calderón-Segura, M.E.; Sekelsky, J. ATM/CHK2 and ATR/Chk1 pathways respond to DNA damage induced by Movento® 240SC and Envidor® 240SC keto-enol insecticides in the germarium of Drosophila melanogaster. Toxics 2023, 11, 754. [Google Scholar] [CrossRef]
- Reinhardt, H.C.; Yaffe, M.B. Kinases that control the cell cycle in response to DNA damage: Chk1, CHK2, and MK2. Curr. Opin. Cell Biol. 2009, 21, 245–255. [Google Scholar] [CrossRef]
- Zuco, V.; Benedetti, V.; Zunino, F. ATM- and ATR-mediated response to DNA damage induced by a novel camptothecin, ST1968. Cancer Lett. 2010, 292, 186–196. [Google Scholar] [CrossRef]
- Wu, X.; Xu, S.; Wang, P.; Wang, Z.-Q.; Chen, H.; Xu, X.; Peng, B. ASPM promotes ATR-CHK1 activation and stabilizes stalled replication forks in response to replication stress. Proc. Natl. Acad. Sci. USA 2022, 119, e2207948119. [Google Scholar] [CrossRef] [PubMed]
- Treszezamsky, A.D.; Kachnic, L.A.; Feng, Z.; Zhang, J.; Tokadjian, C.; Powell, S.N. BRCA1- and BRCA2-deficient cells are sensitive to etoposide-induced DNA double-strand breaks via topoisomerase II. Cancer Res. 2007, 67, 7078–7081. [Google Scholar] [CrossRef]
- Menendez, D.; Anand, J.R.; Murphy, C.C.; Bell, W.J.; Fu, J.; Slepushkina, N.; Buehler, E.; Martin, S.E.; Lal-Nag, M.; Nitiss, J.L.; et al. Etoposide-induced DNA damage is increased in p53 mutants: Identification of ATR and other genes that influence effects of p53 mutations on TOP2-induced cytotoxicity. Oncotarget 2022, 13, 332–346. [Google Scholar] [CrossRef] [PubMed]
- Durinikova, E.; Reilly, N.M.; Buzo, K.; Mariella, E.; Chilà, R.; Lorenzato, A.; Dias, J.M.L.; Grasso, G.; Pisati, F.; Lamba, S.; et al. Targeting the DNA Damage Response Pathways and Replication Stress in Colorectal Cancer. Clin. Cancer Res. 2022, 28, 3874–3889. [Google Scholar] [CrossRef]
- Rossi, R.; Lidonnici, M.R.; Soza, S.; Biamonti, G.; Montecucco, A. The dispersal of replication proteins after etoposide treatment requires the cooperation of Nbs1 with the ataxia telangiectasia Rad3-related/Rhk1 pathway. Cancer Res. 2006, 66, 1675–1683. [Google Scholar] [CrossRef]
- Ponasterone A—Insect Steroid Hormones and Gene Switching. Available online: https://agscientific.com/blog/ponasterone-a-gene-switching.html (accessed on 15 February 2024).
