In Silico and Wet Analysis of BAX Gene G-248A Polymorphism and mRNA Expression in Peptic Ulcer Disease and Gastric Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Cohort
2.2. Rapid Urease Test
2.3. DNA and RNA Isolation
2.4. Genotyping by Restriction Fragment Length Polymorphism (RFLP)
2.5. Expression Analysis by qPCR
2.6. Statistical Analysis
2.7. In Silico Analysis
3. Results
3.1. Genotyping of the BAX Gene
3.1.1. Genotyping of SNP G-248A in the Group of Peptic Ulcer Patients
3.1.2. Genotyping of SNP G-248A in the Group of Gastric Cancer Patients
3.2. BAX Gene Expression Level
3.2.1. Expression of BAX mRNA in Peptic Ulcer Patients
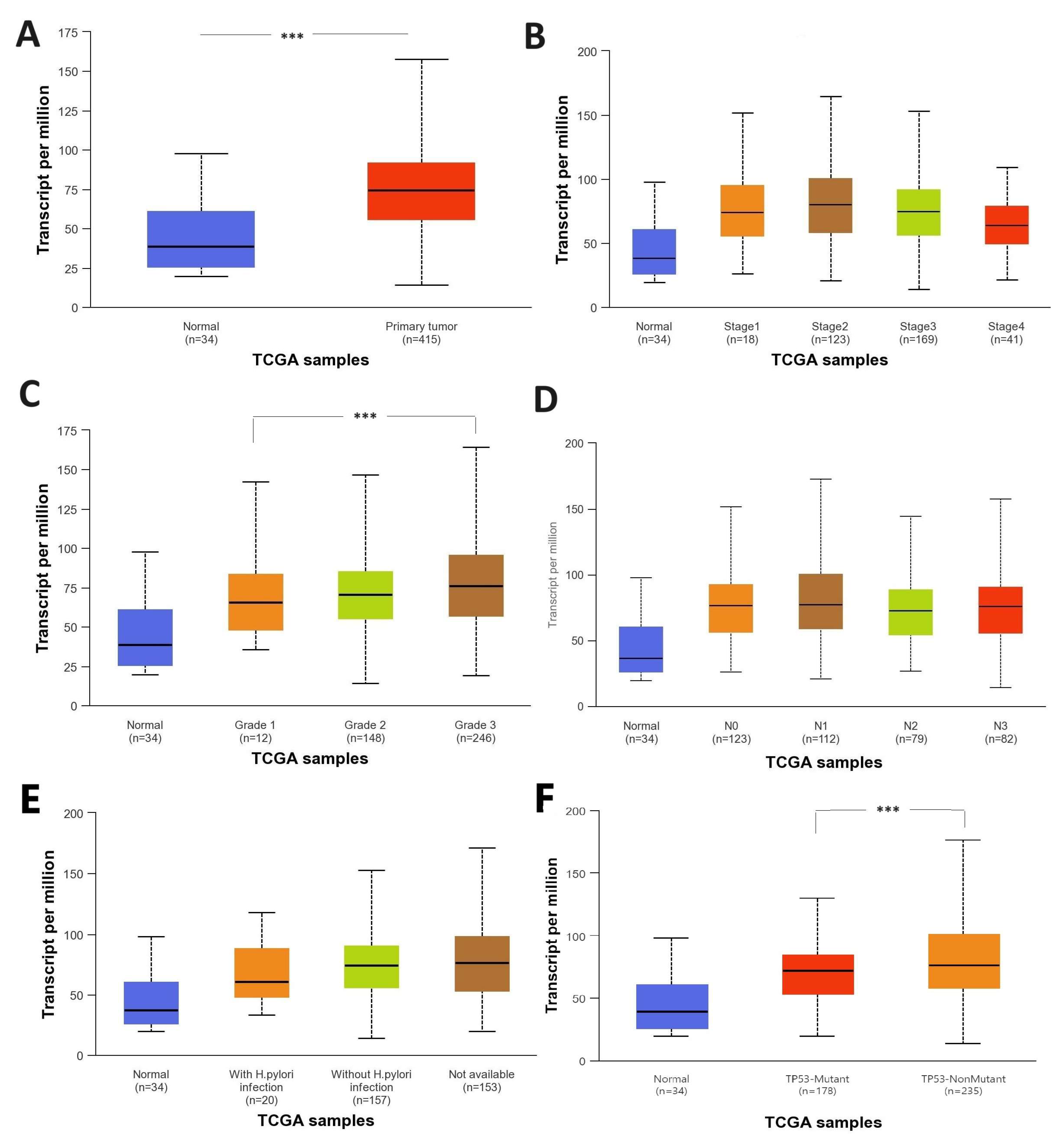
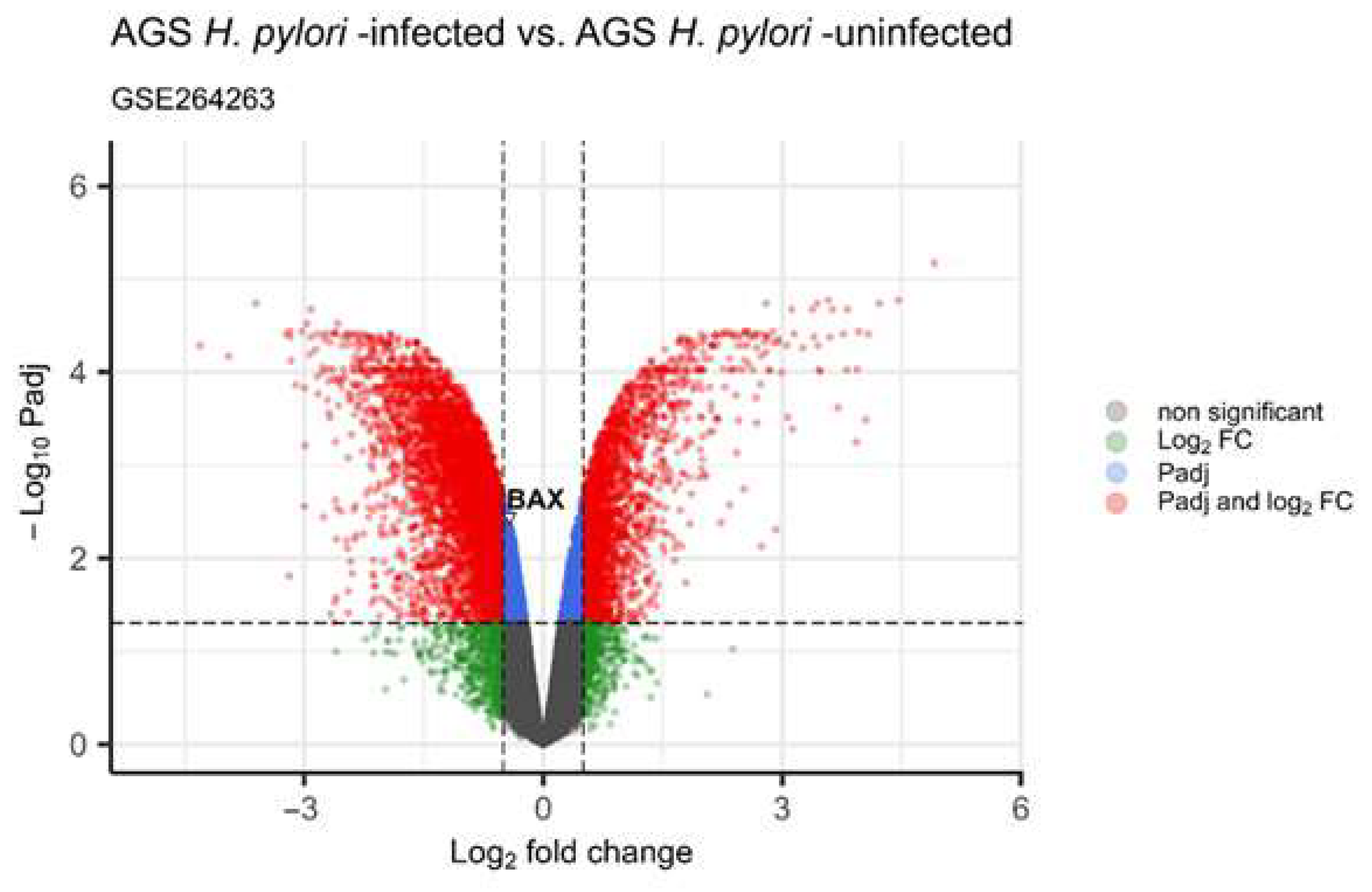
3.2.2. Expression of BAX mRNA in Gastric Cancer Tissue and In Silico Analysis of TCGA- STAD Data and GSE264263 Data
3.2.3. Comparison of BAX Gene Expression Between Peptic Ulcer Patients, Gastric Cancer Patients, and Morphologically Normal Tissue Samples
3.3. Assessment of the Correlation Between SNP G-248A Genotype and BAX Expression
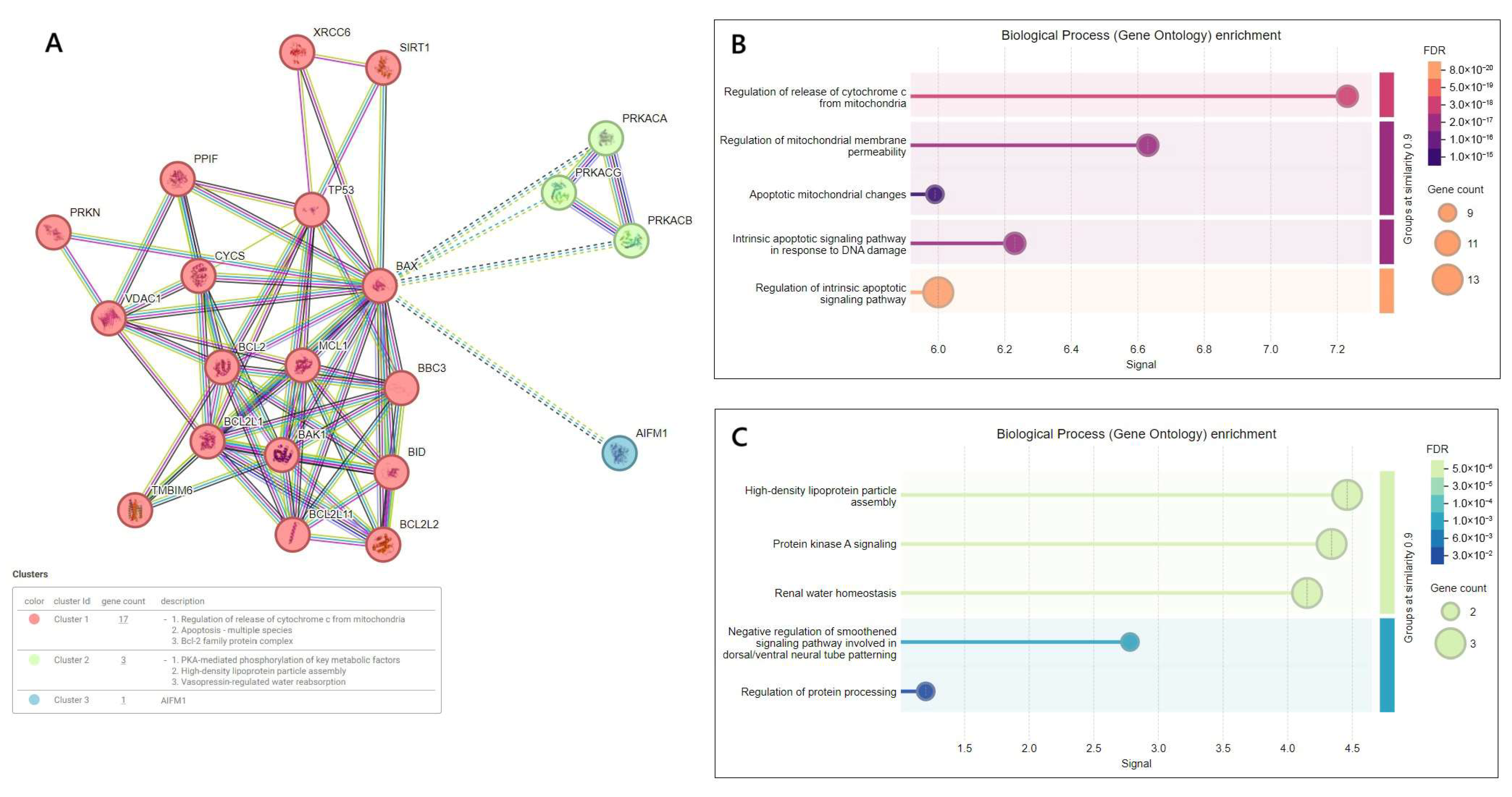
3.4. Interrelation Between BAX Gene Expression Level and Other Target Genes
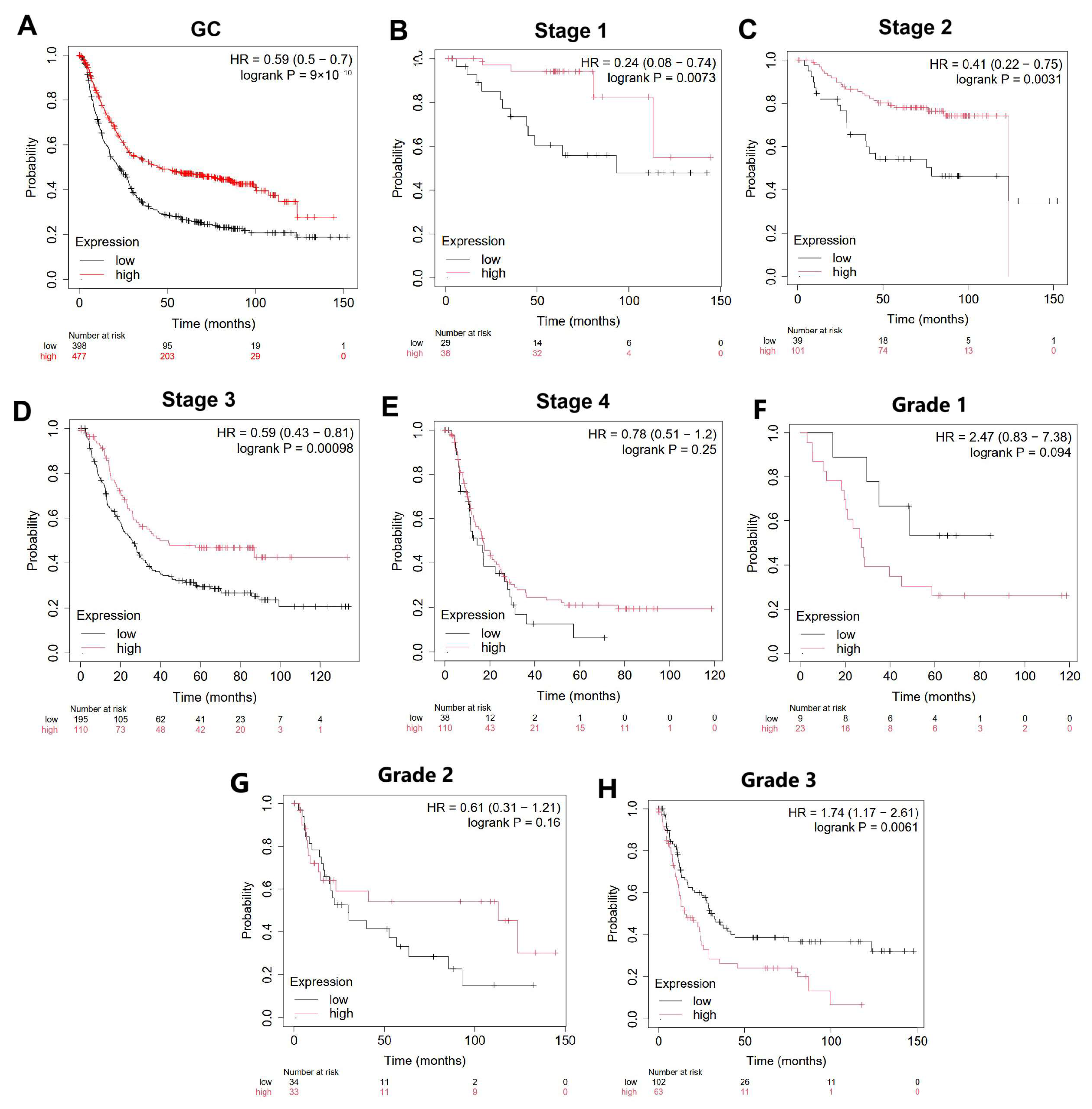
3.5. Prognostic Value of BAX Gene Expression Level in Gastric Cancer
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AGS | gastric adenocarcinoma cell line |
| BAX | B-cell lymphoma 2-associated X protein |
| EBV | Epstein–Barr Virus |
| G1 | Grade 1—well differentiated (low grade) |
| G2 | Grade 2—moderately differentiated (intermediate grade) |
| G3 | Grade 3—poorly differentiated (high grade) |
| GAPDH | glyceraldehyde-3-phosphate dehydrogenase |
| GC | gastric cancer |
| GEO | Gene Expression Omnibus |
| HNSCC | Squamous cell carcinoma of the head and neck |
| HWE | Hardy–Weinberg equilibrium |
| N0 | metastases into regional lymph node |
| N1 | metastases in one to three axillary lymph nodes |
| N2 | metastases in four to nine axillary lymph nodes |
| N3 | metastases in ten or more axillary lymph nodes |
| NSAIDs | non-steroidal anti-inflammatory drugs |
