Toward an Efficient Differentiation of Two Diaporthe Strains Through Mass Spectrometry for Fungal Biotyping
Abstract
1. Introduction
2. Materials and Methods
2.1. Selection of Endophytic Fungi and Culture of Strains
2.2. Fungal Strains Identification: Morphological and Molecular
2.3. Initial Cultivation and Extraction
2.4. Analysis of Organic Extracts with LC-MS
2.5. ESI-TQD-MS/MS of m/z of Interest
3. Results
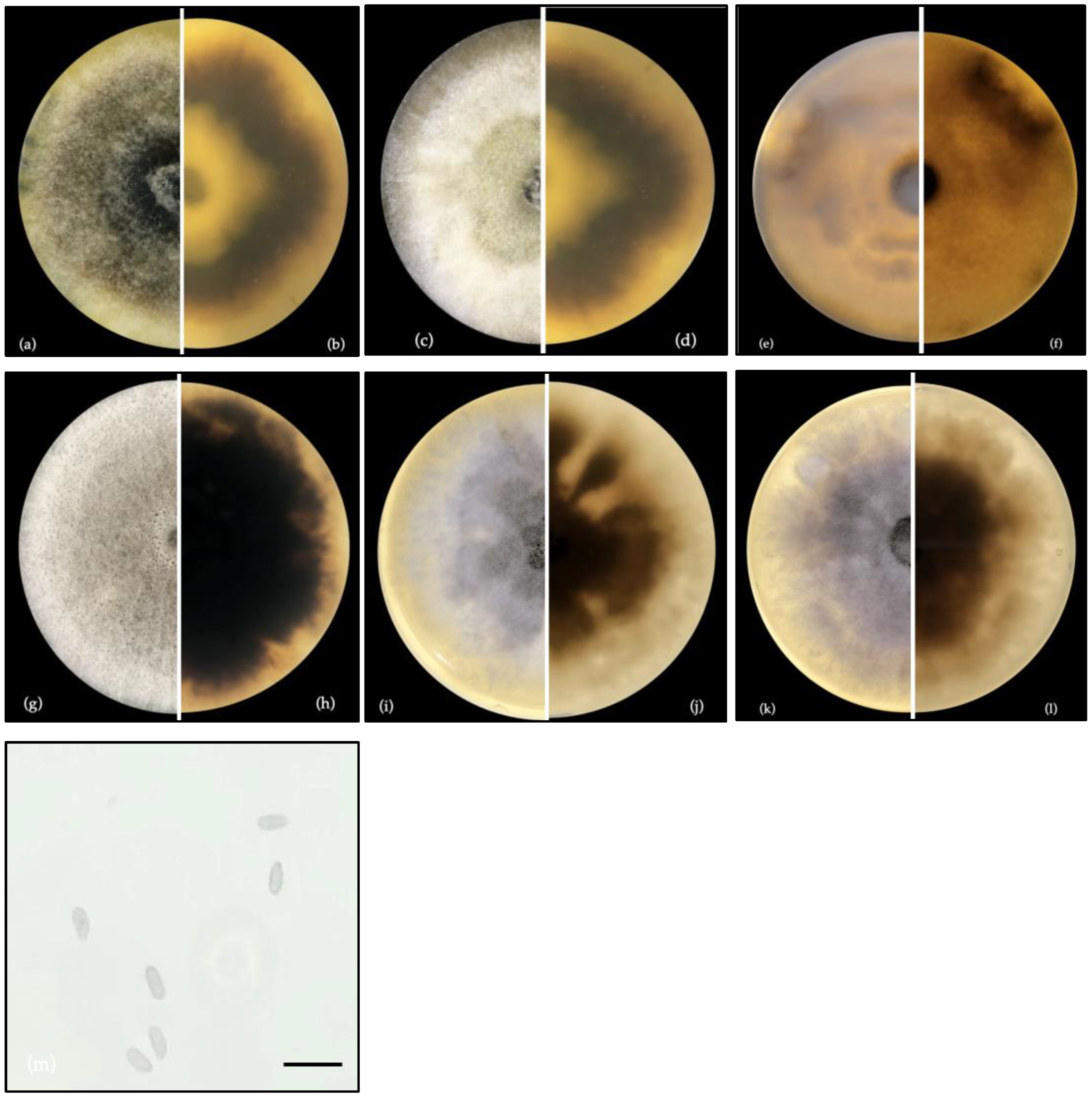
3.1. Morphological Identification and Molecular Characterization of Fungal Strains
3.2. Extraction and UPLC-ESI-MS Profile of Extracts
3.3. Identification of Metabolites with ESI-MS/MS
4. Discussion

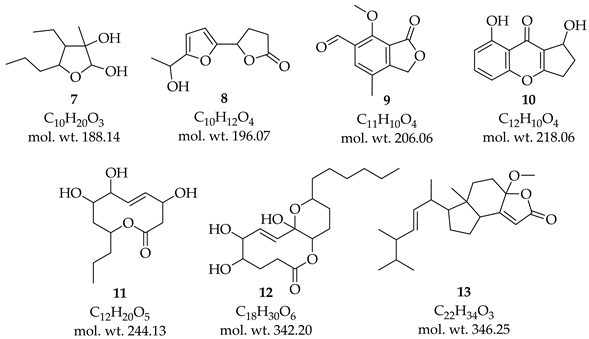
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Strobel, G.A. Endophytes as sources of bioactive products. Microbes Infect. 2003, 5, 535–544. [Google Scholar] [CrossRef] [PubMed]
- Schulz, B.; Boyle, C. The endophytic continuum. Mycol. Res. 2005, 109, 661–686. [Google Scholar] [CrossRef] [PubMed]
- Mattoo, A.J.; Nonzom, S. Endophytic fungi: Understanding complex cross-talks. Symbiosis 2021, 83, 237–264. [Google Scholar] [CrossRef]
- Chagas, F.O.; Pessotti, R.D.C.; Caraballo-Rodríguez, A.M.; Tallarico Pupo, M. Chemical signaling involved in plant-microbe interactions. Chem. Soc. Rev. 2018, 47, 1652–1704. [Google Scholar] [CrossRef]
- Demain, A.L.; Fang, A. The natural functions of secondary metabolites. Adv. Biochem. Eng. Biotechnol. 2000, 69, 1–39. [Google Scholar] [CrossRef]
- Hyde, K.D.; Soytong, K. The fungal endophyte dilemma. Fungal Divers 2008, 33, 163–173. [Google Scholar]
- Wilson, D. Endophyte: The Evolution of a Term, and Clarification of Its Use and Definition. Oikos 1995, 73, 274. [Google Scholar] [CrossRef]
- Hyde, K.D.; Xu, J.; Rapior, S.; Jeewon, R.; Lumyong, S.; Niego, A.G.T.; Abeywickrama, P.D.; Aluthmuhandiram, J.V.S.; Brahamanage, R.S.; Brooks, S.; et al. The amazing potential of fungi: 50 ways we can exploit fungi industrially. Fungal Divers 2019, 97, 1–136. [Google Scholar] [CrossRef]
- Cortés-Sánchez, A.d.J.; Mosqueda-Olivares, T. Una mirada a los organismos fúngicos: Fábricas versátiles de diversos metabolitos secundarios de interés biotecnológico. Química Viva 2013, 12, 64–90. [Google Scholar]
- Yahr, R.; Schoch, C.L.; Dentinger, B.T.M. Scaling up discovery of hidden diversity in fungi: Impacts of barcoding approaches. Philos. Trans. B 2016, 371, 20150336. [Google Scholar] [CrossRef]
- Raja, H.A.; Miller, A.N.; Pearce, C.J.; Oberlies, N.H. Fungal Identification Using Molecular Tools: A Primer for the Natural Products Research Community. J. Nat. Prod. 2017, 80, 756–770. [Google Scholar] [CrossRef]
- Gomes, R.R.; GLienke, C.; Videira, S.I.R.; Lombard, L.; Groenewald, J.Z.; Crous, P.W. Diaporthe: A genus of endophytic, saprobic and plant pathogenic fungi. Persoonia 2013, 31, 1–41. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Liu, F.; Duan, W.; Crous, P.W.; Cai, L. Diaporthe is paraphyletic. IMA Fungus 2017, 8, 153–187. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Manawasinghe, I.S.; Huang, Y.; Shu, Y.; Phillips, A.J.L.; Dissanayake, A.J.; Hyde, K.D.; Xiang, M.; Luo, M. Endophytic Diaporthe Associated With Citrus grandis cv. Tomentosa in China. Front. Microbiol. 2021, 11, 609387. [Google Scholar] [CrossRef] [PubMed]
