A Theoretical Study on the Efficacy and Mechanism of Combined YAP-1 and PARP-1 Inhibitors in the Treatment of Glioblastoma Multiforme Using Peruvian Maca Lepidium meyenii
Abstract
1. Introduction
2. Computational Details
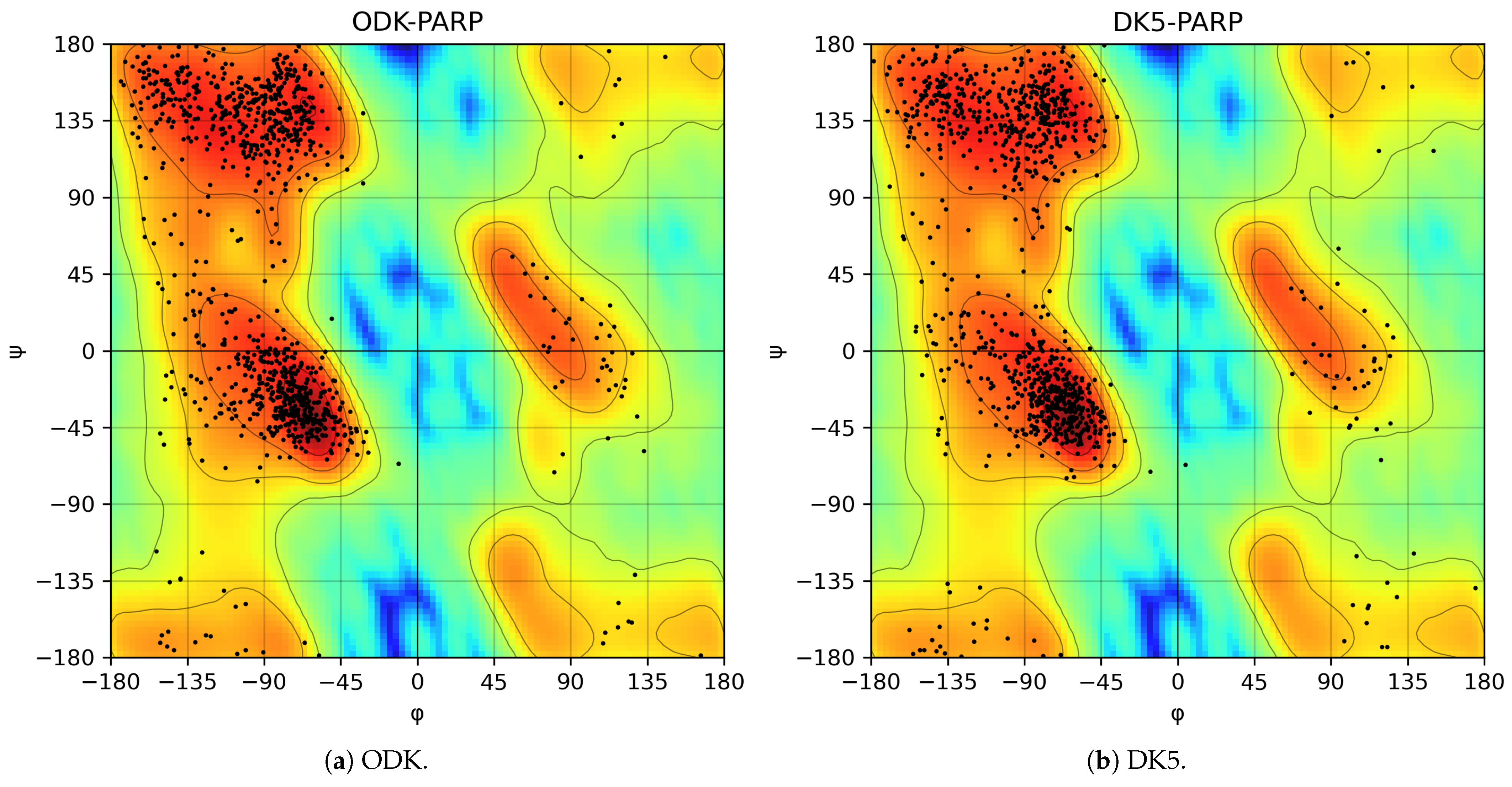
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Deo, S.V.S.; Sharma, J.; Kumar, S. GLOBOCAN 2020 Report on Global Cancer Burden: Challenges and Opportunities for Surgical Oncologists. Ann. Surg. Oncol. 2022, 29, 6497–6500. [Google Scholar] [CrossRef] [PubMed]
- Davis, M.E. Epidemiology and Overview of Gliomas. Semin. Oncol. Nurs. 2018, 34, 420–429. [Google Scholar] [CrossRef] [PubMed]
- Khabibov, M.; Garifullin, A.; Boumber, Y.; Khaddour, K.; Fernandez, M.; Khamitov, F.; Khalikova, L.; Kuznetsova, N.; Kit, O.; Kharin, L. Signaling pathways and therapeutic approaches in glioblastoma multiforme (Review). Int. J. Oncol. 2022, 60, 69. [Google Scholar] [CrossRef]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus Concomitant and Adjuvant Temozolomide for Glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA A Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Garrido, W.; Rocha, J.D.; Jaramillo, C.; Fernandez, K.; Oyarzun, C.; Martin, R.S.; Quezada, C. Chemoresistance in High-Grade Gliomas: Relevance of Adenosine Signalling in Stem-Like Cells of Glioblastoma Multiforme. Curr. Drug Targets 2014, 15, 931–942. [Google Scholar] [CrossRef] [PubMed]
- Angom, R.S.; Nakka, N.M.R.; Bhattacharya, S. Advances in Glioblastoma Therapy: An Update on Current Approaches. Brain Sci. 2023, 13, 1536. [Google Scholar] [CrossRef]
- Grochans, S.; Cybulska, A.M.; Simińska, D.; Korbecki, J.; Kojder, K.; Chlubek, D.; Baranowska-Bosiacka, I. Epidemiology of Glioblastoma Multiforme-Literature Review. Cancers 2022, 14, 2412. [Google Scholar] [CrossRef] [PubMed]
- Patel, M.; Siddiqui, F.; Jin, J.Y.; Mikkelsen, T.; Rosenblum, M.; Movsas, B.; Ryu, S. Salvage reirradiation for recurrent glioblastoma with radiosurgery: Radiographic response and improved survival. J. Neuro-Oncol. 2008, 92, 185–191. [Google Scholar] [CrossRef]
- Cha, J.; Kang, S.G.; Kim, P. Strategies of Mesenchymal Invasion of Patient-derived Brain Tumors: Microenvironmental Adaptation. Sci. Rep. 2016, 6, 24912. [Google Scholar] [CrossRef] [PubMed]
- van den Bent, M.J. Interobserver variation of the histopathological diagnosis in clinical trials on glioma: A clinician’s perspective. Acta Neuropathol. 2010, 120, 297–304. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Li, A.; Lu, H.; Luo, R.; Zhang, M.; Li, Z. TAZ promotes temozolomide resistance by upregulating MCL-1 in human glioma cells. Biochem. Biophys. Res. Commun. 2015, 463, 638–643. [Google Scholar] [CrossRef] [PubMed]
- Khaddour, K.; Johanns, T.; Ansstas, G. The Landscape of Novel Therapeutics and Challenges in Glioblastoma Multiforme: Contemporary State and Future Directions. Pharmaceuticals 2020, 13, 389. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.C.; Ou, C.H.; Huang, Y.C.; Hou, P.C.; Creighton, C.J.; Lin, Y.S.; Hu, C.Y.; Lin, S.C. YAP-1 overexpression contributes to the development of enzalutamide resistance by induction of cancer stemness and lipid metabolism in prostate cancer. Oncogene 2021, 40, 2407–2421. [Google Scholar] [CrossRef] [PubMed]
- Gilabert, M.; Launay, S.; Ginestier, C.; Bertucci, F.; Audebert, S.; Pophillat, M.; Toiron, Y.; Baudelet, E.; Finetti, P.; Noguchi, T.; et al. Poly(ADP-Ribose) Polymerase 1 (PARP1) Overexpression in Human Breast Cancer Stem Cells and Resistance to Olaparib. PLoS ONE 2014, 9, e104302. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, Y.; A, G.; Qu, C.; Chen, J. Pan-Cancer Analysis of PARP1 Alterations as Biomarkers in the Prediction of Immunotherapeutic Effects and the Association of Its Expression Levels and Immunotherapy Signatures. Front. Immunol. 2021, 12, 721030. [Google Scholar] [CrossRef]
- Schlegelmilch, K.; Mohseni, M.; Kirak, O.; Pruszak, J.; Rodriguez, J.R.; Zhou, D.; Kreger, B.T.; Vasioukhin, V.; Avruch, J.; Brummelkamp, T.R.; et al. YAP-1 Acts Downstream of alpha-Catenin to Control Epidermal Proliferation. Cell 2011, 144, 782–795. [Google Scholar] [CrossRef]
- Zhu, T.; Zheng, J.Y.; Huang, L.L.; Wang, Y.H.; Yao, D.F.; Dai, H.B. Human PARP1 substrates and regulators of its catalytic activity: An updated overview. Front. Pharmacol. 2023, 14, 1137151. [Google Scholar] [CrossRef]
- Guichet, P.O.; Masliantsev, K.; Tachon, G.; Petropoulos, C.; Godet, J.; Larrieu, D.; Milin, S.; Wager, M.; Karayan-Tapon, L. Fatal correlation between YAP-1 expression and glioma aggressiveness: Clinical and molecular evidence. J. Pathol. 2018, 246, 205–216. [Google Scholar] [CrossRef]
- Orr, B.A.; Bai, H.; Odia, Y.; Jain, D.; Anders, R.A.; Eberhart, C.G. Yes-Associated Protein 1 Is Widely Expressed in Human Brain Tumors and Promotes Glioblastoma Growth. J. Neuropathol. Exp. Neurol. 2011, 70, 568–577. [Google Scholar] [CrossRef] [PubMed]
- Murnyák, B.; Kouhsari, M.C.; Hershkovitch, R.; Kálmán, B.; Marko-Varga, G.; Klekner, Á.; Hortobágyi, T. PARP1expression and its correlation with survival is tumour molecular subtype dependent in glioblastoma. Oncotarget 2017, 8, 46348–46362. [Google Scholar] [CrossRef]
- Saunders, J.T.; Holmes, B.; Benavides-Serrato, A.; Ku3, S.; Nishimura, R.N.; Gera, J. Targeting the YAP-TEAD interaction interface for therapeutic intervention in glioblastoma. J. Neuro-Oncol. 2021, 152, 217–231. [Google Scholar] [CrossRef]
- Hu, X.; Zhang, Y.; Yu, H.; Zhao, Y.; Sun, X.; Li, Q.; Wang, Y. The role of YAP-1 in survival prediction, immune modulation, and drug response: A pan-cancer perspective. Front. Immunol. 2022, 13, 1012173. [Google Scholar] [CrossRef] [PubMed]
- Drexler, R.; Küchler, M.; Wagner, K.C.; Reese, T.; Feyerabend, B.; Kleine, M.; Oldhafer, K.J. The clinical relevance of the Hippo pathway in pancreatic ductal adenocarcinoma. J. Cancer Res. Clin. Oncol. 2020, 147, 373–391. [Google Scholar] [CrossRef]