| OS | overall survival |
| PPI | protein–protein interaction |
| PUD | Peptic ulcer disease |
| SNP | single nucleotide polymorphism |
| TCGA-STAD | The Cancer Genome Atlas Stomach Adenocarcinoma |
| TIMER2 | Tumor IMmune Estimation Resource 2.0 |
| UALCAN | University of ALabama at Birmingham CANcer data analysis Portal |
References
- Narayanan, M.; Reddy, K.M.; Marsicano, E. Peptic Ulcer Disease and Helicobacter pylori infection. Mo. Med. 2018, 115, 219–224. [Google Scholar] [PubMed]
- Fashner, J.; Gitu, A.C. Diagnosis and treatment of Peptic Ulcer Disease and H. pylori infection. Am. Fam. Physician 2015, 91, 236–242. [Google Scholar]
- Kuna, L.; Jakab, J.; Smolic, R.; Raguz-Lucic, N.; Vcev, A.; Smolic, M. Peptic Ulcer Disease: A brief review of conventional therapy and herbal treatment options. J. Clin. Med. 2019, 8, 179. [Google Scholar] [CrossRef]
- Sai Teja, D.; Srivarsha, G.A.; Qadrie, Z.L. Peptic Ulcer Disease: An overview. IJPPR Human J. 2018, 12, 8–26. [Google Scholar]
- Yildiz, Y.; Yaylim, I.; Ozkan, N.; Arikan, S.; Turan, S.; Kucucuk, S.; Coskunpinar, E.; Isbir, T. Bax promoter G(-248)A polymorphism in a Turkish clinical breast cancer patients: A case-control study. Am. J. Mol. Biol. 2013, 3, 10–16. [Google Scholar] [CrossRef]
- Bentires-Alj, M.; Dejardin, E.; Viatour, P.; Van Lint, C.; Froesch, B.; Reed, J.C.; Merville, M.P.; Bours, V. Inhibition of the NF-κB transcription factor increases Bax expression in cancer cell lines. Oncogene 2001, 20, 2805–2813. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Chen, X.; Zheng, Y.; Liu, Q.; Chen, H.; Cai, Y.; Cao, L.; Lai, X.; Pan, L.; Li, Y.; et al. Prognostic value and susceptibility of BAX rs4645878 polymorphism in cancer: A systematic review and meta-analysis. Medicine 2018, 97, e11591. [Google Scholar] [CrossRef]
- Rampino, N.; Yamamoto, H.; Ionov, Y.; Li, Y.; Sawai, H.; Reed, J.C.; Perucho, M. Somatic frameshift mutations in the BAX gene in colon cancers of the microsatellite mutator phenotype. Science 1997, 275, 967–969. [Google Scholar] [CrossRef]
