How Does Breast Cancer Heterogeneity Determine Changes in Tumor Marker Levels in Saliva?
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
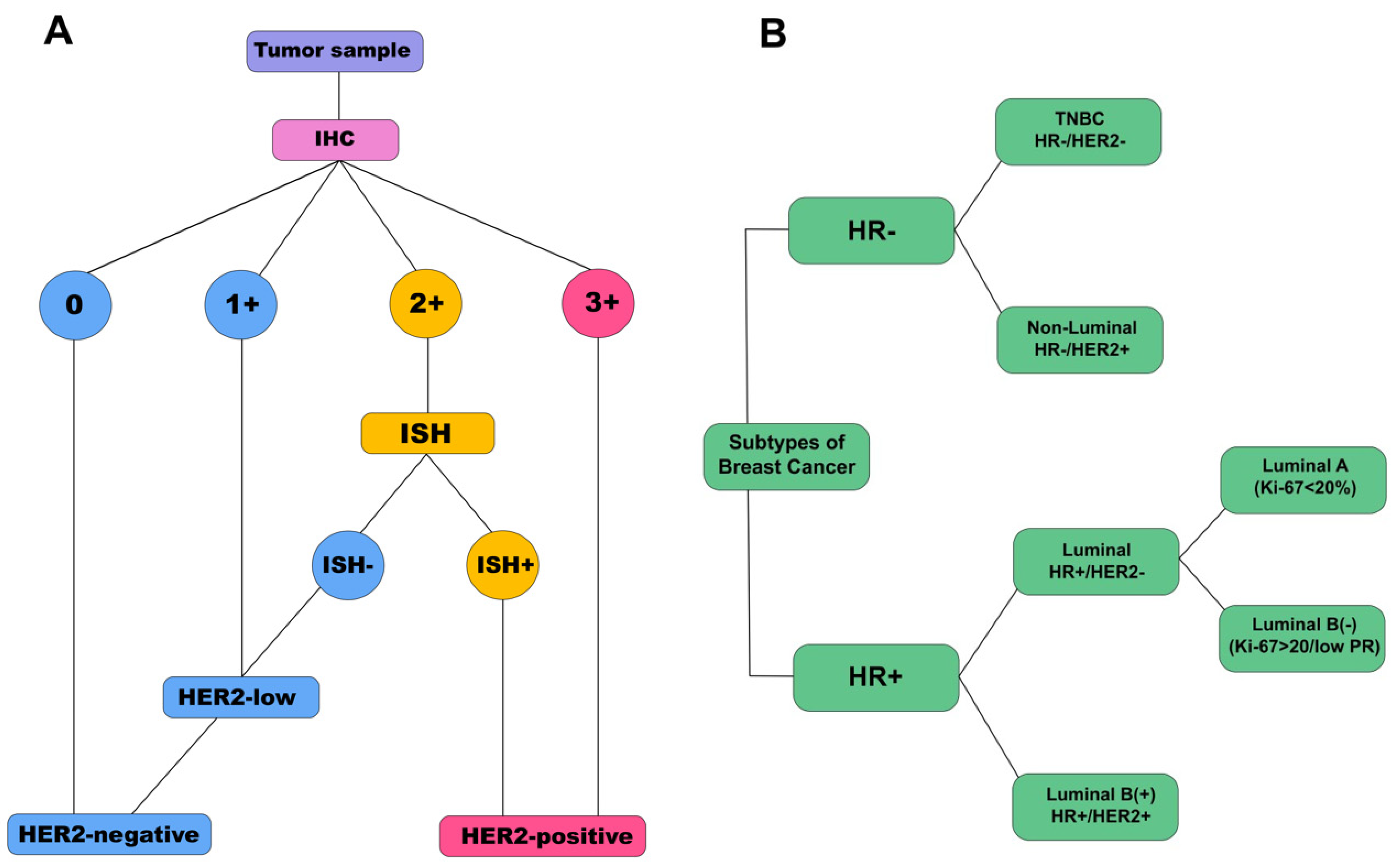
2.2. Immunohistochemical Analysis
2.3. Collection, Storage, and Pre-Treatment of Saliva Samples
2.4. Saliva Enzyme Immunoassay
2.5. Statistical Analysis
3. Results
3.1. Salivary Tumor Marker Levels in Breast Cancer and Healthy Controls
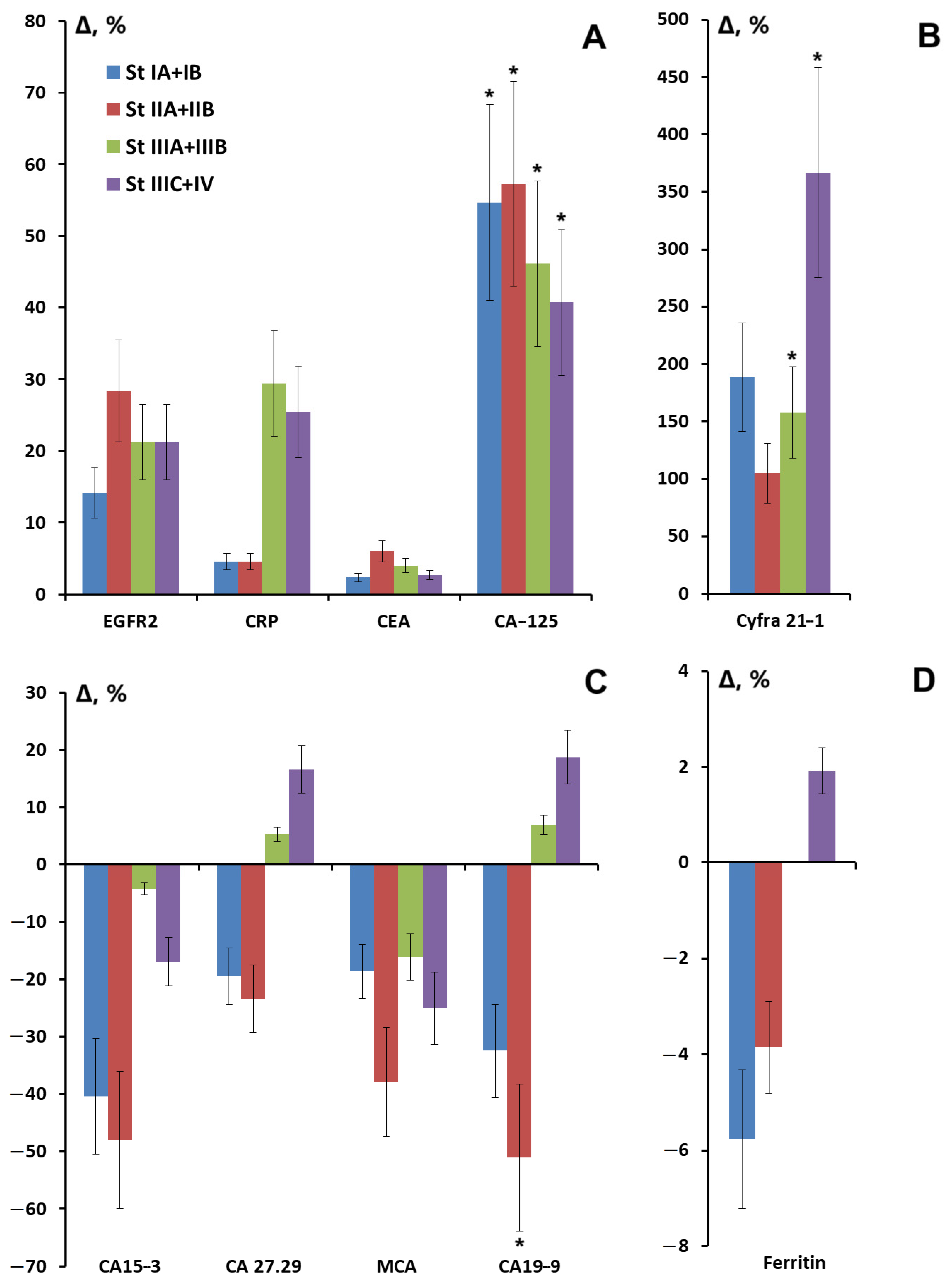
3.2. Salivary Tumor Marker Levels Depending on Breast Cancer Stage
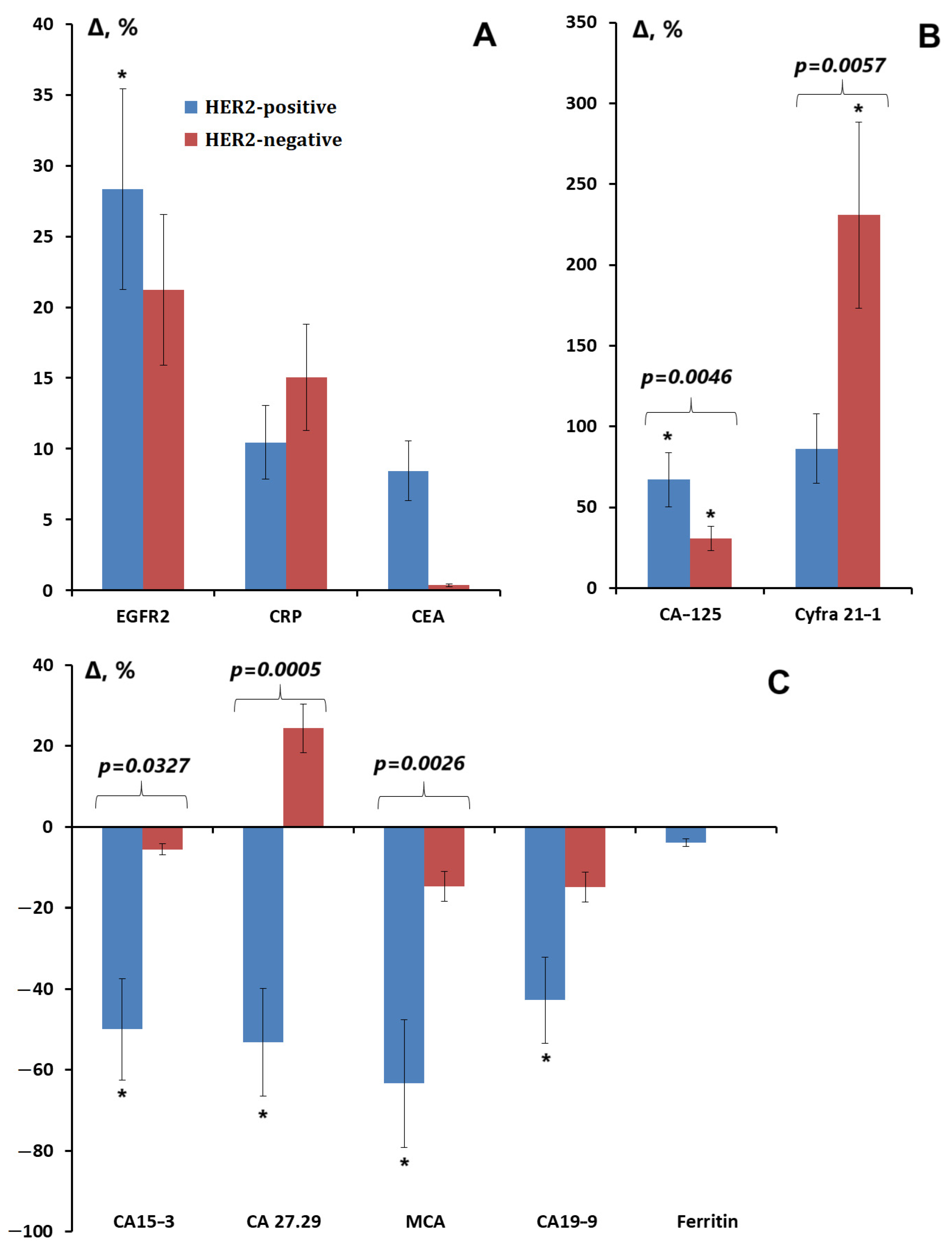
3.3. Salivary Tumor Marker Levels Depending on the Expression of ER, PR, and HER2 Receptors; Proliferative Activity Marker Ki-67; and the Degree of Differentiation of Breast Cancer
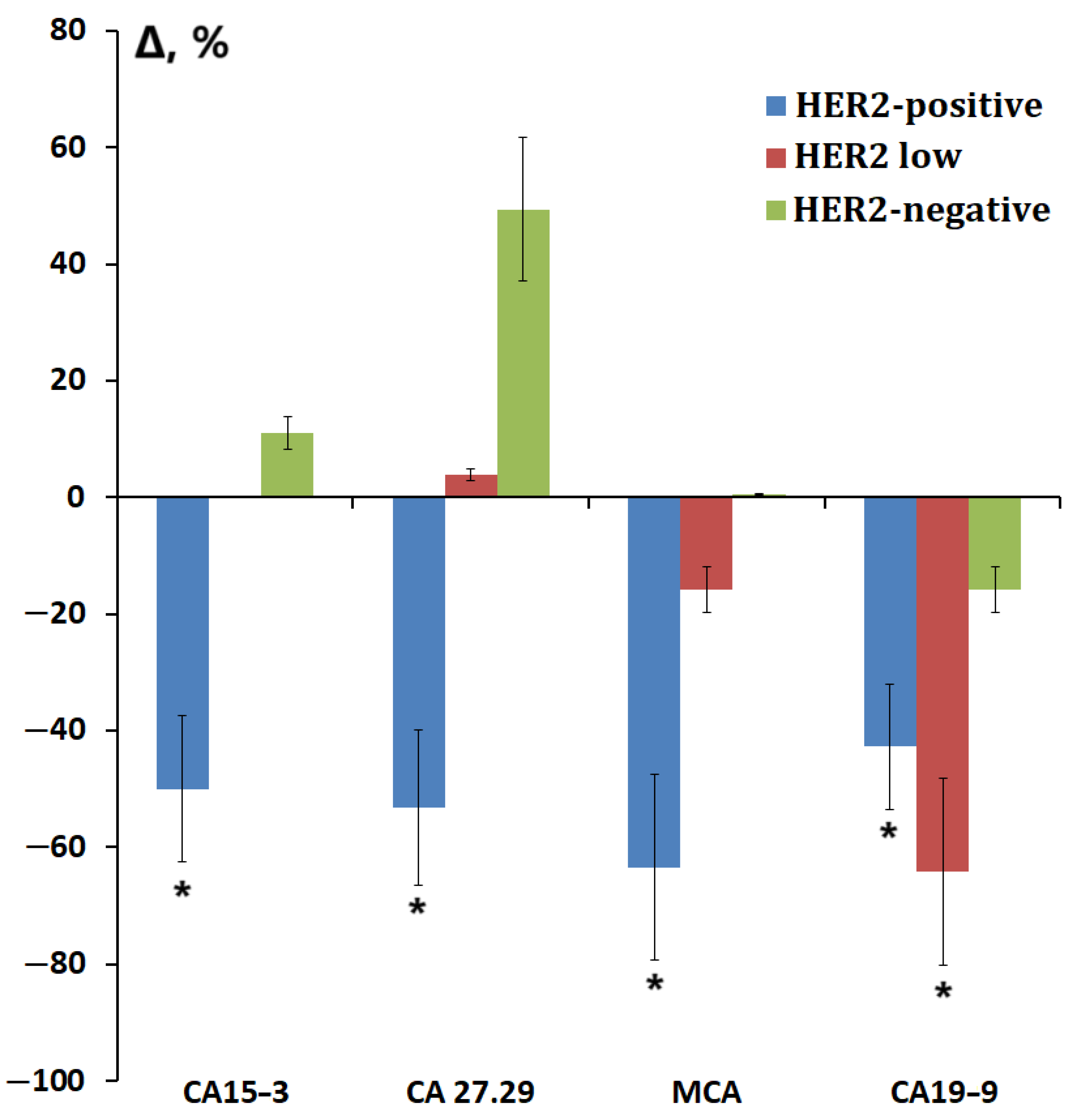
3.4. Salivary Tumor Marker Levels Depending on the Molecular Biological Subtype of Breast Cancer
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dai, X.; Zhang, X.; Lu, P. Toward a holistic view of multiscale breast cancer molecular biomarkers. Biomark. Med. 2019, 13, 1509–1533. [Google Scholar]
- Porto-Mascarenhas, E.C.; Assad, D.X.; Chardin, H.; Gozal, D.; De Luca Canto, G.; Acevedo, A.C.; Guerra, E.N.S. Salivary biomarkers in the diagnosis of breast cancer: A review. Crit. Rev. Oncol. Hematol. 2017, 110, 62–73. [Google Scholar] [PubMed]
- Nonaka, T.; Wong, D.T.W. Saliva Diagnostics. Annu. Rev. Anal. Chem. 2022, 15, 107–121. [Google Scholar]
- Koopaie, M.; Kolahdooz, S.; Fatahzadeh, M.; Manifar, S. Salivary biomarkers in breast cancer diagnosis: A systematic review and diagnostic meta-analysis. Cancer Med. 2022, 11, 2644–2661. [Google Scholar]
- López-Jornet, P.; Aznar, C.; Ceron, J.; Asta, T. Salivary biomarkers in breast cancer: A cross-sectional study. Support Care Cancer 2021, 29, 889–896. [Google Scholar]
- Pink, R.; Simek, J.; Vondrakova, J.; Faber, E.; Michl, P.; Pazdera, J.; Indrak, K. Saliva as a diagnostic medium. Biomed. Pap. Med. Fac. Palacky Univ. Olomouc 2009, 153, 103–110. [Google Scholar]
- Assad, D.X.; Mascarenhas, E.C.P.; Normando, A.G.C.; Chardin, H.; Barra, G.B.; Pratesi, R.; Nóbrega, Y.K.D.M.; Acevedo, A.C.; Guerra, E.N.S. Correlation between salivary and serum CA15-3 concentrations in patients with breast cancer. Mol. Clin. Oncol. 2020, 13, 155–161. [Google Scholar]