- Norphanphoun, C.; Gentekaki, E.; Hongsanan, S.; Jayawardena, R.; Senanayake, I.C.; Manawasinghe, I.S.; Abeywickrama, P.D.; Bhunjun, C.S.; Hyde, K.D. Diaporthe: Formalizing the species-group concept. Mycosphere 2022, 13, 752–819. [Google Scholar] [CrossRef]
- Monkai, J.; Hongsanan, S.; Bhat, D.J.; Dawoud, T.M.; Lumyong, S. Integrative Taxonomy of Novel Diaporthe Species Associated with Medicinal Plants in Thailand. J. Fungi 2023, 9, 603. [Google Scholar] [CrossRef]
- Calla-Quispe, E.; Fuentes-Rivera, H.L.; Ramírez, P.; Martel, C.; Ibañez, A.J. Mass Spectrometry: A Rosetta Stone to Learn How Fungi Interact and Talk. Life 2020, 10, 89. [Google Scholar] [CrossRef]
- Maciá-Vicente, J.G.; Shi, Y.-N.; Cheikh-Ali, Z.; Grün, P.; Glynou, K.; Haghi Kia, S.; Piepenbring, M.; Bode, H.B. Metabolomics-based chemotaxonomy of root endophytic fungi for natural products discovery. Environ. Microbiol. 2018, 3, 1253–1270. [Google Scholar] [CrossRef]
- Hufsky, F.; Scheubert, K.; Sebastian, B. New kids on the block: Novel informatics methods for natural product discovery. Nat. Prod. Rep. 2014, 31, 807–817. [Google Scholar] [CrossRef]
- Bayona, L.M.; Verpoorte, R.; Klinkhamer, P.G.L.; Choi, Y.-H. Thin-Layer Chromatography | Metabolomics. In Encyclopedia of Analytical Science, 3rd ed.; Worsfold, P., Poole, C., Townshend, A., Miró, M., Eds.; Academic Press: Cambridge, MA, USA, 2019; Volume 10, pp. 59–75. ISBN 9780081019849. [Google Scholar]
- Spraker, J.E.; Luu, G.T.; Sanchez, L.M. Imaging mass spectrometry for natural products discovery: A review of ionization methods. Nat. Prod. Rep. 2020, 37, 150–162. [Google Scholar] [CrossRef]
- Smedsgaard, J.; Frisvad, J.C. Using direct electrospray mass spectrometry in taxonomy and secondary metabolite profiling of crude fungal extracts. J. Microbiol. Methods 1996, 25, 5–17. [Google Scholar] [CrossRef]
- Frisvad, J.C. Media and growth conditions for induction of secondary metabolite production. Methods Mol. Biol. 2012, 944, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Ćilerdžić, J.L.; Sofrenić, I.V.; Tešević, V.V.; Brčeski, I.D.; Duletić-Laušević, S.N.; Vukojević, J.B.; Stajić, M.M. Neuroprotective Potential and Chemical Profile of Alternatively Cultivated Ganoderma lucidum Basidiocarps. Chem. Biodivers. 2018, 15, e1800036. [Google Scholar] [CrossRef] [PubMed]
- Westphal, K.R.; Heidelbach, S.; Zeuner, E.J.; Riisgaard-Jensen, M.; Nielsen, M.E.; Vestergaard, S.Z.; Bekker, N.S.; Skovmark, J.; Olesen, C.K.; Thomsen, K.H.; et al. The effects of different potato dextrose agar media on secondary metabolite production in Fusarium. Int. J. Food Microbiol. 2021, 347, 109171. [Google Scholar] [CrossRef]
- Tiwari, P.; Bae, H. Endophytic Fungi: Key Insights, Emerging Prospects, and Challenges in Natural Product Drug Discovery. Microorganisms 2022, 10, 360. [Google Scholar] [CrossRef]
- Bode, H.B.; Bethe, B.; Höfs, R.; Zeeck, A. Big Effects from Small Changes: Possible Ways to Explore Nature’s Chemical Diversity. ChemBioChem 2002, 3, 619–627. [Google Scholar] [CrossRef]
- Hanson, J.R. The Chemistry of Growing Fungi. In The Chemistry of Fungi; Royal Society of Chemistry: London, UK, 2008; pp. 18–31. ISBN 9781847558329. [Google Scholar]