- Fernandes, A.T.; Rocha, N.P.; Vendrame, E.; Russomano, F.; Grinsztejn, B.J.; Friedman, R.K.; Pinto, A.C.; Klumb, E.M.; Avvad, E.; Macedo, J.; et al. Polymorphism in apoptotic BAX (-248G>A) gene but not in anti-apoptotic BCL2 (-938C>A) gene and its protein and mRNA expression are associated with cervical intraepithelial neoplasia. Apoptosis 2015, 20, 1347–1357. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Wang, L.; Qing, Y.; Li, C.; Ren, T.; Li, Q.; Li, M.; Zhang, S.; Shan, J.; Wang, G.; et al. Polymorphisms of BCL2 and BAX genes associate with outcomes in advanced non-small cell lung cancer patients treated with platinum-based chemotherapy. Sci. Rep. 2015, 5, 17766. [Google Scholar] [CrossRef]
- Sun, L.; Wei, L.; Wei, L.; Li, D. Correlation between Bax gene polymorphisms and esophagus cancer. Oncol. Lett. 2018, 16, 7097–7101. [Google Scholar] [CrossRef]
- Wang, S.; Chen, X.; Zhang, X.; Wen, K.; Chen, X.; Gu, J.; Li, J.; Wang, Z. Pro-apoptotic gene BAX is a pan-cancer predictive biomarker for prognosis and immunotherapy efficacy. Aging 2024, 16, 11289–11317. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT. Method 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.V.S.K.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Chandrashekar, D.S.; Karthikeyan, S.K.; Korla, P.K.; Patel, H.; Shovon, A.R.; Athar, M.; Netto, G.J.; Qin, Z.S.; Kumar, S.; Manne, U.; et al. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia 2022, 25, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef] [PubMed]
- Posta, M.; Győrffy, B. Analysis of a large cohort of pancreatic cancer transcriptomic profiles to reveal the strongest prognostic factors. Clin. Transl. Sci. 2023, 16, 1479–1491. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Żebrowska-Nawrocka, M.; Wosiak, A.; Pietrzak, J.; Jeleń, A.; Krygier, A.; Szmajda-Krygier, D.; Sałagacka-Kubiak, A.; Balcerczak, E. NFKB2 gene expression in patients with peptic ulcer diseases and gastric cancer. Mol. Biol. Rep. 2020, 47, 2015–2021. [Google Scholar] [CrossRef]