- Cui, Y.; Yang, M.; Zhu, J.; Zhang, H.; Duan, Z.; Wang, S.; Liao, Z.; Liu, W. Developments in diagnostic applications of saliva in human organ diseases. Med. Nov. Technol. Devices 2022, 13, 100115. [Google Scholar]
- Agha-Hosseini, F.; Mirzaii-Dizgah, I.; Rahimi, A. Correlation of serum and salivary CA15-3 levels in patients with breast cancer. Med. Oral Patol. Oral Cir. Bucal. 2009, 14, e521–e524. [Google Scholar]
- Streckfus, F.C. Salivary Biomarkers to Assess Breast Cancer Diagnosis and Progression: Are We There Yet? In Saliva and Salivary Diagnostics; IntechOpen: Rijeka, Croatia, 2019. [Google Scholar] [CrossRef]
- Chen, W.; Zhang, Z.; Zhang, S.; Zhu, P.; Ko, J.K.; Yung, K.K. MUC1: Structure, Function, and Clinic Application in Epithelial Cancers. Int. J. Mol. Sci. 2021, 22, 6567. [Google Scholar] [CrossRef]
- Einama, T.; Yamagishi, Y.; Takihata, Y.; Suzuki, T.; Yamasaki, T.; Hirose, Y.; Kobayashi, K.; Yonamine, N.; Fujinuma, I.; Tsunenari, T.; et al. Co-expression of mesothelin and CA125/MUC16 is a prognostic factor for breast cancer, especially in luminal-type breast cancer patients. Biomark. Res. 2021, 9, 78. [Google Scholar]
- Khorrami, S.; Tavakoli, M.; Safari, E. Clinical Value of Serum S100A8/A9 and CA15-3 in the Diagnosis of Breast Cancer. Iran. J. Pathol. 2019, 14, 104–112. [Google Scholar] [PubMed]
- You, D.; Kim, H.; Jeong, Y.; Yoon, S.Y.; Lo, E.; Kim, S.; Lee, J.E. Tumorigenicity of EGFR- and/or HER2-Positive Breast Cancers Is Mediated by Recruitment of Tumor-Associated Macrophages. Int. J. Mol. Sci. 2023, 24, 1443. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Lai, H.; Pan, X.; Lin, Q.; Qin, Y.; Liu, F.; Fang, M.; Huang, W.; Wei, C. Development and Validation of a Nomogram for Predicting Survival Based on Ferritin and Transferrin Ratio in Breast Cancer Patients. Cancer Control 2024, 31, 10732748241261553. [Google Scholar]
- Holm, J.B.; Baggesen, E.; Cronin-Fenton, D.; Frystyk, J.; Bruun, J.M.; Christiansen, P.; Borgquist, S. Circulating C-reactive protein levels as a prognostic biomarker in breast cancer across body mass index groups. Sci. Rep. 2024, 14, 14486. [Google Scholar]
- Sergeeva, N.S.; Karmakova, T.A.; Polyak, M.A.; Alentov, I.I.; Kaprin, A.D. Systemic Inflammatory Response as a Prognostic Factor in Breast Cancer. Part I. Tumor-Promoting Inflammation. Serum Inflammatory Markers. Ann. Russ. Acad. Med. Sci. 2022, 77, 345–353. [Google Scholar]