- Venugopalan, A.; Srivastava, S. Endophytes as in vitro production platforms of high value plant secondary metabolites. Biotechnol. Adv. 2015, 33, 873–887. [Google Scholar] [CrossRef]
- Chepkirui, C.; Stadler, M. The genus Diaporthe: A rich source of diverse and bioactive metabolites. Mycol. Prog. 2017, 16, 477–494. [Google Scholar] [CrossRef]
- Nagarajan, K.; Tong, W.-Y.; Leong, C.-R.; Tan, W.-N. Potential of Endophytic Diaporthe sp. as a New Source of Bioactive Compounds. J. Microbiol. Biotechnol. 2021, 31, 493–500. [Google Scholar] [CrossRef]
- Xu, T.-C.; Lu, Y.-H.; Wang, J.-F.; Song, Z.-Q.; Hou, Y.-G.; Liu, S.-S.; Liu, C.-S.; Wu, S.-H. Bioactive Secondary Metabolites of the Genus Diaporthe and Anamorph Phomopsis from Terrestrial and Marine Habitats and Endophytes: 2010–2019. Microorganisms 2021, 9, 217. [Google Scholar] [CrossRef]
- Hilário, S.; Gonçalves, M.F.M. Endophytic Diaporthe as Promising Leads for the Development of Biopesticides and Biofertilizers for a Sustainable Agriculture. Microorganisms 2022, 10, 2453. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Khan, B.; Dai, Q.; Lin, J.; Kang, L.; Rajput, N.A.; Yan, W.; Liu, G. Potential of Secondary Metabolites of Diaporthe Species Associated with Terrestrial and Marine Origins. J. Fungi 2023, 9, 453. [Google Scholar] [CrossRef]
- Jiang, L.; Ma, Q.; Li, A.; Sun, R.; Tang, G.; Huang, X.; Pu, H. Bioactive secondary metabolites produced by fungi of the genus Diaporthe (Phomopsis): Structures, biological activities, and biosynthesis. Arab. J. Chem. 2023, 16, 105062. [Google Scholar] [CrossRef]
- Santos, L.; Alves, A.; Alves, R. Evaluating multi-locus phylogenies for species boundaries determination in the genus Diaporthe. PeerJ 2017, 2017, e3120. [Google Scholar] [CrossRef]
- Aumentado, H.D.; Balendres, M.A. Novel species and new records of Diaporthe causing eggplant leaf and fruit blight in the Philippines. Mycol. Prog. 2024, 23, 23. [Google Scholar] [CrossRef]
- Aumentado, H.D.; Balendres, M.A. Diaporthe melongenae sp. nov, a new fungal species causing leaf blight in eggplant. J. Phytopathol. 2024, 172, e13246. [Google Scholar] [CrossRef]
- Quiles Melero, I.; Peláez, T.; Rezusta López, A.; García-Rodríguez, J. Aplicación de la espectrometría de masas en micología. Enferm. Infecc. Microbiol. Clin. 2016, 34, 26–30. [Google Scholar] [CrossRef]
- Xu, C.; Xu, K.; Yuan, X.-L.; Ren, G.-W.; Wang, X.-Q.; Li, W.; Deng, N.; Wang, X.-F.; Zhang, P. Characterization of diketopiperazine heterodimers as potential chemical markers for discrimination of two dominant black aspergilli, Aspergillus niger and Aspergillus tubingensis. Phytochemistry 2020, 176, 112399. [Google Scholar] [CrossRef]
- Frisvad, J.C.; Andersen, B.; Thrane, U. The use of secondary metabolite profiling in chemotaxonomy of filamentous fungi. Mycol. Res. 2008, 112, 231–240. [Google Scholar] [CrossRef]
- Polizzotto, R.; Andersen, B.; Martini, M.; Grisan, S.; Assante, G.; Musetti, R. A polyphasic approach for the characterization of endophytic Alternaria strains isolated from grapevines. J. Microbiol. Methods 2012, 88, 162–171. [Google Scholar] [CrossRef]
- Figueroa, M.; Jarmusch, A.K.; Raja, H.A.; El-Elimat, T.; Kavanaugh, J.S.; Horswill, A.R.; Cooks, R.G.; Cech, N.B.; Oberlies, N.H. Polyhydroxyanthraquinones as Quorum Sensing Inhibitors from the Guttates of Penicillium restrictum and Their Analysis by Desorption Electrospray Ionization Mass Spectrometry. J. Nat. Prod. 2014, 77, 1351–1358. [Google Scholar] [CrossRef] [PubMed]