- Yu, F.X.; Guan, K.L. The Hippo pathway: Regulators and regulations. Genes Dev. 2013, 27, 355–371. [Google Scholar] [CrossRef]
- Manukyan, A.; Kowalczyk, I.; Melhuish, T.A.; Lemiesz, A.; Wotton, D. Analysis of transcriptional activity by the Myt1 and Myt1l transcription factors. J. Cell. Biochem. 2018, 119, 4644–4655. [Google Scholar] [CrossRef] [PubMed]
- Barry, E.R.; Simov, V.; Valtingojer, I.; Venier, O. Recent Therapeutic Approaches to Modulate the Hippo Pathway in Oncology and Regenerative Medicine. Cells 2021, 10, 2715. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Hu, J.W.; He, X.R.; Jin, W.L.; He, X.Y. Statins: A repurposed drug to fight cancer. J. Exp. Clin. Cancer Res. 2021, 40, 241. [Google Scholar] [CrossRef] [PubMed]
- Read, R.D. Repurposing the drug verteporfin as anti-neoplastic therapy for glioblastoma. Neuro-Oncology 2022, 24, 708–710. [Google Scholar] [CrossRef]
- Brodowska, K.; Al-Moujahed, A.; Marmalidou, A.; Meyer zu Horste, M.; Cichy, J.; Miller, J.W.; Gragoudas, E.; Vavvas, D.G. The clinically used photosensitizer Verteporfin (VP) inhibits YAP-TEAD and human retinoblastoma cell growth in vitro without light activation. Exp. Eye Res. 2014, 124, 67–73. [Google Scholar] [CrossRef] [PubMed]
- D’Amours, D.; Desnoyers, S.; D’Silva, I.; Poirier, G.G. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 1999, 342 Pt 2, 249–268. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wang, Z.; Hou, Y.; Xi, J.; He, Z.; Lu, H.; Du, Z.; Zhong, S.; Yang, Q. A PARP1-related prognostic signature constructing and PARP-1 inhibitors screening for glioma. Front. Cell Dev. Biol. 2022, 10, 916415. [Google Scholar] [CrossRef]
- Pazzaglia, S.; Pioli, C. Multifaceted Role of PARP-1 in DNA Repair and Inflammation: Pathological and Therapeutic Implications in Cancer and Non-Cancer Diseases. Cells 2019, 9, 41. [Google Scholar] [CrossRef] [PubMed]
- Ossovskaya, V.; Koo, I.C.; Kaldjian, E.P.; Alvares, C.; Sherman, B.M. Upregulation of Poly (ADP-Ribose) Polymerase-1 (PARP1) in Triple-Negative Breast Cancer and Other Primary Human Tumor Types. Genes Cancer 2010, 1, 812–821. [Google Scholar] [CrossRef]
- Rojo, F.; García-Parra, J.; Zazo, S.; Tusquets, I.; Ferrer-Lozano, J.; Menendez, S.; Eroles, P.; Chamizo, C.; Servitja, S.; Ramírez-Merino, N.; et al. Nuclear PARP-1 protein overexpression is associated with poor overall survival in early breast cancer. Ann. Oncol. 2012, 23, 1156–1164. [Google Scholar] [CrossRef] [PubMed]
- Hanna, C.; Kurian, K.M.; Williams, K.; Watts, C.; Jackson, A.; Carruthers, R.; Strathdee, K.; Cruickshank, G.; Dunn, L.; Erridge, S.; et al. Pharmacokinetics, safety, and tolerability of olaparib and temozolomide for recurrent glioblastoma: Results of the phase I OPARATIC trial. Neuro-Oncology 2020, 22, 1840–1850. [Google Scholar] [CrossRef]
- Kleinberg, L.; Ye, X.; Supko, J.; Stevens, G.H.; Shu, H.K.; Mikkelsen, T.; Lieberman, F.; Lesser, G.; Lee, E.; Grossman, S. A Multi-Site Phase I Trial of Veliparib with Standard Radiation and Temozolomide in Patients with Newly Diagnosed Glioblastoma Multiforme (GBM). J. Neuro-Oncol. 2023, 165, 499–507. [Google Scholar] [CrossRef]
- Shin, D.; Jeon, S.H.; Piao, J.; Park, H.J.; Tian, W.J.; Moon, D.G.; Ahn, S.T.; Jeon, K.H.; Zhu, G.Q.; Park, I.; et al. Efficacy and Safety of Maca (Lepidium meyenii) in Patients with Symptoms of Late-Onset Hypogonadism: A Randomized, Double-Blind, Placebo-Controlled Clinical Trial. World J. Men’s Health 2023, 41, 692. [Google Scholar] [CrossRef]
- Flores, H.E.; Walker, T.S.; Guimarães, R.L.; Bais, H.P.; Vivanco, J.M. Andean Root and Tuber Crops: Underground Rainbows. HortScience 2003, 38, 161–167. [Google Scholar] [CrossRef]
- Carvalho, F.V.; Ribeiro, P.R. Structural diversity, biosynthetic aspects, and LC-HRMS data compilation for the identification of bioactive compounds of Lepidium meyenii. Food Res. Int. 2019, 125, 108615. [Google Scholar] [CrossRef]
- Caicai, K.; Limin, H.; Liming, Z.; Zhiqiang, Z.; Yongwu, Y. Isolation, purification and antioxidant activity of polysaccharides from the leaves of maca (Lepidium meyenii). Int. J. Biol. Macromol. 2018, 107, 2611–2619. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhu, F. Chemical composition and health effects of maca (Lepidium meyenii). Food Chem. 2019, 288, 422–443. [Google Scholar] [CrossRef] [PubMed]
- da Silva Leitão Peres, N.; Cabrera Parra Bortoluzzi, L.; Medeiros Marques, L.L.; Formigoni, M.; Fuchs, R.H.B.; Droval, A.A.; Reitz Cardoso, F.A. Medicinal effects of Peruvian maca (Lepidium meyenii): A review. Food Funct. 2020, 11, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, F.V.; Fonseca Santana, L.; Diogenes, A. da Silva, V.; Costa, S.L.; Zambotti-Villelae, L.; Colepicolo, P.; Ferraz, C.G.; Ribeiro, P.R. Combination of a multiplatform metabolite profiling approach and chemometrics as a powerful strategy to identify bioactive metabolites in Lepidium meyenii(Peruvian maca). Food Chem. 2021, 364, 130453. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Jin, W.; Cui, Y.; Ao, M.; Liu, H.; Xu, H.; Yu, L. Protective effects of macamides from Lepidium meyenii Walp. against corticosterone-induced neurotoxicity in PC12 cells. RSC Adv. 2019, 9, 23096–23108. [Google Scholar] [CrossRef] [PubMed]
- Tao, H.; Shi, H.; Wang, M.; Xu, Y. Macamide B suppresses lung cancer progression potentially via the ATM signaling pathway. Oncol. Lett. 2023, 25, 115. [Google Scholar] [CrossRef] [PubMed]
- Sneha, P.; Doss, C.G.P. Molecular Dynamics: New Frontier in Personalized Medicine. Adv. Protein Chem. Struct. Biol. 2016, 102, 181–224. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, D.R.; Berrisford, J.M.; Conroy, M.J.; Gutmanas, A.; Anyango, S.; Choudhary, P.; Clark, A.R.; Dana, J.M.; Deshpande, M.; Dunlop, R.; et al. PDBe: Improved findability of macromolecular structure data in the PDB. Nucleic Acids Res. 2019, 48, D335–D343. [Google Scholar] [CrossRef] [PubMed]