- Zhuge, X.; Zhou, H.; Chen, L.; Chen, H.; Chen, X.; Guo, C. The association between serum ferritin levels and malignant intraductal papillary mucinous neoplasms. BMC Cancer 2021, 21, 1253. [Google Scholar]
- Bel’skaya, L.V.; Dyachenko, E.I. Salivary Biomarkers in Breast Cancer: From Salivaomics to Salivaoncoomics. Front. Biosci. 2024, 29, 253. [Google Scholar]
- Ilić, I.R.; Stojanović, N.M.; Radulović, N.S.; Živković, V.V.; Randjelović, P.J.; Petrović, A.S.; Božić, M.; Ilić, R.S. The Quantitative ER Immunohistochemical Analysis in Breast Cancer: Detecting the 3 + 0, 4 + 0, and 5 + 0 Allred Score Cases. Medicina 2019, 55, 461. [Google Scholar] [CrossRef]
- Wolff, A.C.; Hammond, M.E.H.; Allison, K.H.; Harvey, B.E.; Mangu, P.B.; Bartlett, J.M.S.; Bilous, M.; Ellis, I.O.; Fitzgibbons, P.; Hanna, W.; et al. Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer: American Society of Clinical Oncology/College of American Pathologists Clinical Practice Guideline Focused Update. J. Clin. Oncol. 2018, 36, 2105–2122. [Google Scholar]
- Stalhammar, G.; Robertson, S.; Wedlund, L.; Lippert, M.; Rantalainen, M.; Bergh, J.; Hartman, J. Digital image analysis of Ki67 in hot spots is superior to both manual Ki67 and mitotic counts in breast cancer. Histopathology 2018, 72, 974–989. [Google Scholar] [CrossRef] [PubMed]
- Zavalishina, L.E.; Oliushina, E.M.; Andreeva, Y.Y.; Kuznetsova, O.A.; Moskvina, L.V.; Frank, G.A. Updated CAP guidelines for determining molecular biological subtypes of breast cancer. Arkhiv. Patol. 2023, 85, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Rehak, N.N.; Cecco, S.A.; Csako, G. Biochemical composition and electrolyte balance of “unstimulated” whole human saliva. Clin. Chem. Lab. Med. 2000, 38, 335–343. [Google Scholar] [PubMed]
- Patel, N.; Belcher, J.; Thorpe, G.; Forsyth, N.R.; Spiteri, M.A. Measurement of C-reactive protein, procalcitonin and neutrophil elastase in saliva of COPD patients and healthy controls: Correlation to self-reported wellbeing parameters. Respir. Res. 2015, 16, 62. [Google Scholar] [CrossRef]
- Ouellet-Morin, I.; Danese, A.; Williams, B.; Arseneault, L. Validation of a high-sensitivity assay for C-reactive protein in human saliva. Brain Behav. Immun. 2011, 25, 640–646. [Google Scholar]