- Fox Ramos, A.E.; Evanno, L.; Poupon, E.; Champy, P.; Beniddir, M.A. Natural products targeting strategies involving molecular networking: Different manners, one goal. Nat. Prod. Rep. 2019, 36, 960–980. [Google Scholar] [CrossRef] [PubMed]
- Kelman, M.J.; Renaud, J.B.; Seifert, K.A.; Mack, J.; Yeung, K.K.-C.; Sumarah, M.W. Chemotaxonomic Profiling of Canadian Alternaria Populations Using High-Resolution Mass Spectrometry. Metabolites 2020, 10, 238. [Google Scholar] [CrossRef] [PubMed]
- Cicaloni, V.; Salvini, L.; Vitalini, S.; Garzoli, S. Chemical Profiling and Characterization of Different Cultivars of Cannabis sativa L. Inflorescences by SPME-GC-MS and UPLC-MS. Separations 2022, 9, 90. [Google Scholar] [CrossRef]
- Molinar, E.; Rios, N.; Spadafora, C.; Elizabeth Arnold, A.; Coley, P.D.; Kursar, T.A.; Gerwick, W.H.; Cubilla-Rios, L. Coibanoles, a new class of meroterpenoids produced by Pycnoporus sanguineus. Tetrahedron Lett. 2012, 53, 919–922. [Google Scholar] [CrossRef]
- Humber, R.A. Fungi: Preservation of cultures. In Manual of Techniques in Insect Pathology; Lacey, L.A., Ed.; Academic Press: Cambridge, MA, USA, 1997; pp. 269–279. ISBN 9780124325555. [Google Scholar]
- Delgado Gómez, L.M.; Torres-Mendoza, D.; Hernández-Torres, K.; Ortega, H.E.; Cubilla-Rios, L. Identification of Secondary Metabolites from the Mangrove-Endophyte Lasiodiplodia iranensis F0619 by UPLC-ESI-MS/MS. Metabolites 2023, 13, 912. [Google Scholar] [CrossRef]
- Frisvad, J.C. Fungal Chemotaxonomy. In Biosynthesis and Molecular Genetics of Fungal Secondary Metabolites, Volume 2; Zeilinger, S., Martín, J.-F., García-Estrada, C., Eds.; Springer: New York, NY, USA, 2015; pp. 103–121. ISBN 978-1-4939-2531-5. [Google Scholar]
- Vidal, A.; Parada, R.; Mendoza, L.; Cotoras, M. Endophytic fungi isolated from plants growing in central andean precordillera of Chile with antifungal activity against Botrytis cinerea. J. Fungi 2020, 6, 149. [Google Scholar] [CrossRef]
- Sun, M.-H.; Gao, L.; Liu, X.-Z.; Wang, J.-L. Fungal sporulation in two-stage cultivation. Mycosystema 2009, 28, 64–72. [Google Scholar]
- Sun, W.; Huang, S.; Xia, J.; Zhang, X.; Li, Z. Morphological and molecular identification of Diaporthe species in south-western China, with description of eight new species. MycoKeys 2021, 77, 65–95. [Google Scholar] [CrossRef]
- Tekpinar, A.D.; Kalmer, A. Utility of various molecular markers in fungal identification and phylogeny. Nov. Hedwigia 2019, 109, 187–224. [Google Scholar] [CrossRef]
- Arnold, A.E.; Mejía, L.C.; Kyllo, D.; Rojas, E.I.; Maynard, Z.; Robbins, N.; Herre, E.A. Fungal endophytes limit pathogen damage in a tropical tree. Proc. Natl. Acad. Sci. USA 2003, 100, 15649–15654. [Google Scholar] [CrossRef] [PubMed]
- Rohlfs, M.; Albert, M.; Keller, N.P.; Kempken, F. Secondary chemicals protect mould from fungivory. Biol. Lett. 2007, 3, 523–525. [Google Scholar] [CrossRef] [PubMed]
- Rateb, M.E.; Ebel, R. Secondary metabolites of fungi from marine habitats. Nat. Prod. Rep. 2011, 28, 290–344. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.; Verma, A.; Rai, N.; Singh, A.K.; Singh, S.K.; Kumar, B.; Kumar, R.; Gautam, V. Mass Spectrometry-Based Technology and Workflows for Studying the Chemistry of Fungal Endophyte Derived Bioactive Compounds. ACS Chem. Biol. 2021, 16, 2068–2086. [Google Scholar] [CrossRef]