- Noble, W.S. How does multiple testing correction work? Nat. Biotechnol. 2009, 27, 1135–1137. [Google Scholar] [CrossRef] [PubMed]
- Savage, K.I.; Gorski, J.J.; Barros, E.M.; Irwin, G.W.; Manti, L.; Powell, A.J.; Pellagatti, A.; Lukashchuk, N.; McCance, D.J.; McCluggage, W.G.; et al. Identification of a BRCA1-mRNA splicing complex required for efficient DNA repair and maintenance of genomic stability. Mol. Cell 2014, 54, 445–459. [Google Scholar] [CrossRef]
- Choi, E.-H.; Yoon, S.; Hahn, Y.; Kim, K.P. Cellular dynamics of Rad51 and Rad54 in response to postreplicative stress and DNA damage in HeLa cells. Mol. Cells 2017, 40, 143–150. [Google Scholar] [CrossRef]
- Scully, R.; Chen, J.; Plug, A.; Xiao, Y.; Weaver, D.; Feunteun, J.; Ashley, T.; Livingston, D.M. Association of BRCA1 with RAD51 in mitotic and meiotic cells. Cell 1997, 88, 265–275. [Google Scholar] [CrossRef]
- Zhang, Y.; Hunter, T. Roles of Chk1 in cell biology and cancer therapy. Int. J. Cancer 2014, 134, 1013–1023. [Google Scholar] [CrossRef]
- Gorgoulis, V.G.; Halazonetis, T.D.; Negrini, S. Genomic instability—An evolving hallmark of cancer. Nat. Rev. Mol. Cell Biol. 2010, 11, 220–228. [Google Scholar]
- Zhou, Q.; Hong, Y.; Zhan, Q. Role for Kruppel-like factor 4 in determining the outcome of p53 response to DNA damage. Cancer Res. 2009, 69, 8284–8292. [Google Scholar] [CrossRef] [PubMed]
- Panier, S.; Boulton, S.J. Double-strand break repair: 53BP1 comes into focus. Nat. Rev. Mol. Cell Biol. 2014, 15, 7–18. [Google Scholar] [CrossRef]
- Zhu, W.; Li, X.; Zhang, Y.; Tang, F.; Xu, W. KLF4 facilitates chromatin accessibility remodeling in porcine early embryos. Sci. China Life Sci. 2024, 67, 96–112. [Google Scholar] [CrossRef]
- Yoon, H.S.; Ghaleb, A.M.; Nandan, M.O.; Hisamuddin, I.M.; Dalton, W.B.; Yang, V.W. Krüppel-like factor 4 prevents centrosome amplification following γ-irradiation-induced DNA damage. Oncogene 2005, 24, 4017–4025. [Google Scholar] [CrossRef]
- Deng, C.-X. BRCA1: Cell cycle checkpoint, genetic instability, DNA damage response and cancer evolution. Nucleic Acids Res. 2006, 34, 1416–1426. [Google Scholar] [CrossRef] [PubMed]
- Stok, C.; Kok, Y.P.; van den Tempel, N.; van Vugt, M.A.T.M. Shaping the BRCAness mutational landscape by alternative double-strand break repair, replication stress and mitotic aberrancies. Nucleic Acids Res. 2021, 49, 4239–4257. [Google Scholar] [CrossRef]
- Yoshida, K.; Miki, Y. Role of BRCA1 and BRCA2 as regulators of DNA repair, transcription, and cell cycle in response to DNA damage. Cancer Sci. 2004, 95, 866–871. [Google Scholar] [CrossRef]
- Zhou, Z.; Huang, F.; Shrivastava, I.; Zhu, R.; Luo, A.; Hottiger, M.; Bahar, I.; Liu, Z.; Cristofanilli, M.; Wan, Y. New insight into the significance of KLF4 PARylation in genome stability, carcinogenesis, and therapy. EMBO Mol. Med. 2020, 12, e12391. [Google Scholar] [CrossRef]
- Montecucco, A.; Zanetta, F.; Biamonti, G. Molecular mechanisms of etoposide. Excli. J. 2015, 14, 95–108. [Google Scholar]
- Narayanaswamy, P.B.; Tkachuk, S.; Haller, H.; Dumler, I.; Kiyan, Y. Chk1 and RAD51 activation after DNA damage is regulated via urokinase receptor/TLR4 signaling. Cell Death Dis. 2016, 7, e2363. [Google Scholar] [CrossRef] [PubMed]