- Varadi, M.; Berrisford, J.; Deshpande, M.; Nair, S.S.; Gutmanas, A.; Armstrong, D.; Pravda, L.; Al-Lazikani, B.; Anyango, S.; Barton, G.J.; et al. PDBe-KB: A community-driven resource for structural and functional annotations. Nucleic Acids Res. 2019, 48, D344–D353. [Google Scholar] [CrossRef]
- Mesrouze, Y.; Bokhovchuk, F.; Izaac, A.; Meyerhofer, M.; Zimmermann, C.; Fontana, P.; Schmelzle, T.; Erdmann, D.; Furet, P.; Kallen, J.; et al. Adaptation of the bound intrinsically disordered protein YAP to mutations at the YAP: TEAD interface. Protein Sci. 2018, 27, 1810–1820. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figur11, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2021, 50, D439–D444. [Google Scholar] [CrossRef] [PubMed]
- Varadi, M.; Bertoni, D.; Magana, P.; Paramval, U.; Pidruchna, I.; Radhakrishnan, M.; Tsenkov, M.; Nair, S.; Mirdita, M.; Yeo, J.; et al. AlphaFold Protein Structure Database in 2024: Providing structure coverage for over 214 million protein sequences. Nucleic Acids Res. 2023, 52, D368–D375. [Google Scholar] [CrossRef] [PubMed]
- Meng, E.C.; Goddard, T.D.; Pettersen, E.F.; Couch, G.S.; Pearson, Z.J.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Tools for structure building and analysis. Protein Sci. 2023, 32, e4792. [Google Scholar] [CrossRef]
- Burley, S.K.; Berman, H.M.; Bhikadiya, C.; Bi, C.; Chen, L.; Costanzo, L.D.; Christie, C.; Duarte, J.M.; Dutta, S.; Feng, Z.; et al. Protein Data Bank: The single global archive for 3D macromolecular structure data. Nucleic Acids Res. 2018, 47, D520–D528. [Google Scholar] [CrossRef]
- Burley, S.K.; Berman, H.M.; Kleywegt, G.J.; Markley, J.L.; Nakamura, H.; Velankar, S. Protein Data Bank (PDB): The Single Global Macromolecular Structure Archive. In Protein Crystallography; Springer: New York, NY, USA, 2017; pp. 627–641. [Google Scholar] [CrossRef]
- Langelier, M.F.; Planck, J.L.; Roy, S.; Pascal, J.M. Structural Basis for DNA Damage–Dependent Poly(ADP-ribosyl)ation by Human PARP-1. Science 2012, 336, 728–732. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2023 update. Nucleic Acids Res. 2022, 51, D1373–D1380. [Google Scholar] [CrossRef]
- PubChem. Macamide 1—Pubchem.ncbi.nlm.nih.gov. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/Macamide-1 (accessed on 12 November 2024).
- PubChem. Macamide 2—Pubchem.ncbi.nlm.nih.gov. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/Macamide-2 (accessed on 12 November 2024).
- PubChem. N-((3-Methoxyphenyl)methyl)octadeca-9,12,15-trienamide, (9Z,12Z,15Z)—Pubchem.ncbi.nlm.nih.gov. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/73353637 (accessed on 12 November 2024).
- PubChem. Stearic Acid—Pubchem.ncbi.nlm.nih.gov. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/Stearic-Acid (accessed on 12 November 2024).
- PubChem. Pentahomomethionine—Pubchem.ncbi.nlm.nih.gov. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/Pentahomomethionine (accessed on 12 November 2024).
- PubChem. Phytosphingosine—Pubchem.ncbi.nlm.nih.gov. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/Phytosphingosine (accessed on 12 November 2024).
- PubChem. N-Benzylpentadecanamide—Pubchem.ncbi.nlm.nih.gov. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/N-Benzylpentadecanamide (accessed on 12 November 2024).
- PubChem. Veliparib—Pubchem.ncbi.nlm.nih.gov. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/Veliparib (accessed on 12 November 2024).
- PubChem. Olaparib—Pubchem.ncbi.nlm.nih.gov. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/Olaparib (accessed on 12 November 2024).
- PubChem. Verteporfin|C41H42N4O8—Chemspider.com. Available online: https://www.chemspider.com/Chemical-Structure.4515032.html (accessed on 12 November 2024).
- Pence, H.E.; Williams, A. ChemSpider: An Online Chemical Information Resource. J. Chem. Educ. 2010, 87, 1123–1124. [Google Scholar] [CrossRef]
- Abraham, M.; Alekseenko, A.; Basov, V.; Bergh, C.; Briand, E.; Brown, A.; Doijade, M.; Fiorin, G.; Fleischmann, S.; Gorelov, S.; et al. GROMACS 2024.3 Manual. Zenodo, 2024. Available online: https://zenodo.org/records/13457083 (accessed on 1 December 2024). [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Tirado-Rives, J. Potential energy functions for atomic-level simulations of water and organic and biomolecular systems. Proc. Natl. Acad. Sci. USA 2005, 102, 6665–6670. [Google Scholar] [CrossRef] [PubMed]
- Dodda, L.S.; Cabeza de Vaca, I.; Tirado-Rives, J.; Jorgensen, W.L. LigParGen web server: An automatic OPLS-AA parameter generator for organic ligands. Nucleic Acids Res. 2017, 45, W331–W336. [Google Scholar] [CrossRef] [PubMed]
- Dodda, L.S.; Vilseck, J.Z.; Tirado-Rives, J.; Jorgensen, W.L. 1.14*CM1A-LBCC: Localized Bond-Charge Corrected CM1A Charges for Condensed-Phase Simulations. J. Phys. Chem. B 2017, 121, 3864–3870. [Google Scholar] [CrossRef] [PubMed]
- Dewar, M.J.S.; Zoebisch, E.G.; Healy, E.F.; Stewart, J.J.P. Development and use of quantum mechanical molecular models. 76. AM1: A new general purpose quantum mechanical molecular model. J. Am. Chem. Soc. 1985, 107, 3902–3909. [Google Scholar] [CrossRef]
- Yanai, T.; Tew, D.P.; Handy, N.C. A new hybrid exchange–correlation functional using the Coulomb-attenuating method (CAM-B3LYP). Chem. Phys. Lett. 2004, 393, 51–57. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian˜16 Revision C.01; Gaussian Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Xu, Y.; Wang, S.; Hu, Q.; Gao, S.; Ma, X.; Zhang, W.; Shen, Y.; Chen, F.; Lai, L.; Pei, J. CavityPlus: A web server for protein cavity detection with pharmacophore modelling, allosteric site identification and covalent ligand binding ability prediction. Nucleic Acids Res. 2018, 46, W374–W379. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2009, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef]
- Gnuplot: An Interactive Plotting Program. Available online: http://www.gnuplot.info/docs_4.0/gnuplot.html (accessed on 12 November 2024).