- Andolfi, A.; Boari, A.; Evidente, M.; Cimmino, A.; Vurro, M.; Ash, G.; Evidente, A. Gulypyrones A and B and Phomentrioloxins B and C Produced by Diaporthe gulyae, a Potential Mycoherbicide for Saffron Thistle (Carthamus lanatus). J. Nat. Prod. 2015, 78, 623–629. [Google Scholar] [CrossRef]
- Evidente, A.; Rodeva, R.; Andolfi, A.; Stoyanova, Z.; Perrone, C.; Motta, A. Phytotoxic polyketides produced by Phomopsis foeniculi, a strain isolated from diseased Bulgarian fennel. Eur. J. Plant Pathol. 2011, 130, 173–182. [Google Scholar] [CrossRef]
- Hu, M.; Yang, X.-Q.; Wan, C.-P.; Wang, B.-Y.; Yin, H.-Y.; Shi, L.-J.; Wu, Y.-M.; Yang, Y.-B.; Zhou, H.; Ding, Z.-T. Potential antihyperlipidemic polyketones from endophytic Diaporthe sp. JC-J7 in Dendrobium nobile. RSC Adv. 2018, 8, 41810–41817. [Google Scholar] [CrossRef]
- Tan, Q.; Yan, X.; Lin, X.; Huang, Y.; Zheng, Z.; Song, S.; Lu, C.; Shen, Y. Chemical Constituents of the Endophytic Fungal Strain Phomopsis sp. NXZ-05 of Camptotheca acuminata. Helv. Chim. Acta 2007, 90, 1811–1817. [Google Scholar] [CrossRef]
- Wu, S.-H.; Chen, Y.-W.; Shao, S.-C.; Wang, L.-D.; Li, Z.-Y.; Yang, L.-Y.; Li, S.-L.; Huang, R. Ten-Membered Lactones from Phomopsis sp., an Endophytic Fungus of Azadirachta indica. J. Nat. Prod. 2008, 71, 731–734. [Google Scholar] [CrossRef]
- Chen, C.-J.; Liu, X.-X.; Zhang, W.-J.; Zang, L.-Y.; Wang, G.; Weng Ng, S.; Tan, R.-X.; Ge, H.-M. Sesquiterpenoids isolated from an endophytic fungus Diaporthe sp. RSC Adv. 2015, 5, 17559–17565. [Google Scholar] [CrossRef]
- Shi, C.; Yuan, L.; Zhao, P.-J. Two new lactone derivatives from an endophyte Diaporthe sp. XZ-07 cultivated on Camptotheca acuminata. Zhongguo Zhong Yao Za Zhi 2016, 41, 1860–1863. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Huang, Y.; Fang, M.; Wang, J.; Zheng, Z.; Su, W. Cytotoxic and antimicrobial metabolites from marine lignicolous fungi, Diaporthe sp. FEMS Microbiol. Lett. 2005, 251, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Bungihan, M.E.; Tan, M.A.; Kitajima, M.; Kogure, N.; Franzblau, S.G.; dela Cruz, T.E.E.; Takayama, H.; Nonato, M.G. Bioactive metabolites of Diaporthe sp. P133, an endophytic fungus isolated from Pandanus amaryllifolius. J. Nat. Med. 2011, 65, 606–609. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-Y.; Wang, M.-Z.; Huang, Y.-J.; Shen, Y.-M. Secondary metabolites from Phomopsis sp. A123. Mycology 2010, 1, 254–261. [Google Scholar] [CrossRef]
- Lin, X.; Lu, C.-H.; Shen, Y.-M. One New Ten-membered Lactone from Phomopsis sp. B27, an Endophytic Fungus of Annona squamosa. Chin. J. Nat. Med. 2008, 6, 391–394. [Google Scholar] [CrossRef]
- Ito, A.; Maeda, H.; Tonouchi, A.; Hashimoto, M. Relative and absolute structure of phomolide C. Biosci. Biotechnol. Biochem. 2015, 79, 1067–1069. [Google Scholar] [CrossRef]
- Li, G.; Kusari, S.; Kusari, P.; Kayser, O.; Spiteller, M. Endophytic Diaporthe sp. LG23 Produces a Potent Antibacterial Tetracyclic Triterpenoid. J. Nat. Prod. 2015, 78, 2128–2132. [Google Scholar] [CrossRef]



| Culture Media | Period of Incubation | F0728 | F0891 |
|---|---|---|---|