- Yao, Q.; Weigel, B.; Kersey, J. Synergism between etoposide and 17-AAG in leukemia cells: Critical roles for Hsp90, FLT3, topoisomerase II, Chk1, and RAD51. Clin. Cancer Res. 2007, 13, 1591–1600. [Google Scholar] [CrossRef] [PubMed]
- Yarden, R.I.; Pardo-Reoyo, S.; Sgagias, M.; Cowan, K.H.; Brody, L.C. BRCA1 regulates the G2/M checkpoint by activating Chk1 kinase upon DNA damage. Nat. Genet. 2002, 30, 285–289. [Google Scholar] [CrossRef]
- Meyer, F.; Becker, S.; Classen, S.; Parplys, A.C.; Mansour, W.Y.; Riepen, B.; Timm, S.; Ruebe, C.; Jasin, M.; Wikman, H.; et al. Prevention of DNA replication stress by Chk1 leads to chemoresistance despite a DNA repair defect in homologous recombination in breast cancer. Cells 2020, 9, 238. [Google Scholar] [CrossRef]
- Théard, D.; Coisy, M.; Ducommun, B.; Concannon, P.; Darbon, J.-M. Etoposide and Adriamycin but not Genistein can activate the checkpoint kinase Chk2 independently of ATM/ATR. Biochem. Biophys. Res. Commun. 2001, 289, 1199–1204. [Google Scholar] [CrossRef]
- Smith, J.; Mun Tho, L.; Xu, N.; Gillespie, D.A. The ATM–Chk2 and ATR–Chk1 pathways in DNA damage signaling and cancer. Adv. Cancer Res. 2010, 108, 73–112. [Google Scholar] [PubMed]
- Choi, H.; Roh, J. Role of KLF4 in the regulation of apoptosis and cell cycle in rat granulosa cells during the periovulatory period. Int. J. Mol. Sci. 2018, 20, 87. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Johns, D.C.; Geiman, D.E.; Marban, E.; Dang, D.T.; Hamlin, G.; Sun, R.; Yang, V.W. Krüppel-like factor 4 (gut-enriched krüppel-like factor) inhibits cell proliferation by blocking G1/S progression of the cell cycle. J. Biol. Chem. 2001, 276, 30423–30428. [Google Scholar] [CrossRef]
- Li, S.; Wang, L.; Wang, Y.; Zhang, C.; Hong, Z.; Han, Z. The synthetic lethality of targeting cell cycle checkpoints and PARPs in cancer treatment. J. Hematol. Oncol. 2022, 15, 147. [Google Scholar] [CrossRef]
- Weber, A.M.; Ryan, A.J. ATM and ATR as therapeutic targets in cancer. Pharmacol. Ther. 2015, 149, 124–138. [Google Scholar] [CrossRef]
- Evans, P.M.; Liu, C. Roles of Krüppel-like factor 4 in normal homeostasis, cancer and stem cells. Acta Biochim. Biophys. Sin. 2008, 40, 554–564. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rubinstein, M.H.; Conroy, A.; Pezzuto, E.L.; Al Qoronz, H.; Wertimer, P.; Hagos, E.G. Krüppel-like Factor 4-Deficient Cells Are Sensitive to Etoposide-Induced DNA Damage. Curr. Issues Mol. Biol. 2025, 47, 217. https://doi.org/10.3390/cimb47040217
Rubinstein MH, Conroy A, Pezzuto EL, Al Qoronz H, Wertimer P, Hagos EG. Krüppel-like Factor 4-Deficient Cells Are Sensitive to Etoposide-Induced DNA Damage. Current Issues in Molecular Biology. 2025; 47(4):217. https://doi.org/10.3390/cimb47040217
Chicago/Turabian StyleRubinstein, Maxwell H., Aidan Conroy, Elisabeth L. Pezzuto, Hadeel Al Qoronz, Patrick Wertimer, and Engda G. Hagos. 2025. "Krüppel-like Factor 4-Deficient Cells Are Sensitive to Etoposide-Induced DNA Damage" Current Issues in Molecular Biology 47, no. 4: 217. https://doi.org/10.3390/cimb47040217
APA StyleRubinstein, M. H., Conroy, A., Pezzuto, E. L., Al Qoronz, H., Wertimer, P., & Hagos, E. G. (2025). Krüppel-like Factor 4-Deficient Cells Are Sensitive to Etoposide-Induced DNA Damage. Current Issues in Molecular Biology, 47(4), 217. https://doi.org/10.3390/cimb47040217