- Laskowski, R.A.; Jabłońska, J.; Pravda, L.; Vařeková, R.S.; Thornton, J.M. PDBsum: Structural summaries of PDB entries. Protein Sci. 2017, 27, 129–134. [Google Scholar] [CrossRef]
- Jurrus, E.; Engel, D.; Star, K.; Monson, K.; Brandi, J.; Felberg, L.E.; Brookes, D.H.; Wilson, L.; Chen, J.; Liles, K.; et al. Improvements to the APBS biomolecular solvation software suite. Protein Sci. 2017, 27, 112–128. [Google Scholar] [CrossRef] [PubMed]
- Kumari, R.; Ku3, R.; Lynn, A. g_mmpbsa–A GROMACS Tool for High-Throughput MM-PBSA Calculations. J. Chem. Inf. Model. 2014, 54, 1951–1962. [Google Scholar] [CrossRef] [PubMed]
- Baker, N.A.; Sept, D.; Joseph, S.; Holst, M.J.; McCammon, J.A. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA 2001, 98, 10037–10041. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Shi, S.; Yi, J.; Wang, N.; He, Y.; Wu, Z.; Peng, J.; Deng, Y.; Wang, W.; Wu, C.; et al. ADMETlab 3.0: An updated comprehensive online ADMET prediction platform enhanced with broader coverage, improved performance, API functionality and decision support. Nucleic Acids Res. 2024, 52, W422–W431. [Google Scholar] [CrossRef] [PubMed]

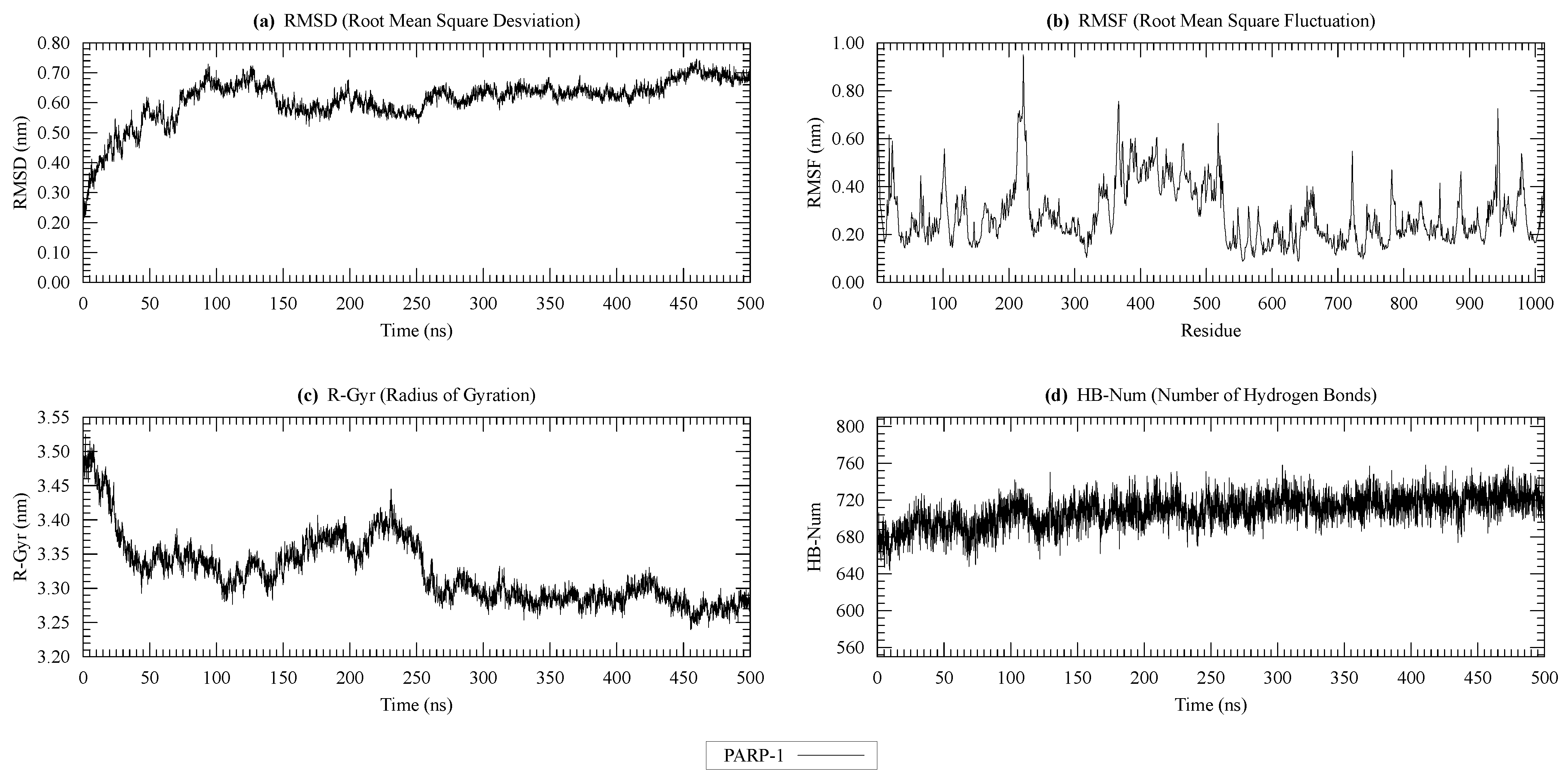
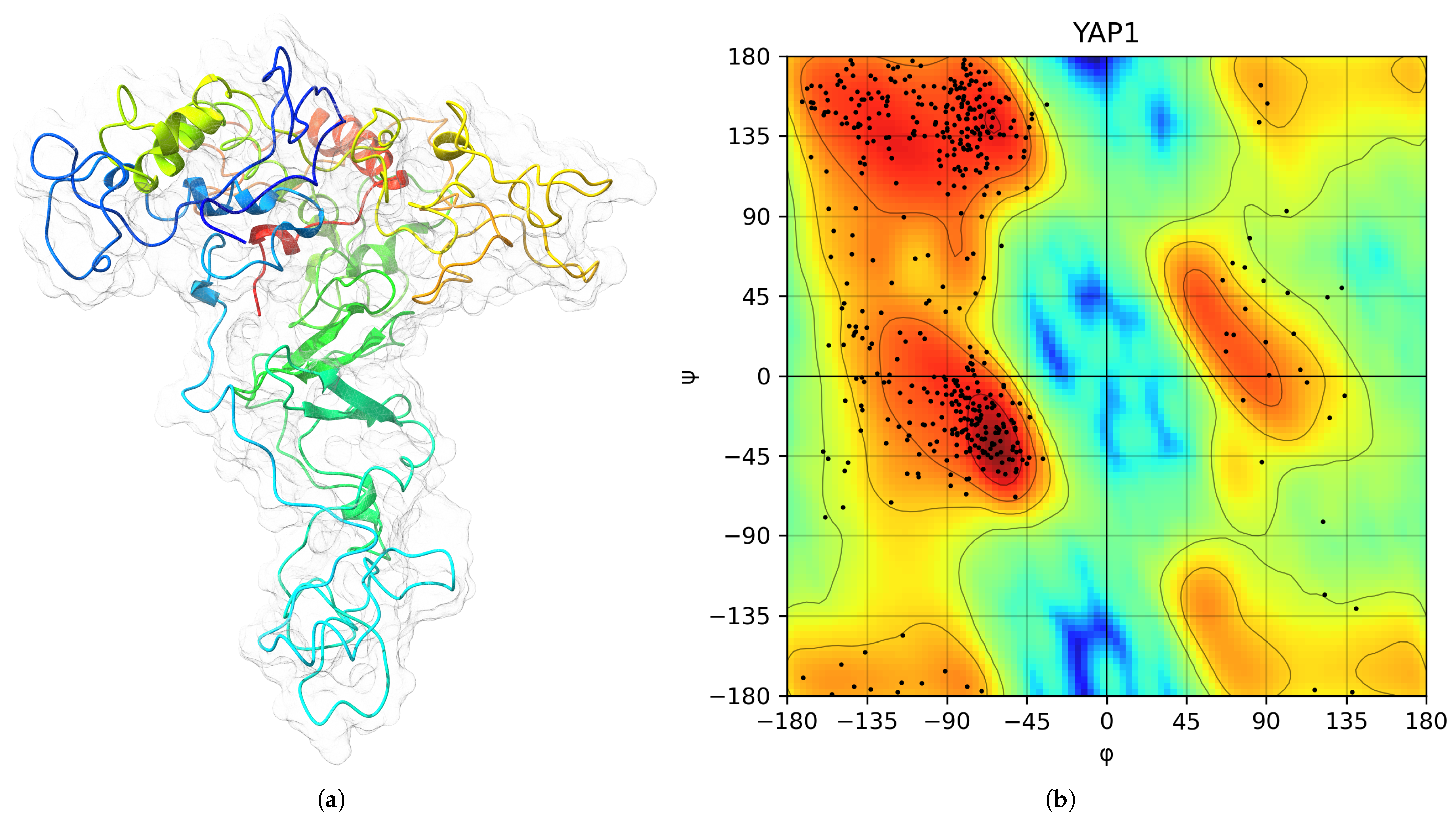

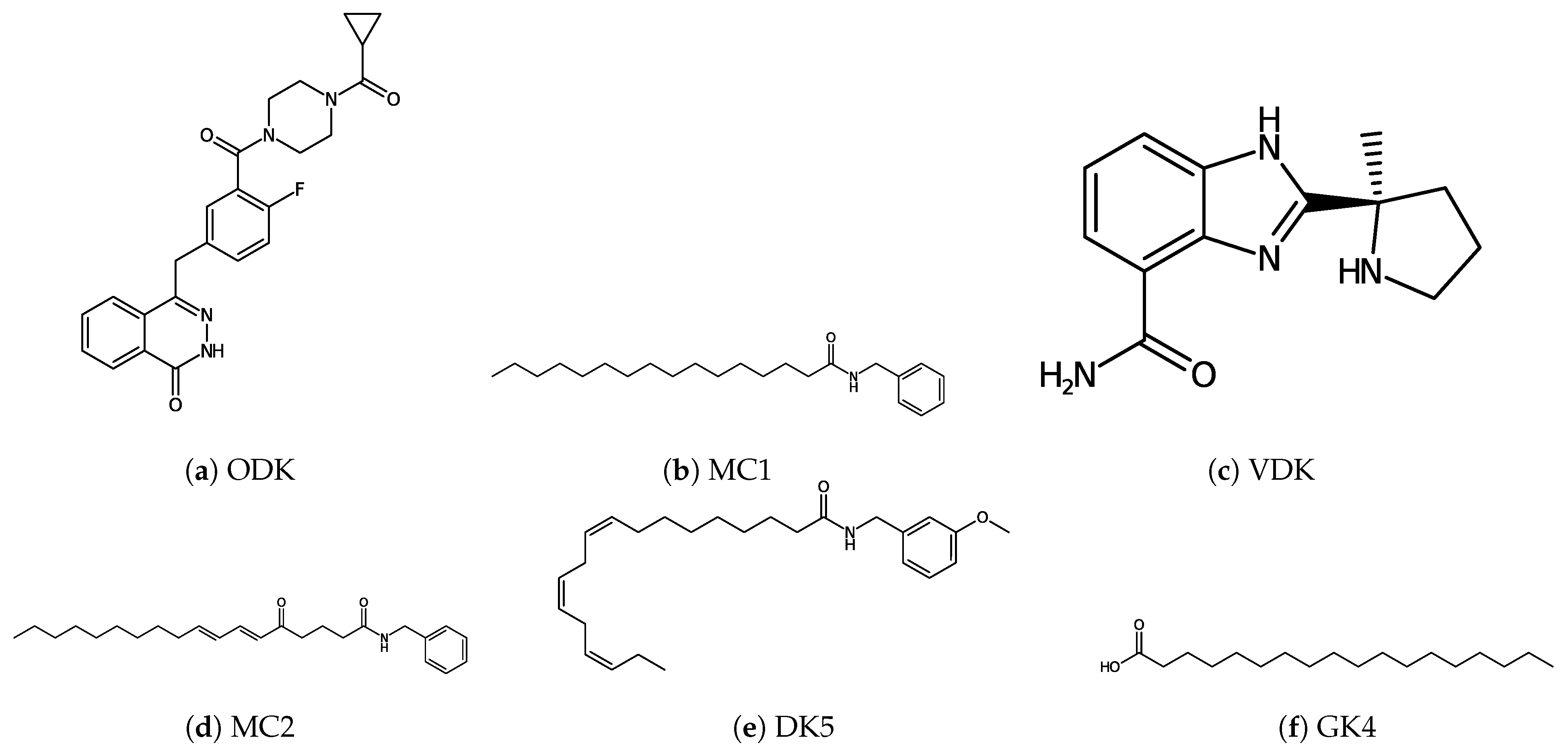
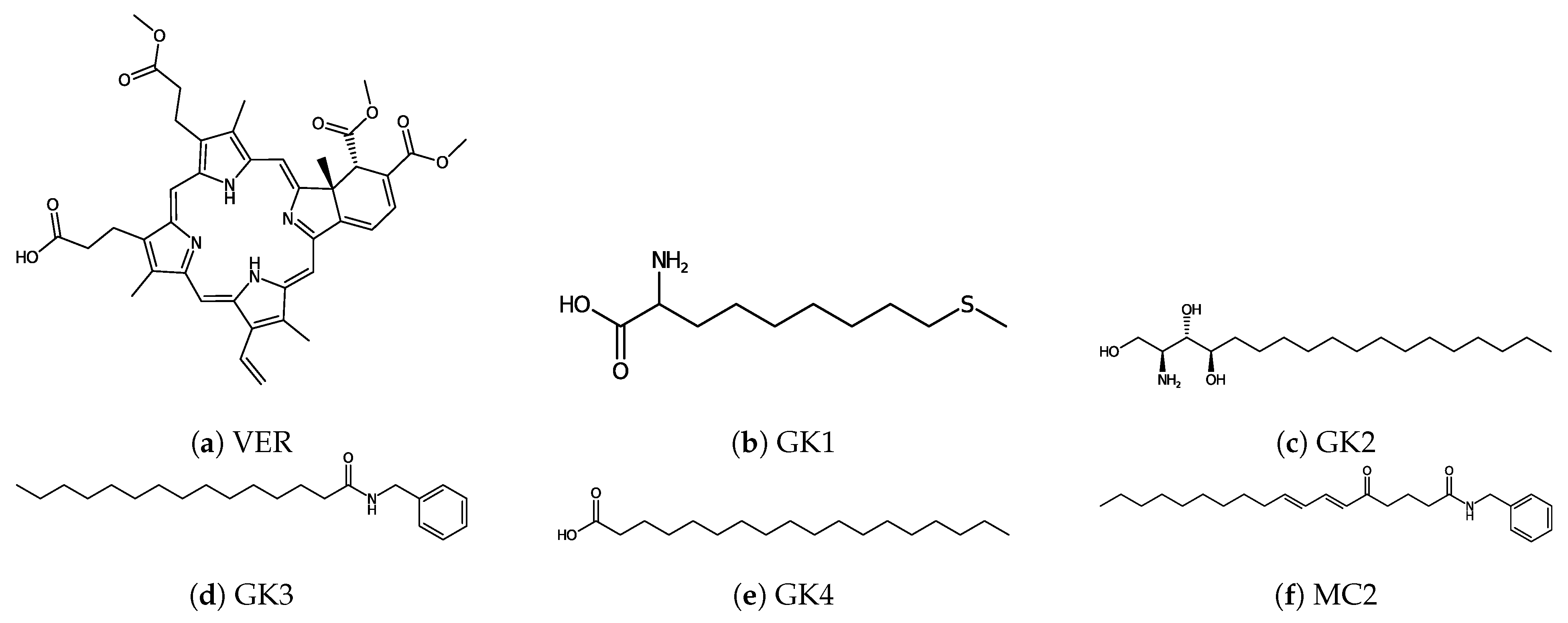

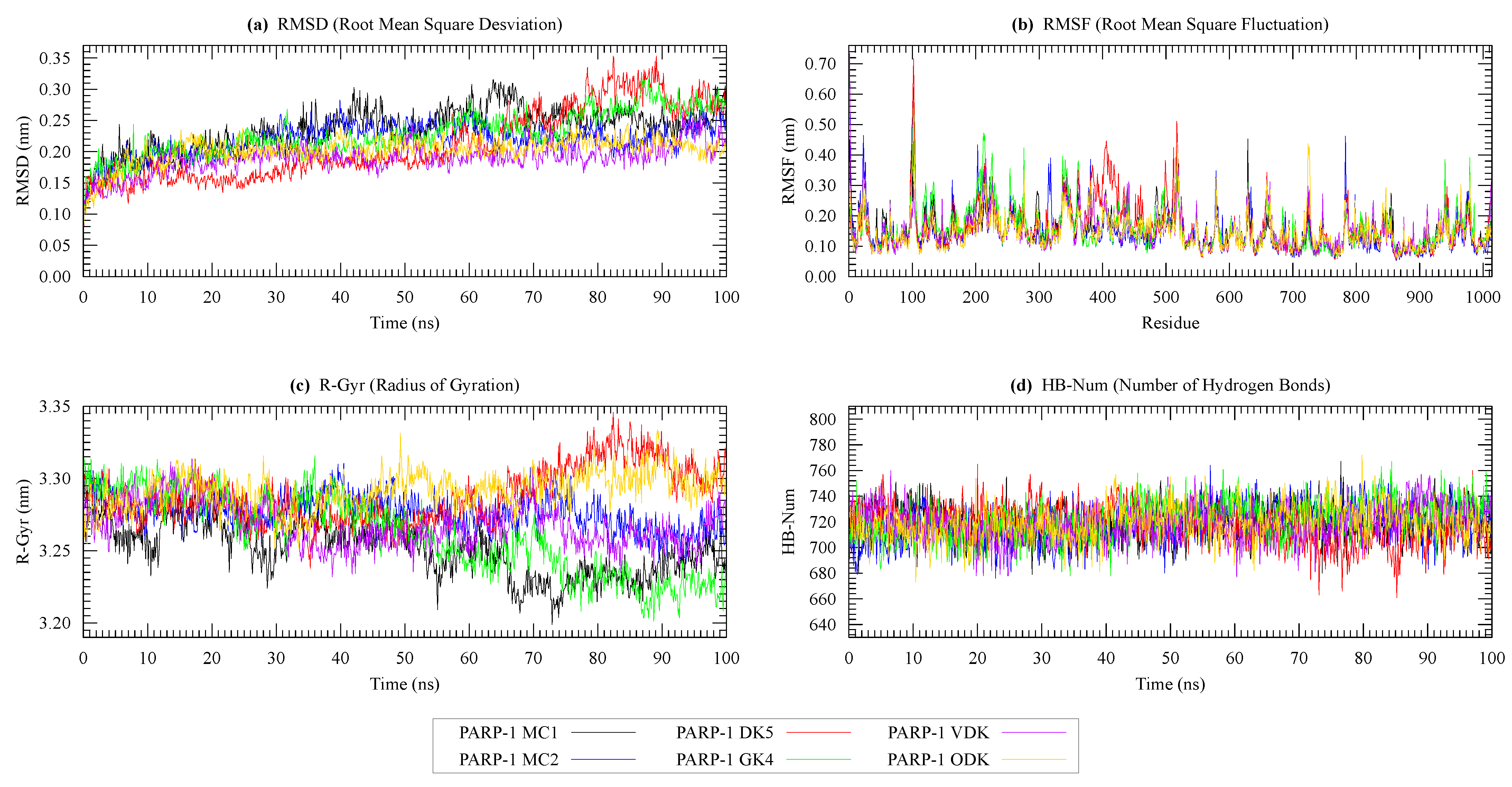
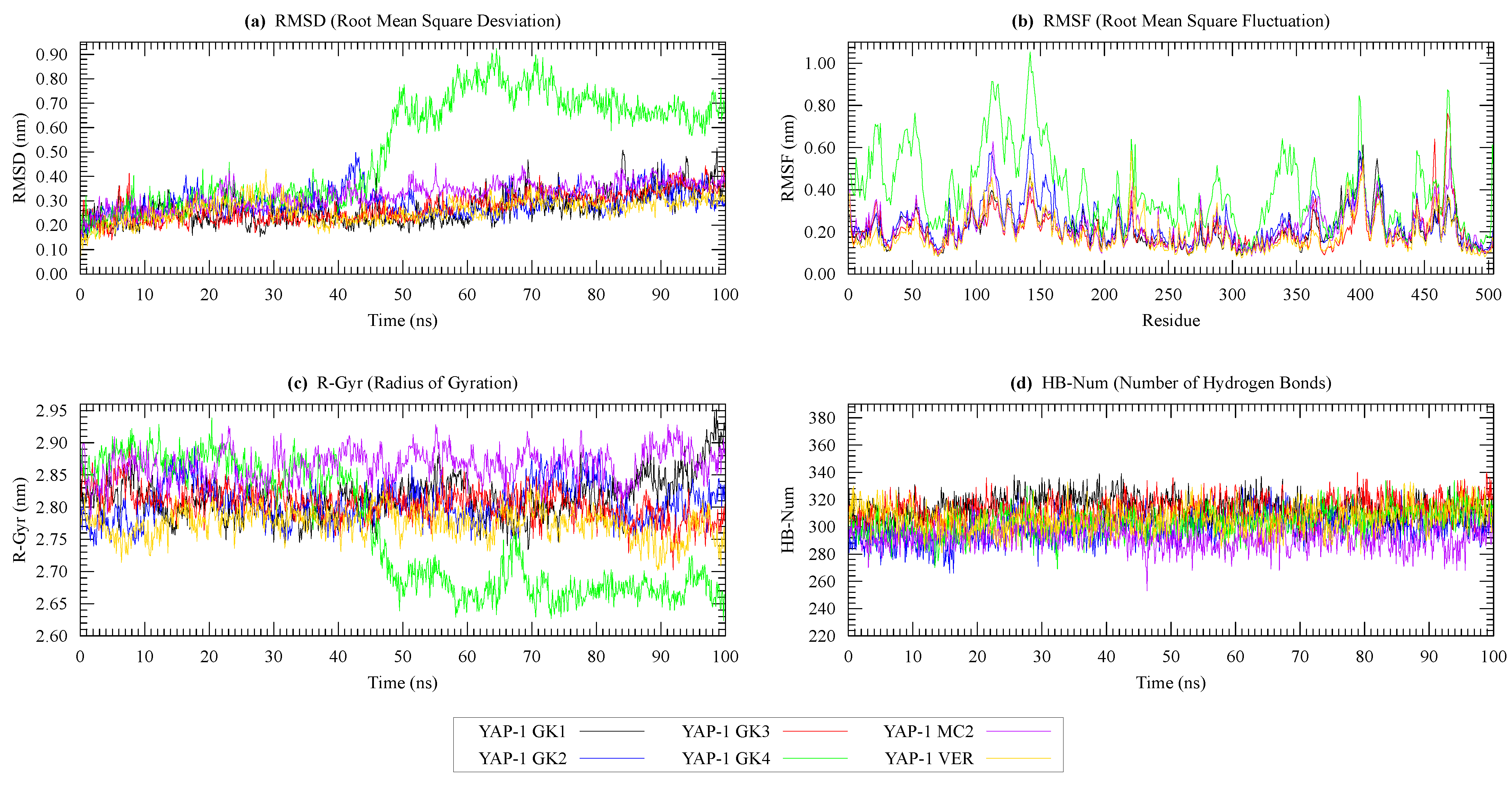

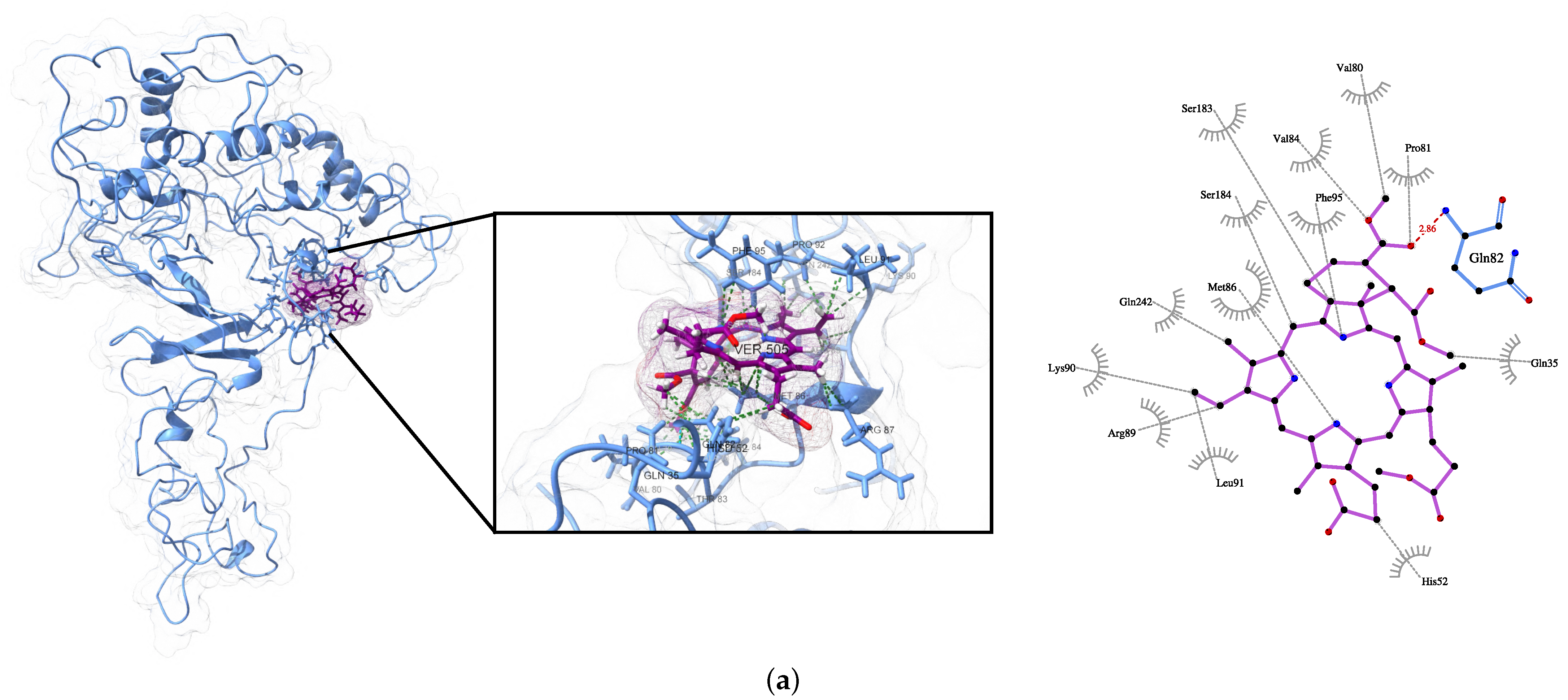
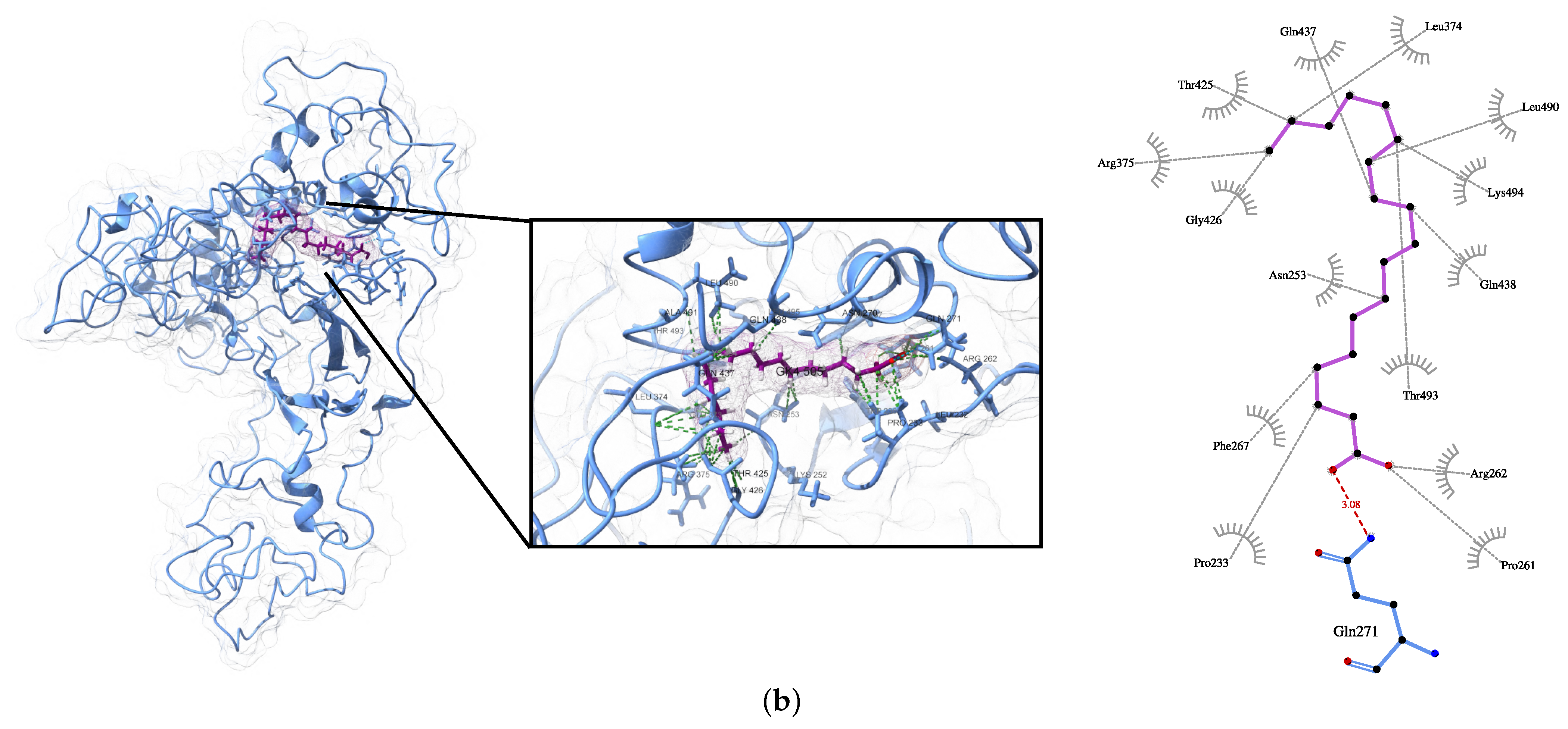


| Protein | Surface Area () | Volume () | DrugScore | Druggability | |
|---|---|---|---|---|---|