| MEA | 7 days | 163 (100) **; 177 (56.8); 211 (28.4); 213 (34.6); 215 (58.8); 307 (13.2); 325 (14.3) | 159 (46.6); 180 (87.6); 183 (100); 193 (80.3); 207 (32.1); 211 (69.4); 229 (19.0); 245 (17.9); 251 (14.8) 268 (14.8); 279 (13.0); 331 (10.2); 340 (11.2); 384 (11.2) |
| 15 days | 163 (23.3); 177 (100); 191 (52.3); 207 (13.6); 213 (10.8) | 159 (37.9); 185 (100); 211 (25.3); 213 (26.7); 279 (15.0) 307 (11.4); 321 (68.5); 343 (65.4); 351 (16.3); 365 (12.3) | |
| 22 days | 163 (16.5); 177 (100); 191 (60.6); 205 (15.8); 207 (16.1) | 159 (23.4); 185 (100); 207 (58.0); 213 (33.1); 243 (23.7); 251 (20.0); 321 (44.0); 343 (32.9) | |
| 30 days | 175 (48.1); 177 (35.7); 179 (68.6); 187 (100); 196 (39.8); 213 (25.5); 233 (39.3); 277 (24.5); 317 (39.0); 333 (19.0); 390 (10.5) | 159 (17.6); 180 (22.7); 185 (100); 207 (40.5); 213 (28.3); 243 (19.8); 321 (43.4); 343 (43.1); 365 (12.8); 379 (12.7) | |
| PDA | 7 days | 160 (100); 163 (52.0); 171 (51.5); 177 (50.1); 180 (37.9); 189 (35.2); 215 (46.8); 227 (14.6); 345 (10.4) | 180 (27.1); 195 (89.8); 213 (58.7); 295 (18.0); 331 (12.5); 348 (19.0); 351 (18.0; 353 (100) |
| 15 days | 160 (79.3); 163 (94.5); 171 (44.36); 177 (100); 189 (72.02); 191 (40.1); 213 (47.5); 215 (58.1); 259 (19.9); 309 (28.1); 325 (18.6) | 180 (50.4); 195 (53.3); 211 (40.2); 213 (34.3); 268 (24.2); 295 (12.0) 351 (21.0); 353 (100) | |
| 22 days | 160 (52.4); 163 (100); 177 (62.7); 189 (30.3); 191 (19.4); 211 (14.2); 213 (21.2); 215 (20.3); 309 (16.5) | 165 (23.5); 180 (27.1) 185 (34.1); 193 (58.0); 195 (92.2); 213 (51.9); 233 (23.9); 268 (40.5); 351 (28.7); 353 (100) | |
| 30 days | 163 (100); 177 (57.5); 191 (89.5); 207 (76.3); 213 (96.6); 215 (64.0); 229 (45.7); 259 (58.0); 275 (29.7); 307 (20.7); 359 (23.5); 413 (17.3); 429 (10.2) | 165 (33.8); 183 (21.8); 193 (62.2); 195 (57.0); 211 (16.6); 213 (28.8); 237 (21.4); 268 (21.2); 351 (24.2); 353 (100) | |
| SDA | 7 days | 160 (100); 163 (39.7); 171 (41.4); 189 (41.5); 211 (24.4); 215 (64.0) 265 (25.1); 309 (16.5); 325 (10.9); 337 (12.5); 353 (11.3) | 180 (23.5); 197 (47.8); 211 (100); 245 (58.3); 261 (13.3) |
| 15 days | 160 (100); 163 (21.7); 164 (29.8); 175 (36.0); 189 (48.9); 211 (23.6); 215 (26.5) | 164 (27.4); 180 (50.0; 188 (73.4); 197 (45.4); 211 (100); 245 (57.6); 261 (19.7); 353 (31.8) | |
| 22 days | 160 (100); 163 (51.0); 171 (37.2); 175 (36.0); 183 (34.3); 189 (52.1); 193 (41.8); 211 (30.4); 213 (34.0); 215 (80.7); 228 (29.0); 265 (20.0); 309 (25.0) | 164 (47.0); 180 (45.0); 197 (34.7); 211 (100); 245 (64.8); 261 (18.7); 353 (33.2) | |
| 30 days | 160 (100); 163 (51.7); 171 (32.4); 177 (35.3); 189 (52.8); 211 (27.3); 213 (31.0); 215 (70.0); 265 (26.5); 309 (22.7) | 164 (57.0); 180 (44.8); 185 (30.6); 197 (38.5); 211 (100); 245 (61.8); 353 (33.8) |
| Culture Media | Period of Incubation | F0728 | F0891 |
|---|---|---|---|
| MEA | 7 days | 169 (55.7) **; 179 (57.6); 193 (58.3); 195 (100); 209 (66.1); 213 (20.9); 235 (15.7); 279 (16.8); 349 (20.1); 357 (18.0); 371 (11.5) | 155 (100); 173 (20.0); 199 (19.7); 217 (12.5); 249 (82.5); 329 (27.2); 385 (11.5); 505 (16.7) |
| 15 days | 169 (20.9); 193 (17.2); 209 (30.1); 211 (27.7); 213 (16.7); 345 (19.8); 363 (100); 365 (50.1); 378 (34.9); 394 (38.0) 396 (50.1) | 155 (100); 173 (92.7); 185 (34.9); 199 (49.8); 213 (30.2); 227 (20.9); 249 (29.2); 319 (21.2); 329 (26.7); 337 (15.3); 505 (12.9); 709 (23.1) | |