- Jung, S.Y.; Papp, J.C.; Sobel, E.M.; Pellegrini, M.; Yu, H.; Zhang, Z.F. Genetically Predicted C-Reactive Protein Associated With Postmenopausal Breast Cancer Risk: Interrelation With Estrogen and Cancer Molecular Subtypes Using Mendelian Randomization. Front. Oncol. 2021, 10, 630994. [Google Scholar]
- Roque, A.T.; Gambeloni, R.Z.; Felitti, S.; Ribeiro, M.L.; Santos, J.C. Inflammation-induced oxidative stress in breast cancer pa-tients. Med. Oncol. 2015, 32, 263. [Google Scholar]
- Lee, T.; Teng, T.Z.J.; Shelat, V.G. Carbohydrate antigen 19-9—Tumor marker: Past, present, and future. World J. Gastrointest. Surg. 2020, 12, 468–490. [Google Scholar]
- Huebner, H.; Häberle, L.; Müller, V.; Schrader, I.; Lorenz, R.; Forstbauer, H.; Fink, V.; Schochter, F.; Bekes, I.; Mahner, S.; et al. MUC1 (CA27.29) before and after Chemotherapy and Prognosis in High-Risk Early Breast Cancer Patients. Cancers 2022, 14, 1721. [Google Scholar] [CrossRef]
- Nath, S.; Mukherjee, P. MUC1: A multifaceted oncoprotein with a key role in cancer progression. Trends Mol. Med. 2014, 20, 332–342. [Google Scholar] [CrossRef]
- Pang, Z.; Dong, X.; Deng, H.; Wang, C.; Liao, X.; Liao, C.; Liao, Y.; Tian, W.; Cheng, J.; Chen, G.; et al. MUC1 triggers lineage plasticity of Her2 positive mammary tumors. Oncogene 2022, 41, 3064–3078. [Google Scholar] [CrossRef] [PubMed]
- Yue, T.; Partyka, K.; Maupin, K.A.; Hurley, M.; Andrews, P.; Kaul, K.; Moser, A.J.; Zeh, H.; Brand, R.E.; Haab, B.B. Identification of blood-protein carriers of the CA 19-9 antigen and characterization of prevalence in pancreatic diseases. Proteomics 2011, 11, 3665–3674. [Google Scholar] [PubMed]
- Lehmann, B.D.; Bauer, J.A.; Chen, X.; Sanders, M.E.; Chakravarthy, A.B.; Shyr, Y.; Pietenpol, J.A. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Investig. 2011, 121, 2750–2767. [Google Scholar] [PubMed]
- Tarantino, P.; Viale, G.; Press, M.F.; Hu, X.; Penault-Llorca, F.; Bardia, A.; Batistatou, A.; Burstein, H.J.; Carey, L.A.; Cortes, J.; et al. ESMO expert consensus statements (ECS) on the definition, diagnosis, and management of HER2-low breast cancer. Ann. Oncol. 2023, 34, 645–659. [Google Scholar]
- Sengupta, A.; Valdramidou, D.; Huntley, S.; Hicks, S.J.; Carrington, S.D.; Corfield, A.P. Distribution of MUC1 in the normal human oral cavity is localized to the ducts of minor salivary glands. Arch. Oral Biol. 2001, 46, 529–538. [Google Scholar]