| PARP-1 | 1 | 7220.25 | 14,216.62 | 8047.00 | Strong |
| 2 | 6021.25 | 10,959.88 | 5759.00 | Strong | |
| 3 | 4917.75 | 9253.25 | 5613.00 | Strong | |
| 4 | 3259.75 | 6120.75 | 3390.00 | Strong | |
| 5 | 2481.25 | 4174.62 | 2678.00 | Strong | |
| 6 | 2584.75 | 5497.62 | 2651.00 | Strong | |
| YAP-1 | 1 | 1063.50 | 1767.75 | 2340.00 | Strong |
| 2 | 1496.00 | 2576.00 | 2111.00 | Strong | |
| 3 | 370.75 | 532.50 | 284.00 | Medium | |
| 4 | 553.25 | 822.38 | 82.00 | Medium | |
| 5 | 456.50 | 592.62 | −59.00 | Medium | |
| 6 | 715.75 | 856.62 | −129.00 | Medium |
| Protein | Inhibitor | Energy (kcal/mol) |
|---|---|---|
| PARP-1 | MC1 | −8.10 |
| MC2 | −8.73 | |
| DK5 | −8.99 | |
| GK4 | −6.76 | |
| ODK | −11.13 | |
| VDK | −9.34 | |
| YAP-1 | GK1 | −5.40 |
| GK2 | −7.16 | |
| GK3 | −7.92 | |
| GK4 | −6.58 | |
| MC2 | −9.16 | |
| VER | −10.40 |
| Energy Component (kcal/mol) | Poli-(ADP-Ribose)–Polimerase-1 (PARP1) | |||||
|---|---|---|---|---|---|---|
| MC1 | MC2 | DK5 | GK4 | ODK | VDK | |
| Van der Waals Energy | −48.92 ± 2.47 | −55.00 ± 2.03 | −53.89 ± 2.55 | −45.82 ± 2.22 | −54.15 ± 2.31 | −41.51 ± 2.33 |
| Electrostatic Energy | −3.17 ± 3.30 | 0.13 ± 2.19 | −0.31 ± 2.43 | −3.37 ± 2.82 | −9.91 ± 2.96 | −1.74 ± 4.29 |
| Polar Solvation Energy | 18.68 ± 3.07 | 29.11 ± 3.57 | 20.81 ± 3.07 | 20.27 ± 4.10 | 25.45 ± 2.19 | 16.89 ± 4.56 |
| SASA Energy | −5.24 ± 0.26 | −5.65 ± 0.20 | −5.81 ± 0.25 | −5.43 ± 0.24 | −5.15 ± 0.20 | −3.76 ± 0.17 |
| SAV Energy | −50.75 ± 5.48 | −53.37 ± 5.63 | −54.72 ± 5.35 | −49.83 ± 5.06 | −47.34 ± 5.70 | −37.99 ± 3.08 |