| 22 days | 167 (42.5); 169 (59.6); 193 (44.7); 209 (59.7); 211 (67.9); 217 (100); 249 (31.2); 345 (66.2); 363 (59.6); 365 (24.5); 379 (20.2); 396 (23.7) | 155 (70.0); 173 (100); 185 (38.9); 199 (59.6); 213 (30.0); 227 (25.8); 241 (27.0); 249 (24.4); 269 (22.1); 279 (19.9); 319 (27.5); 329 (53.3); 489 (13.2); 505 (17.1) | |
| 30 days | 165 (20.3); 206 (16.9); 251 (25.7); 291 (74.1); 363 (100); 379 (25.1); 415 (74.9); 469 (14.3); 499 (12.2); 612 (25.0) | 155 (95.1); 173 (100); 185 (43.4); 199 (72.0); 213 (57.0); 225 (35.0); 241 (48.8); 249 (36.0); 255 (33.9); 269 (32.8); 283 (26.4); 319 (79.0); 329 (63.4); 337 (42.9); 353 (26.6); 473 (26.0); 489 (17.6); 505 (40.8); 533 (27.6); 537 (17.4); 547 (14.4); 577 (14.0) | |
| PDA | 7 days | 162 (18.4); 169 (13.0); 193 (34.0); 195 (30.3); 209 (27.0); 285 (14.3); 363 (100); 365 (18.2) | 327 (16.2); 329 (100); 330 (13.1) |
| 15 days | 169 (29.6); 179 (100); 193 (38.8); 195 (75.3); 197 (62.4); 209 (34.5); 217 (19.7); 249 (18.9); 347 (14.2); 357 (14.3) | 235 (25.6); 249 (100); 251 (49.2); 327 (16.6); 329 (98.9) | |
| 22 days | 169 (82.0); 195 (100); 209 (68.3); 213 (25.9); 248 (23.5); 331 (21.8); 347 (56.2); 385 (17.8); 407 (38.2); 417 (19.0) | 235 (32.7); 249 (100); 251 (74.2); 327 (13.6); 329 (78.5) | |
| 30 days | 179 (31.0); 193 (49.6); 197 (95.5); 209 (24.5); 217 (100); 235 (20.0) 249 (32.3); 337 (11.0) | 235 (69.0); 249 (100); 251 (21.1); 267 (20.9); 329 (72.2) | |
| SDA | 7 days | 165 (25.4); 193 (13.6); 204 (15.3); 209 (100); 211 (14.6) | 165 (100); 193 (10.6); 251 (11.1); 346 (21.5) |
| 15 days | 165 (12.9); 193 (15.6); 204 (11.0); 209 (100); 211 (45.1) | 155 (11.5); 178 (10.6); 259 (18.2); 275 (10.0); 329 (83.3); 346 (35.5); 390 (10.0) | |
| 22 days | 165 (27.5); 193 (27.0); 204 (100) | 173 (10.0); 259 (10.0); 281 (10.0); 329 (42.9); 346 (10.0); 390 (10.0) | |
| 30 days | 165 (26.3); 193 (19.2); 204 (100); 209 (92.6); 211 (28.4) | 173 (10.0); 259 (10.0); 329 (46.8); 346 (10.0) |
| Culture Media | Time of Culture | F0728 | F0891 | ||
|---|---|---|---|---|---|
| ESI (+) | ESI (-) | ESI (+) | ESI (-) | ||
| 7 days | 211 (28.4) | -------- | 211 (69.4) | -------- | |
| MEA | 15 days | 213 (10.8) | 213 (16.7) | 213 (26.7) | 213 (30.2) |
| 22 days | -------- | 249 (31.2) | -------- | 249 (24.4) | |
| 30 days | 213 (25.5) | -------- | 213 (28.3) | -------- | |
| 7 days | 180 (37.9) | -------- | 180 (27.1) | -------- | |
| PDA | 15 days | 213 (47.5) | 249 (18.9) | 213 (34.3) | 249 (100) |
| 22 days | 213 (21.2) | -------- | 213 (51.9) | -------- | |
| 30 days | 213 (96.6) | 235 (20.0); 249 (32.3) | 213 (28.8) | 235 (69.0); 249 (100) | |
| 7 days | 211 (24.4) | 193 (13.6) | 211 (100) | 193 (10.6) | |
| SDA | 15 days | 211 (23.6) | -------- | 211 (100) | -------- |
| 22 days | 211 (30.4) | -------- | 211 (100) | -------- | |
| 30 days | 211 (27.3) | -------- | 211 (100) | -------- | |
| Compound | Precursor Ion | Product Ions (m/z) and Lost Fragments |
|---|---|---|
| 1 [ESI (+)] | 211 [M + H]+ | 196 (4) * [-CH3]; 183 (5) [-CO]; 141 (35) [-C4H6O]; 123 (70) [-C4H6O/H2O]; 97 (20) [-C4H6O/CO2]; 91 (52) [-C4H6O/H2O/OHCH3]; 70 (73); 57 (92) |