- Mirmajidi, S.H.; Najafi, M.; Mirmajidi, S.T.; Nasri Nasrabadi, N. Study of regulatory promoter polymorphism (-248 G>A) of Bax; gene in patients with gastric cancer in the northern provinces of Iran. Gastroenterol. Hepatol. Bed Bench 2016, 9, 36–44. [Google Scholar]
- Ocaña, M.G.; Valle-Garay, E.; Montes, A.H.; Meana, A.; Cartón, J.A.; Fierer, J.; Celada, A.; Asensi, V. Bax gene G(-248)A promoter polymorphism is associated with increased lifespan of the neutrophils of patients with osteomyelitis. Genet. Med. 2007, 9, 249–255. [Google Scholar] [CrossRef]
- Bhatt, D.; Verma, A.K.; Bharti, P.S.; Goyal, Y.; Alsahli, M.A.; Almatroudi, A.; Rahmani, A.H.; Almatroodi, S.; Joshi, P.C.; Alam, M.M.; et al. BCL-2 (-938C>A), BAX (-248G>A), and HER2 Ile655Val Polymorphisms and Breast Cancer Risk in Indian Population. J. Oncol. 2021, 2021, 8865624. [Google Scholar] [CrossRef]
- Olbromski, P.J.; Bogacz, A.; Bukowska, M.; Kamiński, A.; Moszyński, R.; Pawlik, P.; Szeliga, A.; Kotrych, K.; Czerny, B. Analysis of the Polymorphisms and Expression Levels of the BCL2, BAX and c-MYC Genes in Patients with Ovarian Cancer. Int. J. Mol. Sci. 2023, 24, 16309. [Google Scholar] [CrossRef]
- Javid, J.; Mir, R.; Julka, P.; Ray, P.C.; Saxena, A. BAX G(-248)A Gene Polymorphism and Its Association with Risk of Non-Small Cell Lung Cancer—A Case Control Study. Open J. Apoptosis 2015, 4, 47–58. [Google Scholar] [CrossRef]
- Chen, K.; Hu, Z.; Wang, L.E.; Sturgis, E.M.; El-Naggar, A.K.; Zhang, W.; Wei, Q. Single-nucleotide polymorphisms at the TP53-binding or responsive promoter regions of BAX and BCL2 genes and risk of squamous cell carcinoma of the head and neck. Carcinogenesis 2007, 28, 2008–2012. [Google Scholar] [CrossRef]
- Chatterjee, K.; De, S.; Deb Roy, S.; Sahu, S.K.; Chakraborty, A.; Ghatak, S.; Das, N.; Mal, S.; Roy Chattopadhyay, N.; Das, P.; et al. BAX-248 G>A and BCL2-938 C>A Variant Lowers the Survival in Patients with Nasopharyngeal Carcinoma and Could be Associated with Tissue-Specific Malignancies: A Multi-Method Approach. Asian Pac. J. Cancer Prev. 2021, 22, 1171–1181. [Google Scholar] [CrossRef]