- Yan, W.; Apweiler, R.; Balgley, B.M.; Boontheung, P.; Bundy, J.L.; Cargile, B.J.; Cole, S.; Fang, X.; Gonzalez-Begne, M.; Griffin, T.J.; et al. Systematic comparison of the human saliva and plasma proteomes. Proteom. Clin. Appl. 2009, 3, 116–134. [Google Scholar]
- Morgado, M.; Sutton, M.N.; Simmons, M.; Warren, C.R.; Lu, Z.; Constantinou, P.E.; Liu, J.; Francis, L.L.; Conlan, R.S.; Bast, R.C., Jr.; et al. Tumor necrosis factor-α and interferon-γ stimulate MUC16 (CA125) expression in breast, endometrial and ovarian cancers through NFκB. Oncotarget 2016, 7, 14871–14884. [Google Scholar] [CrossRef]
- Mathew, S.; Ali, A.; Abdel-Hafiz, H.; Fatima, K.; Suhail, M.; Archunan, G.; Begum, N.; Jahangir, S.; Ilyas, M.; Chaudhary, A.G.; et al. Biomarkers for virus-induced hepatocellular carcinoma (HCC). Infect. Genet. Evol. 2014, 26, 327–339. [Google Scholar]
- Zhang, Z.; Tian, C.; Shi, Q.; Hao, J.; Zhao, N.; Liu, Z. Diagnostic Value of CYFRA 21-1 in the Cerebrospinal Fluid for Leptomeningeal Metastasis. Dis. Markers 2017, 2017, 2467870. [Google Scholar] [CrossRef]
- Farahani, H.; Amri, J.; Alaee, M.; Mohaghegh, F.; Rafiee, M. Serum and Saliva Levels of Cancer Antigen 15-3, Carcinoembryonic Antigen, Estradiol, Vaspin, and Obestatin as Biomarkers for the Diagnosis of Breast Cancer in Postmenopausal Women. Lab. Med. 2020, 51, 620–627. [Google Scholar]
- Weinberg, F.; Peckys, D.B.; de Jonge, N. EGFR Expression in HER2-Driven Breast Cancer Cells. Int. J. Mol. Sci. 2020, 21, 9008. [Google Scholar] [CrossRef] [PubMed]
- Hynes, N.E.; Lane, H.A. ERBB receptors and cancer: The complexity of targeted inhibitors. Nat. Rev. Cancer 2005, 5, 341–354. [Google Scholar] [PubMed]
- Lemmon, M.A.; Schlessinger, J.; Ferguson, K.M. The EGFR family: Not so prototypical receptor tyrosine kinases. Cold Spring Harb. Perspect. Biol. 2014, 6, a020768. [Google Scholar]
- Chow, L.W.-C.; Lie, E.F.; Toi, M. Advances in EGFR/HER2-directed clinical research on breast cancer. Adv. Cancer Res. 2020, 147, 375–428. [Google Scholar] [PubMed]
- Streckfus, C.F.; Arreola, D.; Edwards, C.; Bigler, L. Salivary protein profiles among her2/neu-receptor-positive and -negative breast cancer patients: Support for using salivary protein profiles for modeling breast cancer progression. J. Oncol. 2012, 2012, 413256. [Google Scholar]