| 2 [ESI (+)] | 213 [M + H]+ | 195 (30) [-H2O]; 185 (2) [-CO]; 181 (5) [-HOCH3]; 177 (5) [-2H2O]; 167 (12) [-C2H6O]; 153 (55) [-CH3OH/CO]; 149 (35) [-C2H6O/H2O]; 109 (38) [-C4H9O/OCH3]; 107 (60) [-C2H4O/CO2/H2O]; 95 (100) [-C4H8O/C2H6O]; 69 (73) [-C4H8O/C2H6O/C2H2]; 57 (92) |
| 3 [ESI (-)] | 193 [M − H]− | 175 (55) [-H2O]; 149 (38) [-CO2]: 147 (52) [-OCH3/CH3]; 131 (30) [-H2O/CO2]; 107 (23) [-C4H7/OCH3] |
| 4 [ESI (+)] | 215 [M + H]+ | 171 (4) [-CO2]; 153 (5) [-CO2/H2O]; 151 (22) [-CO/2H2O]; 139 (100) [-C2H2O/H2O2]; 97 (18) [-C3H6/C2H2O/H2O2] |
| 5 [ESI (-)] | 235 [M − H]− | 193 (8) [-C3H6]; 191 (100) [-CHO/CH3]; 190 (31) [-C3H9]; 149 (5) [-C3H6/CHO/CH3]; 146 (2) [-C4H12/CHO]; 123 (8) [-C6H8O2] |
| 6 [ESI (-)] | 249 [M − H]− | 205 (100) [-CO2]; 203 (30) [-CO/H2O]; 189 (12) [-CO2/H2O]; 189 (12) [-CO2/H2O]; 187 (6) [-CO/2H2O]; 163 (8); 135 (12) |
| 7 [ESI (+)] | 189 [M + H]+ | 174 (100) [-CH3]; 159 (38) [-C2H6]; 158 (5) [-CH3O]; 146 (5) [-C3H7]; 131 (12) [-C4H10] |
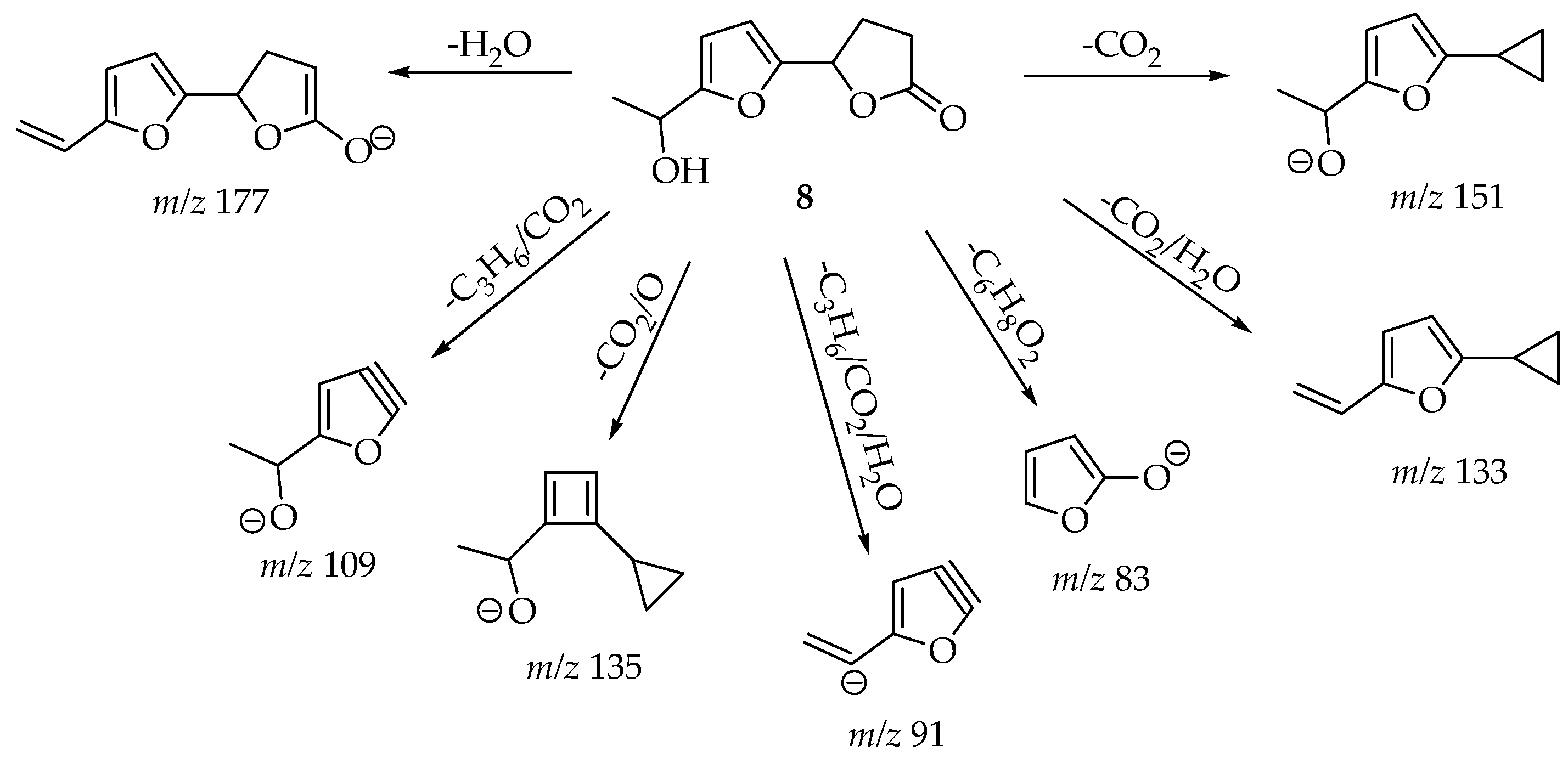
| 8 [ESI (-)] | 195 [M − H]− | 177 (6) [-H2O]; 151 (100) [-CO2]; 135 (32) [-CO2/O]; 133 (6) [-CO2/H2O]; 109 (30) [-C3H6/CO2]; 91 (8) [-C3H6/CO2/H2O]; 83 (8) [-C5H8/CO2] |
| 9 [ESI (+)] | 207 [M + H]+ | 192 (100) [-CH3]; 191 (4) [-O]; 174 (4) [-CH3]; 164 (6) [-CH3/CO]; 163 (5) [-CO2] |
| 10 [ESI (-)] | 217 [M − H]− | 173 (68) [-C2H4O/H+]; 158 (100) [-C2H3O2/H+] |
| 11 [ESI (+)] | 245 [M + H]+ | 159 (18) [-C5H10O]; 153 (16) [-C2H4/CO/2H2O]; 151 (62) [-C2H6/CO/2H2O]; 139 (100) [-C3H8/CO2/H2O] |
| 12 [ESI (+)] | 343 [M + H]+ | 231 (18) [-C8H16]; 213 (14) [-C8H16/H2O]; 183 (14) [-C9H18O/H2O]; 157 (25) [-C9H18O/CO2]; 155 (25) [-C9H18O/H2O/CO]; 149 (48) [-C9H18O/H2O/H2O2]; 123 (42) [-C9H18O/H2O2/CO2]; 121 (60) [-C9H18O/H2O/H2O2/CO]; 119 (84) [-C9H18O/3H2O/CO]; 107 (72) [-C9H18O/H2O2/C2H2O/H2O]; 105 (76) [-C9H18O/H2O2/CO2/H2O]; 95 (78) [-C9H18O/H2O/H2O2/C3H2O]; 93 (100) [-C9H18O/H2O/H2O2/C3H4O] |
| 13 [ESI (-)] | 345 [M − H]− | 327 (6) [-H2O]; 301 (13) [-CO2]; 285 (16) [-CO/CH3OH]; 276 (36) [-C5H9]; 260 (16) [-C5H10/CH3]; 259 (60) [-C5H11/CH3]; 257 (27) [-C3H8/CO2]; 215 (100) [-C5H11/CH3/CO2]; 214 (62) [-C5H12/CH3/CO2] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hernández-Torres, K.; Torres-Mendoza, D.; Navarro-Velasco, G.; Cubilla-Rios, L. Toward an Efficient Differentiation of Two Diaporthe Strains Through Mass Spectrometry for Fungal Biotyping. Curr. Issues Mol. Biol. 2025, 47, 53. https://doi.org/10.3390/cimb47010053
Hernández-Torres K, Torres-Mendoza D, Navarro-Velasco G, Cubilla-Rios L. Toward an Efficient Differentiation of Two Diaporthe Strains Through Mass Spectrometry for Fungal Biotyping. Current Issues in Molecular Biology. 2025; 47(1):53. https://doi.org/10.3390/cimb47010053
Chicago/Turabian StyleHernández-Torres, Kathleen, Daniel Torres-Mendoza, Gesabel Navarro-Velasco, and Luis Cubilla-Rios. 2025. "Toward an Efficient Differentiation of Two Diaporthe Strains Through Mass Spectrometry for Fungal Biotyping" Current Issues in Molecular Biology 47, no. 1: 53. https://doi.org/10.3390/cimb47010053
APA StyleHernández-Torres, K., Torres-Mendoza, D., Navarro-Velasco, G., & Cubilla-Rios, L. (2025). Toward an Efficient Differentiation of Two Diaporthe Strains Through Mass Spectrometry for Fungal Biotyping. Current Issues in Molecular Biology, 47(1), 53. https://doi.org/10.3390/cimb47010053