- Cardoso-Duarte, L.C.A.; Fratelli, C.F.; Pereira, A.S.R.; de Souza, J.N.G.; Freitas, R.S.; de Morais, R.M.; Sobrinho, A.B.; Sousa Silva, C.M.; de Oliveira, J.R.; Oliveira, D.M.; et al. BAX gene (−248 G > A) polymorphism in a sample of patients diagnosed with thyroid cancer in the Federal District, Brazil. Int. J. Biol. Markers 2021, 36, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Xu, Y.; Dou, Y.; Xu, D. Helicobacter pylori and gastric cancer: Mechanisms and new perspectives. J. Hematol. Oncol. 2025, 18, 10. [Google Scholar] [CrossRef] [PubMed]
- Iimura, M.; Nakamura, T.; Shinozaki, S.; Iizuka, B.; Inoue, Y.; Suzuki, S.; Hayashi, N. Bax is downregulated in inflamed colonic mucosa of ulcerative colitis. Gut 2000, 47, 228–235. [Google Scholar] [CrossRef] [PubMed]
- Bartchewsky, W., Jr.; Martini, M.R.; Squassoni, A.C.; Alvarez, M.C.; Ladeira, M.S.; Salvatore, D.M.; Trevisan, M.A.; Pedrazzoli, J., Jr.; Ribeiro, M.L. Effects of Helicobacter pylori infection on the expressions of Bax and Bcl-2 in patients with chronic gastritis and gastric cancer. Dig. Dis. Sci. 2010, 55, 111–116. [Google Scholar] [CrossRef]
- Liu, H.F.; Liu, W.W.; Wang, G.A.; Teng, X.C. Effect of Helicobacter pylori infection on Bax protein expression in patients with gastric precancerous lesions. World J. Gastroenterol. 2005, 11, 5899–5901. [Google Scholar] [CrossRef]
- Zhang, H.; Fang, D.C.; Wang, R.Q.; Yang, S.M.; Liu, H.F.; Luo, Y.H. Effect of Helicobacter pylori infection on expression of Bcl-2 family members in gastric adenocarcinoma. World J. Gastroenterol. 2004, 10, 227–230. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhao, M.; Shi, F.; Zheng, S.; Xiong, L.; Zheng, L. A review of signal pathway induced by virulent protein CagA of Helicobacter pylori. Front. Cell. Infect. Microbiol. 2023, 13, 1062803. [Google Scholar] [CrossRef]
- Mandal, M.; Olson, D.J.; Sharma, T.; Vadlamudi, R.K.; Kumar, R. Butyric acid induces apoptosis by up-regulating Bax expression via stimulation of the c-Jun N-terminal kinase/activation protein-1 pathway in human colon cancer cells. Gastroenterology 2001, 120, 71–78, Erratum in Gastroenterology 2013, 144, 1576. [Google Scholar] [CrossRef]