- De Abreu Pereira, D.; Areias, V.R.; Franco, M.F.; Benitez, M.C.; do Nascimento, C.M.; de Azevedo, C.M.; Alves, G. Measurement of HER2 in saliva of women in risk of breast cancer. Pathol. Oncol. Res. 2013, 19, 509–513. [Google Scholar]
- Laidi, F.; Bouziane, A.; Lakhdar, A.; Khabouze, S.; Rhrab, B.; Zaoui, F. Salivary expression of soluble HER2 in breast cancer patients with positive and negative HER2 status. OncoTargets Ther. 2014, 7, 1285–1289. [Google Scholar]
| Feature | Luminal A, n = 22 | Luminal B(-), n = 22 | Luminal B(+), n = 22 | Non-Luminal, n = 22 | TNBC, n = 22 |
|---|---|---|---|---|---|
| Age, years | 65.3 [51.7; 69.9] | 64.5 [47.6; 66.6] | 61.0 [48.2; 67.4] | 57.6 [51.3; 68.5] | 65.2 [41.6; 70.2] |
| Clinical Stage | |||||
| Stage IA + IB | 22.7% | 9.1% | 13.6% | 22.7% | 18.2% |
| Stage IIA + IIB | 31.8% | 27.3% | 36.4% | 40.9% | 27.3% |
| Stage IIIA + IIIB | 18.2% | 31.8% | 31.8% | 22.7% | 36.3% |
| Stage IIIC + IV | 27.3% | 31.8% | 18.2% | 13.7% | 18.2% |
| Lymph node status | |||||
| N0 | 36.4% | 22.7% | 59.1% | 54.5% | 40.9% |
| N1–3 | 63.6% | 77.3% | 40.9% | 45.5% | 59.1% |
| Degree of differentiation (G) | |||||
| G I + II | 94.7% | 38.9% | 52.4% | 25.0% | 33.3% |
| G III | 5.3% | 61.1% | 47.6% | 75.0% | 66.7% |
| Marker of proliferative activity Ki-67 | |||||
| <20% | 100% | 13.6% | 18.2% | 4.5% | 13.6% |
| >20% | 0% | 86.4% | 81.8% | 95.5% | 86.4% |
| Tumor Markers | Healhty Controls, n = 30 | Breast Cancer, n = 110 | p-Value |
|---|---|---|---|
| EGFR2, ng/mL | 0.603 [0.474; 0.731] | 0.731 [0.517; 0.944] | 0.0685 |
| CRP, mU/mL | 0.153 [0.118; 0.212] | 0.176 [0.122; 0.312] | 0.2528 |
| CEA, ng/mL | 83.37 [68.14; 94.02] | 86.94 [77.97; 94.32] | 0.4262 |
| CA125, U/mL | 234.51 [100.83; 308.04] | 350.98 [254.90; 448.69] | 0.0001 * |
| CYFRA 21-1, ng/mL | 2.04 [1.03; 7.13] | 4.93 [1.87; 12.51] | 0.0177 * |
| CA15-3, U/mL | 39.6 [21.4; 92.7] | 31.0 [14.1; 83.6] | 0.2741 |
| CA 27.29, U/mL | 3.08 [2.11; 5.22] | 2.66 [1.31; 6.47] | 0.2373 |
| MCA, U/mL | 21.10 [6.24; 75.92] | 16.61 [7.08; 47.11] | 0.3967 |
| CA19-9, U/mL | 49.23 [21.64; 214.64] | 37.59 [16.55; 111.91] | 0.0963 |
| Ferritin, ng/mL | 15.6 [14.1; 18.2] | 15.0 [14.1; 17.1] | 0.8096 |
| Tumor Markers | Estrogen Receptors | Progesterone Receptors | ||
|---|---|---|---|---|
| ER(-), n = 47 (Group 1) | ER(+), n = 63 (Group 2) | PR(-), n = 59 (Group 1) | PR(+), n = 51 (Group 2) | |