| Binding energy | −89.39 ± 7.20 | −84.74 ± 7.31 | −93.89 ± 6.79 | −84.16 ± 6.42 | −91.06 ± 6.95 | −68.15 ± 6.74 |
| Energy Component (kcal/mol) | Yes1 (Associated Transcriptional Regulator YAP-1) | |||||
|---|---|---|---|---|---|---|
| GK1 | GK2 | GK3 | GK4 | MC2 | VER | |
| Van der Waals Energy | −31.33 ± 4.04 | −35.55 ± 4.08 | −47.62 ± 4.62 | −50.62 ± 2.53 | −55.40 ± 2.55 | −64.28 ± 4.50 |
| Electrostatic Energy | −3.14 ± 4.41 | −0.95 ± 2.70 | −5.26 ± 2.97 | −4.18 ± 2.20 | −3.22 ± 2.69 | −9.54 ± 4.75 |
| Polar Solvation Energy | 10.23 ± 2.40 | 17.46 ± 3.23 | 24.67 ± 3.82 | 16.90 ± 3.06 | 26.18 ± 4.48 | 24.57 ± 2.35 |
| SASA Energy | −3.56 ± 0.27 | −4.84 ± 0.39 | −5.89 ± 0.28 | −5.47 ± 0.22 | −5.91 ± 0.24 | −7.53 ± 0.41 |
| SAV Energy | −34.55 ± 4.92 | −43.58 ± 7.03 | −54.32 ± 6.34 | −57.40 ± 4.47 | −58.91 ± 4.92 | −64.59 ± 7.26 |
| Binding energy | −62.36 ± 10.28 | −67.47 ± 9.84 | −88.42 ± 10.86 | −100.77 ± 6.50 | −97.29 ± 7.18 | −121.37 ± 10.75 |
| Property | Model Name | GK1 | GK2 | GK3 | GK4 | MC2 | VER | MC1 | DK5 | ODK | VDK |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Physicochemical | logS | −1.59 | −3.70 | −0.85 | −0.85 | −0.87 | −3.12 | −1.22 | −4.50 | −3.05 | −3.2 |
| logP | 2.558 | 3.197 | 1.699 | 1.674 | 2.308 | 2.712 | 2.389 | 3.112 | 3.009 | 2.844 | |
| logD | 0.900 | 1.000 | 0.682 | 0.944 | 0.52 | 0.865 | 0.659 | 1.312 | 1.112 | 0.998 | |
| Molecular Weight (Da) | 219.13 | 317.29 | 331.29 | 284.27 | 383.28 | 389.50 | 218.40 | 401.30 | 412.80 | 432.20 | |
| QED | 0.583 | 0.329 | 0.370 | 0.336 | 0.207 | 0.512 | 0.471 | 0.62 | 0.543 | 0.487 | |