- Ohmae, T.; Hirata, Y.; Maeda, S.; Shibata, W.; Yanai, A.; Ogura, K.; Yoshida, H.; Kawabe, T.; Omata, M. Helicobacter pylori activates NF-κB via the alternative pathway in B lymphocytes. J. Immunol. 2005, 175, 7162–7169. [Google Scholar] [CrossRef]
- Grimm, T.; Schneider, S.; Naschberger, E.; Huber, J.; Guenzi, E.; Kieser, A.; Reitmeir, P.; Schulz, T.F.; Morris, C.A.; Stürzl, M. EBV latent membrane protein-1 protects B cells from apoptosis by inhibition of BAX. Blood 2005, 105, 3263–3269. [Google Scholar] [CrossRef]
- Lamb, A.; Chen, L.F. The many roads traveled by Helicobacter pylori to NF-κB activation. Gut Microbes 2010, 1, 109–113. [Google Scholar] [CrossRef] [PubMed]
- Rajabi-Moghaddam, M.; Abbaszadeh, H. Gene polymorphisms and risk of head and neck squamous cell carcinoma: A systematic review. Rep. Pract. Oncol. Radiother. 2022, 27, 1058–1076. [Google Scholar] [CrossRef]
- Mrózek, A.; Petrowsky, H.; Sturm, I.; Kraus, J.; Hermann, S.; Hauptmann, S.; Lorenz, M.; Dörken, B.; Daniel, P.T. Combined p53/Bax mutation results in extremely poor prognosis in gastric carcinoma with low microsatellite instability. Cell Death Differ. 2003, 10, 461–467. [Google Scholar] [CrossRef]
- Facundo, A.N.; Magalhães, M.; Nascimento, G.C.; Azulay, R.S.; Santos, R.M.; Freitas, L.A.; Nascimento, A.; Rodrigues, V.P.; Santos, W.C.; Beckman, A.; et al. The expression of VDACs and Bcl2 family genes in pituitary adenomas: Clinical correlations and postsurgical outcomes. Front Endocrinol. 2024, 10, 1481050. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Yang, H.; Qiao, X.; Chen, Y.; Zheng, L.; Lin, J.; Lang, J.; Yu, Q.; Wang, Z. The E3 ubiquitin ligase TRIM17 promotes gastric cancer survival and progression via controlling BAX stability and antagonizing apoptosis. Cell Death Differ. 2023, 10, 2322–2335. [Google Scholar] [CrossRef] [PubMed]