| EGFR2, ng/mL | 0.731 [0.560; 0.902] | 0.774 [0.474; 1.030] | 0.731 [0.560; 0.902] | 0.731 [0.474; 1.115] |
| - | p2-HC = 0.0439 | p1-HC = 0.0303 | - | |
| CRP, mU/mL | 0.135 [0.111; 0.193] | 0.224 [0.148; 0.406] | 0.136 [0.120; 0.218] | 0.215 [0.150; 0.374] |
| p1-2 = 0.0004 | p1-2 = 0.0004, p2-HC = 0.0274 | p1-2 = 0.0040 | p1-2 = 0.0040 | |
| CEA, ng/mL | 84.05 [75.30; 92.60] | 87.18 [79.39; 95.33] | 88.65 [77.75; 93.86] | 85.60 [78.22; 95.00] |
| CA125, U/mL | 385.49 [229.61; 472.16] | 330.59 [258.63; 421.18] | 385.4 [234.31; 460.59] | 330.59 [254.90; 410.20] |
| p1-HC = 0.0002 | p2-HC = 0.0004 | p1-HC = 0.0001 | p2-HC = 0.0014 | |
| CYFRA 21-1, ng/mL | 4.22 [1.77; 9.52] | 5.28 [2.45; 15.48] | 4.22 [1.56; 9.88] | 5.52 [2.89; 14.75] |
| - | p2-HC = 0.0085 | - | p2-HC = 0.0064 | |
| CA15-3, U/mL | 25.11 [13.32; 83.65] | 36.94 [14.95; 84.69] | 25.11 [12.96; 73.75] | 37.86 [18.22; 84.79] |
| CA 27.29, U/mL | 1.94 [1.21; 5.09] | 2.99 [1.40; 6.56] | 2.18 [1.07; 5.09] | 3.06 [1.54; 6.72] |
| MCA, U/mL | 12.99 [5.67; 31.32] | 17.71 [8.01; 56.73] | 12.99 [5.67; 31.32] | 17.78 [8.64; 71.97] |
| CA19-9, U/mL | 41.64 [16.64; 111.91] | 32.55 [16.18; 125.27] | 42.18 [16.27; 116.91] | 28.73 [16.91; 108.91] |
| Ferritin, ng/mL | 15.6 [14.7; 17.5] | 15.0 [14.1; 16.8] | 15.6 [14.7; 16.8] | 14.9 [13.8; 17.1] |
| Tumor Markers | Degree of Differentiation | Proliferative Activity Index | ||
|---|---|---|---|---|
| GI + II, n = 46 (Group 1) | GIII, n = 47 (Group 2) | Ki-67 Low, n = 32 (Group 1) | Ki-67 High, n = 85 (Group 2) | |
| EGFR2, ng/mL | 0.731 [0.517; 1.030] | 0.731 [0.517; 0.902] | 0.795 [0.496; 1.030] | 0.731 [0.560; 0.902] |
| - | - | p1-HC = 0.0407 | - | |
| CRP, mU/mL | 0.206 [0.122; 0.374] | 0.150 [0.118; 0.218] | 0.206 [0.144; 0.390] | 0.160 [0.120; 0.258] |
| p1-2 = 0.0380 | p1-2 = 0.0380 | - | - | |
| CEA, ng/mL | 89.34 [81.65; 97.13] | 84.38 [77.97; 91.68] | 88.97 [77.71; 96.04] | 86.69 [77.97; 93.07] |
| CA125, U/mL | 350.29 [258.24; 504.12] | 350.39 [234.31; 421.18] | 346.27 [278.92; 454.64] | 350.39 [234.31; 448.63] |
| p1-HC = 0.0005 | p2-HC = 0.0005 | p1-HC = 0.0007 | p2-HC = 0.0003 | |
| CYFRA 21-1, ng/mL | 5.26 [1.87; 14.36] | 4.53 [1.56; 9.39] | 4.58 [2.12; 11.69] | 5.07 [1.86; 12.86] |
| p1-HC = 0.0142 | - | p1-HC = 0.0304 | p2-HC = 0.0262 | |