| H-Bond Donors | 3 | 5 | 1 | 1 | 1 | 2 | 4 | 3 | 2 | 1 | |
| H-Bond Acceptors | 3 | 4 | 2 | 2 | 3 | 4 | 3 | 5 | 4 | 4 | |
| Caco-2 Permeability | −5.25 | −5.12 | - | - | - | −4.90 | −4.76 | −4.79 | −4.60 | −4.78 | |
| MDCK Permeability | −4.6 | −4.8 | - | - | - | −4.7 | −4.9 | −4.6 | −4.8 | −4.9 | |
| Absorption | Pgp Inhibitor | No | No | No | No | No | Yes | No | No | Yes | No |
| Pgp Substrate | No | No | Yes | No | Yes | Yes | No | No | No | Yes | |
| HIA | 0.957 | 0.116 | 0.682 | 0.944 | 0.520 | 0.821 | 0.719 | 0.850 | 0.753 | 0.678 | |
| Distribution | PPB | 0.675 | 0.759 | 0.284 | 0.074 | 0.966 | 0.902 | 0.751 | 0.822 | 0.814 | 0.803 |
| VD | 47.06 | 51.90 | 16.00 | 16.00 | 1.70 | 39.80 | 45.00 | 41.20 | 42.70 | 40.60 | |
| Fu | 0.001 | 0.329 | 0.370 | 0.336 | 0.207 | 0.258 | 0.150 | 0.112 | 0.302 | 0.290 | |
| CYP1A2 Inhibitor | Yes | Yes | No | No | Yes | No | Yes | No | Yes | No | |
| Metabolism | CYP2C19 Substrate | No | No | Yes | Yes | No | No | No | No | Yes | Yes |
| CYP3A4 Inhibitor | Yes | Yes | No | Yes | Yes | Yes | No | No | Yes | No | |
| CL (L/h) | 3.15 | 3.57 | 2.80 | 2.85 | 3.32 | 2.99 | 3.42 | 2.71 | 2.85 | 2.77 | |
| t1/2 (h) | 0.215 | 0.260 | 0.186 | 0.171 | 0.176 | 0.190 | 0.210 | 0.230 | 0.215 | 0.221 | |
| Excretion | hERG Blockers | No | No | Yes | No | Yes | Yes | No | No | Yes | No |
| DILI | No | No | No | No | Yes | Yes | No | No | No | Yes | |
| Ames Toxicity | No | No | No | No | No | No | No | Yes | Yes | No | |