- Sohn, S.K.; Jung, J.T.; Kim, D.H.; Kim, J.G.; Kwak, E.K.; Park, T.I.; Shin, D.G.; Sohn, K.R.; Lee, K.B. Prognostic significance of bcl-2, bax, and p53 expression in diffuse large B-cell lymphoma. Am. J. Hematol. 2003, 73, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Kazemi, N.; Shahrestani, S.B. Effect of Saffron Extract on Expression of Bax and Bcl-2 Genes in Gastric Adenocarcinoma Cell Line (AGS). Gene Cell Tissue 2018, 5, e63608. [Google Scholar] [CrossRef]

| BAX G-248A | Peptic Ulcer Patients N = 183 | Healthy Individuals N = 86 | p | q |
|---|---|---|---|---|
| GG | 142 (77.6%) | 63 (73.3%) | 0.2079 | 0.62352 |
| GA | 37 (20.2%) | 23 (26.7%) | ||
| AA | 4 (2.2%) | 0 (0.0%) | ||
| HWE $ (p values) | 0.9053 | 0.3300 | ||
| Gastric Cancer Patients (N = 13) | Healthy Individuals (N = 86) | |||
| GG | 1 (7.7%) | 63 (73.3%) | <0.001 | 0.006 |
| GA | 10 (76.9%) | 23 (26.7%) | ||
| AA | 2 (15.4%) | 0 (0.0%) | ||
| HWE $ (p values) | 0.2545 | 0.3300 |
| BAX G-248A | Peptic Ulcer Patients N = 183 | |||
|---|---|---|---|---|
| Infected N = 90 | Uninfected N = 93 | p | q | |
| GG | 67 (74.5%) | 75 (80.6%) | 0.4393 | 0.6235 |
| GA | 20 (22.2%) | 17 (18.3%) | ||
| AA | 3 (3.3%) | 1 (1.1%) | ||
| H. pylori Infection Severity(N = 90) assessed at | ||||
| (+) N = 47 | (++) or (+++) N = 43 | |||
| GG | 34 (37.8%) | 33 (36.7%) | 0.8306 | 0.8306 |
| GA | 11 (12.2%) | 9 (10.0%) | ||
| AA | 2 (2.2%) | 1 (1.1%) | ||
| Relative BAX mRNA Level | N | Median | Min. | Max. | Lower Quartile | Upper Quartile | p | q | |
|---|---|---|---|---|---|---|---|---|---|
| PEPTIC ULCER | All cases | 36 | 1.2000 | 0.0949 | 14.5829 | 0.2483 | 3.0201 | ||
| Women | 21 | 0.4784 | 0.0949 | 14.5829 | 0.2237 | 2.9887 | 0.5421 | 0.6324 | |
| Men | 15 | 1.6543 | 0.0967 | 9.6657 | 0.4806 | 3.0515 | |||
| Uninfected with H. pylori | 26 | 2.3924 | 0.1014 | 14.5829 | 0.3305 | 3.6205 | 0.0207 | 0.1449 | |
| Infected with H. pylori | 10 | 0.3862 | 0.0949 | 1.9402 | 0.1851 | 1.0043 | |||
| Severity of infection assessed at (+) | 6 | 0.2384 | 0.0949 | 1.9402 | 0.0967 | 0.4806 | 0.2410 | 0.3976 | |
| Severity of infection assessed at (++) or (+++) | 4 | 0.8443 | 0.2021 | 1.8313 | 0.4432 | 1.4178 | |||