| CA15-3, U/mL | 31.36 [14.95; 75.83] | 20.62 [13.32; 86.67] | 33.27 [17.15; 66.09] | 29.19 [13.78; 84.69] |
| CA 27.29, U/mL | 2.88 [1.31; 7.27] | 2.58 [1.33; 5.56] | 2.66 [1.36; 6.64] | 3.06 [1.32; 6.47] |
| MCA, U/mL | 13.15 [7.34; 51.00] | 17.78 [7.04; 44.24] | 16.91 [8.61; 72.32] | 16.48 [6.61; 35.27] |
| CA19-9, U/mL | 35.59 [16.27; 108.91] | 40.64 [21.36; 116.91] | 23.73 [15.14; 97.32] | 42.36 [16.91; 116.91] |
| - | - | p1-HC = 0.0323 | - | |
| Ferritin, ng/mL | 14.7 [13.8; 16.8] | 15.6 [15.0; 17.9] | 15.0 [14.0; 16.9] | 15.4 [14.4; 17.1] |
| p1-2 = 0.0394 | p1-2 = 0.0394 | - | - | |
| Tumor Markers | HER2-Positive | HER2-Negative | |||
|---|---|---|---|---|---|
| Non-Luminal, n = 22 | Luminal B(+), n = 22 | Luminal A, n = 22 | Luminal B(-), n = 22 | TNBC, n = 22 | |
| EGFR2, ng/mL | 25 * p = 0.0131 | 32 | 11 | 25 | 14 |
| CRP, mU/mL | −18 | 35 | 137 * p = 0.0027 | 5 | −7 |
| CEA, ng/mL | 8 | 8 | 5 | 3 | −4 |
| CA125, U/mL | 71 * p < 0.0001 | 59 * p = 0.0014 | 44 * p = 0.0080 | 24 * p = 0.0184 | 15 |
| CYFRA 21-1, ng/mL | 98 | 56 | 328 * p = 0.0198 | 329 * p = 0.0249 | 183 |
| CA15-3, U/mL | −36 | −65 * p = 0.0382 | 19 | −4 | −37 |
| CA 27.29, U/mL | −57 * p = 0.0016 | −25 * p = 0.0348 | 41 | 10 | 19 |
| MCA, U/mL | −60 | −65 * p = 0.0301 | 54 | −12 | −36 |
| CA19-9, U/mL | −34 | −55 * p = 0.0384 | −52 * p = 0.0324 | −4 | 35 |
| Ferritin, ng/mL | −3 | −4 | −6 | −3 | 2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dyachenko, E.I.; Bel’skaya, L.V. How Does Breast Cancer Heterogeneity Determine Changes in Tumor Marker Levels in Saliva? Curr. Issues Mol. Biol. 2025, 47, 216. https://doi.org/10.3390/cimb47040216
Dyachenko EI, Bel’skaya LV. How Does Breast Cancer Heterogeneity Determine Changes in Tumor Marker Levels in Saliva? Current Issues in Molecular Biology. 2025; 47(4):216. https://doi.org/10.3390/cimb47040216
Chicago/Turabian StyleDyachenko, Elena I., and Lyudmila V. Bel’skaya. 2025. "How Does Breast Cancer Heterogeneity Determine Changes in Tumor Marker Levels in Saliva?" Current Issues in Molecular Biology 47, no. 4: 216. https://doi.org/10.3390/cimb47040216
APA StyleDyachenko, E. I., & Bel’skaya, L. V. (2025). How Does Breast Cancer Heterogeneity Determine Changes in Tumor Marker Levels in Saliva? Current Issues in Molecular Biology, 47(4), 216. https://doi.org/10.3390/cimb47040216