| Skin Sensitization | No | No | No | No | No | No | No | No | Yes | No |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Turpo-Peqqueña, A.G.; Luna-Prado, S.; Valencia-Arce, R.J.; Del-Carpio-Carrazco, F.L.; Gómez, B. A Theoretical Study on the Efficacy and Mechanism of Combined YAP-1 and PARP-1 Inhibitors in the Treatment of Glioblastoma Multiforme Using Peruvian Maca Lepidium meyenii. Curr. Issues Mol. Biol. 2025, 47, 40. https://doi.org/10.3390/cimb47010040
Turpo-Peqqueña AG, Luna-Prado S, Valencia-Arce RJ, Del-Carpio-Carrazco FL, Gómez B. A Theoretical Study on the Efficacy and Mechanism of Combined YAP-1 and PARP-1 Inhibitors in the Treatment of Glioblastoma Multiforme Using Peruvian Maca Lepidium meyenii. Current Issues in Molecular Biology. 2025; 47(1):40. https://doi.org/10.3390/cimb47010040
Chicago/Turabian StyleTurpo-Peqqueña, Albert Gabriel, Sebastian Luna-Prado, Renato Javier Valencia-Arce, Fabio Leonardo Del-Carpio-Carrazco, and Badhin Gómez. 2025. "A Theoretical Study on the Efficacy and Mechanism of Combined YAP-1 and PARP-1 Inhibitors in the Treatment of Glioblastoma Multiforme Using Peruvian Maca Lepidium meyenii" Current Issues in Molecular Biology 47, no. 1: 40. https://doi.org/10.3390/cimb47010040
APA StyleTurpo-Peqqueña, A. G., Luna-Prado, S., Valencia-Arce, R. J., Del-Carpio-Carrazco, F. L., & Gómez, B. (2025). A Theoretical Study on the Efficacy and Mechanism of Combined YAP-1 and PARP-1 Inhibitors in the Treatment of Glioblastoma Multiforme Using Peruvian Maca Lepidium meyenii. Current Issues in Molecular Biology, 47(1), 40. https://doi.org/10.3390/cimb47010040