| GASTRIC CANCER | All cases | 13 | 1.4241 | 0.0477 | 14.7230 | 0.3950 | 3.9724 | ||
| Women | 5 | 1.4241 | 0.3950 | 8.2249 | 0.9794 | 1.4743 | 1.0000 | 1.0000 | |
| Men | 8 | 1.3608 | 0.0477 | 14.7230 | 0.3029 | 7.4503 | |||
| TNM Tis or I | 6 | 2.8329 | 0.9794 | 10.9283 | 1.0281 | 8.2249 | 0.1375 | 0.3934 | |
| TNM II or III | 7 | 0.3950 | 0.0477 | 14.7230 | 0.2570 | 1.4743 | |||
| G1 | 6 | 2.8329 | 0.3487 | 10.9283 | 0.9794 | 8.2249 | 0.2840 | 0.3976 | |
| G2 or G3 | 7 | 1.0281 | 0.0477 | 14.7230 | 0.2570 | 1.4743 | |||
| MORPHOLOGICALLY NORMAL TISSUE | All cases | 6 | 1.4382 | 0.0677 | 5.1860 | 0.1523 | 2.8375 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Żebrowska-Nawrocka, M.; Świechowski, R.; Szmajda-Krygier, D.; Lenda, B.; Mirowski, M.; Czaplij, M.; Balcerczak, M.; Balcerczak, E. In Silico and Wet Analysis of BAX Gene G-248A Polymorphism and mRNA Expression in Peptic Ulcer Disease and Gastric Cancer. Curr. Issues Mol. Biol. 2025, 47, 1005. https://doi.org/10.3390/cimb47121005
Żebrowska-Nawrocka M, Świechowski R, Szmajda-Krygier D, Lenda B, Mirowski M, Czaplij M, Balcerczak M, Balcerczak E. In Silico and Wet Analysis of BAX Gene G-248A Polymorphism and mRNA Expression in Peptic Ulcer Disease and Gastric Cancer. Current Issues in Molecular Biology. 2025; 47(12):1005. https://doi.org/10.3390/cimb47121005
Chicago/Turabian StyleŻebrowska-Nawrocka, Marta, Rafał Świechowski, Dagmara Szmajda-Krygier, Bartosz Lenda, Marek Mirowski, Michał Czaplij, Mariusz Balcerczak, and Ewa Balcerczak. 2025. "In Silico and Wet Analysis of BAX Gene G-248A Polymorphism and mRNA Expression in Peptic Ulcer Disease and Gastric Cancer" Current Issues in Molecular Biology 47, no. 12: 1005. https://doi.org/10.3390/cimb47121005
APA StyleŻebrowska-Nawrocka, M., Świechowski, R., Szmajda-Krygier, D., Lenda, B., Mirowski, M., Czaplij, M., Balcerczak, M., & Balcerczak, E. (2025). In Silico and Wet Analysis of BAX Gene G-248A Polymorphism and mRNA Expression in Peptic Ulcer Disease and Gastric Cancer. Current Issues in Molecular Biology, 47(12), 1005. https://doi.org/10.3390/cimb47121005






